Introduction
Hilar cholangiocarcinoma, a malignant liver tumor
that accounts for most cases of extrahepatic cholangiocarcinoma, is
characterized by an occult course and specific location, which
cause difficulties in its diagnosis and treatment. Previous
prognostic studies have revealed that pathologic stage has no
relationship with its prognosis (1). Therefore, a preoperative evaluation
system which has a high correlation to surgical resection rates and
prognoses is needed. In addition, the functions and mechanisms of
this system in the genesis and progression of hilar
cholangiocarcinoma should be elucidated to better distinguish this
disease from intrahepatic types of tumors.
The Wnt signaling pathway regulates cell
proliferation, differentiation, apoptosis and other biological
processes. The activation of the Wnt pathway is closely related to
tumorigenesis and progression of various types of tumors (2,3). Its
transient activation plays important roles in liver development
(4,5), liver cell regeneration (6), liver metabolism (7), oxygen stress (8) and other processes, thereby stimulating
the progression of various chronic liver diseases, such as
hepatitis B virus (HBV) infection (9) and hepatic fibrosis (10). In hepatocellular carcinoma (HCC),
multiple factors in the Wnt pathway, such as Wnt, β-catenin, APC,
Axin and sFRP1, are overexpressed in tumor cells (11,12);
more than 60 target genes of the Wnt pathway are activated by
β-catenin/TCF heterodimer, among which c-myc, c-jun, cyclin D1 and
vascular endothelial growth factor (VEGF) are most commonly
activated (13,14). These aberrantly activated oncogenes,
such as c-myc and cyclin D1, induce uncontrollable proliferation in
liver cells and lead to the genesis of HCC. The aberrant activation
of the Wnt pathway has been proven to closely relate with a subtype
of cholangiocarcinoma (15).
β-catenin gene mutations were detected in a few patients with
cholangiocarcinoma, suggesting that tyrosine
phosphorylation-dependent β-catenin activation or Wnt/Frizzled
dysfunction may contribute to the genesis of cholangiocarcinoma
(16). Elucidating the functions
and mechanisms of the Wnt pathway in cholangiocarcinoma,
particularly hilar cholangiocarcinoma, will benefit the diagnosis
and treatment of this disease.
In the present study, we detected the expression of
the Wnt pathway-related factors, Wnt2, Wnt3, β-catenin and
transcription factor 4 (TCF4), and its target genes, c-myc and
cyclin D1, in 4 cholangiocarcinoma cell lines and blocked the Wnt
pathway by RNA interference (RNAi) in order to explore potential
gene therapy for hilar cholangiocarcinoma.
Materials and methods
Cell lines and main reagents
Human hilar cholangiocarcinoma cell line FRH0201 was
kindly gifted by Professor Xiaopeng Wu at the Department of General
Surgery, Qilu Hospital, China, Shandong University. Human
intrahepatic cholangiocarcinoma cell lines HCCC-9810, SSP-25 and
RBE were purchased from Shanghai Cell Bank, Chinese Academy of
Sciences.
RPMI-1640 culture medium containing fetal bovine
serum (FBS), 0.25% trypsin, 1×105 U/ml penicillin and 10
mg/ml streptomycin were purchased from Gibco-BRL. MTT and DMSO
solutions were produced by Sigma-Aldrich. TRizol, Lipofectamine™
2000, SuperScript™ II RNase H− reverse transcriptase,
dATP, dGTP, dCTP and dTTP were produced by Invitrogen. The primary
rabbit anti-human antibody was purchased from Santa Cruz
Biotechnology, Inc. ECL detection solution, HRP-labeled secondary
marker antibody, IgG Fc HRP-labeled goat anti-rabbit secondary
antibody. The total protein extraction and BCA protein detection
kits were purchased from Nanjing Keygen Biotech Co. (Nanjing,
China).
Cell culture
Cells were cultured in RPMI-1640 medium containing
either 10% FBS (for FRH0201, HCCC-9810 and SSP-25 cells) or 20% FBS
(for RBE cells) at 37°C in 5% CO2. After trypsin
digestion and passage, cells were centrifuged at 1,000 × g for 5
min, placed in a refrigerator at 4°C for 30 min, and at −20°C
overnight, rethawed in water at 37°C, again centrifuged at 1,000 ×
g for 5 min, and cultured at 37°C in 5% CO2.
Reverse transcription-polymerase chain
reaction (RT-PCR)
The mRNA expression levels of the Wnt
pathway-related factors, Wnt2, Wnt3, β-catenin and TCF4, and its
target genes, c-myc and cyclin D1, in FRH0201, HCCC-9810, SSP-25
and RBE cells were detected by RT-PCR. Total RNA was extracted from
cells at a logarithmic growth phase with TRizol reagent according
to the manufacturer’s instructions and electrophoresed on agarose
gel. The 20 μl of RT solution was composed of 4.0 μl of 5X RNA PCR
buffer, 1.0 μl of oligo dT-adaptor primer, 2.0 μl of 10 mM dNTP
mixture, 0.625 μl of RNase inhibitor, 1.0 μl of MV reverse
transcriptase, 9.375 μl of RNase-free DEPC H2O and 2.0
μl of sample RNA. The RT conditions were: 42°C for 30 min, 99°C for
5 min, and 5°C for 5 min.
The sequences of the genes were obtained from
GenBank (http://www.ncbi.nlm.nih.gov/Genbank/). Primers were
designed with Primer Expression 2.0 software (Table I) and synthesized by Invitrogen.
GAPDH was used as internal reference. The 20 μl of PCR solution was
composed of 2.0 μl of 10X buffer, 0.2 μl of Blend Taq®,
3.0 μl of 2 mM dNTP, 1.0 μl of template, 1.0 μl of the upstream
primer, 1.0 μl of the downstream primer, and 11.8 μl of
ddH2O. The PCR conditions were denaturation at 95°C for
5 min, 28 cycles of annealing at 94°C for 30 sec, 59°C for 30 sec,
72°C for 1 min, and 72°C for 10 min, with elongation at 4°C for 1
h. Finally, 1.0 μl of PCR products were electrophoresed on 1.5%
agarose gel at 100 V for 20 to 25 min and assessed with the gel
imaging analysis system. The grey scale value of each well was
determined with an image analyser. The relative mRNA level of
target genes was calculated as the grey scale value of the target
gene/grey scale value of GAPDH.
 | Table ISequences of the RT-PCR primers. |
Table I
Sequences of the RT-PCR primers.
| Gene | Primer sequences | Tm (°C) | GC (°C) | Length (bp) |
|---|
| Wnt2 |
5′-AACGCTGACTGGACAACCG-3′ | 58.9 | 57.9 | 158 |
|
5′-GGGGCTTCCGTTGAGATAAA-3′ | 59.3 | 50 | |
| Wnt3 |
5′-CTGTGACTCGCATCATAAGGG-3′ | 58.1 | 52.4 | 159 |
|
5′-GCCTCGTTGTTGTGCTTGTT-3′ | 58.5 | 50 | |
| TCF4 |
5′-CCCAGACTACTCCGTTCCT-3′ | 53.7 | 57.9 | 143 |
|
5′-GGAAGCCGAAGATACAGG-3′ | 52.5 | 55.6 | |
| β-catenin |
5′-CAAGTGGGTGGTATAGAGG-3′ | 49.7 | 52.6 | 327 |
|
5′-CTGGGTATCCTGATGTGC-3′ | 50.5 | 55.6 | |
| c-myc |
5′-GGGCTTTATCTAACTCGCTGTA-3′ | 56.5 | 45.5 | 217 |
|
5′-GGGCAAAGTTTCGTGGAT-3′ | 55.3 | 50 | |
| Cyclin D1 |
5′-GCGAGGAACAGAAGTGCG-3′ | 57 | 61.1 | 195 |
|
5′-GGATGGAGTTGTCGGTGTAGAT-3′ | 58.1 | 50 | |
| GAPDH |
5′-AACGTGTCAGTGGTGGACCT-3′ | 60.48 | 55 | 400 |
|
5′-AGGGGAGATTCAGTGTGGTG-3′ | 59.96 | 55 | |
Western blot analysis
The protein expression levels of Wnt2, Wnt3,
β-catenin, TCF4, c-myc and cyclin D1 in FRH0201, HCCC-9810, SSP-25
and RBE cells were detected by western blotting. Total protein was
extracted from the cells with the KGP250 protein extraction kit (50
ml of lysis buffer, 250 μl of phosphatase inhibitor, 50 μl of
protease inhibitor, 500 μl of PMSF) and detected with the KGPBCA
protein detection kit (5 ml of 0.5 μg/μl standard protein solution,
50 ml of BCA solution A, 1 ml of BCA solution B) according to the
manufacturers’ instructions. After SDS-PAGE electrophoresis, the
protein was transferred on PVDF, blocked with TTBS (20 mM Tris-HCl,
pH 7.4, 150 mM NaCl, 0.25% Tween-20, 5% fat-free milk powder) and
washed with PBS for 10 min. After adding the primary antibody (at
1:500 dilution), the protein was cultured at room temperature for 1
h, washed with PBS for 10 min, and the secondary antibody was added
(at 1:1,000 dilution). Culturing was carried out for 1 h and then
washing with PBS for 10 min. After adding the ECL solution, the
protein was cultured in the dark for 5 min. The film was scanned
and assessed with the gel imaging analysis system. The grey scale
value of each well was measured with an image analyser. The
relative protein level of the target proteins was calculated as the
grey scale value of the target protein/grey scale value of
GAPDH.
Immunofluorescence assay
The expression of Wnt2 and β-catenin in FRH0201,
HCCC-9810, SSP-25 and RBE cells was detected by immunofluorescence
microscopy. Cells were seeded into 6-well plates at a density of
1×106 cells/well, with 3 wells for each group, then
fixed in acetone at room temperature for 20 min, washed with PBS
for 3 times, 5 min each time, punched with Triton-100 at 37°C for
20 min, washed with PBS, and blocked with non-immune animal serum
at 37°C for 40 min. After removing the serum, cells were added to
the primary Wnt2 or β-catenin antibody (at 1:100 dilution) and
cultured at 4°C overnight. The next day, cells were washed with
PBS, added together with FITC-labeled goat anti-rabbit secondary
antibody and cultured in the dark at 37°C for 30 min. After washing
with PBS, cell nuclei were counterstained with Hoechst at 37°C for
20 min, washed with PBS and blocked with 50% glycerin, then
observed under a fluorescence microscope.
siRNA transfection
Wnt2-siRNA, β-catenin-siRNA and nonsense-siRNA were
synthesized by RiboBio Co. (Guangzhou, China) (Table II). FRH0201 cells were digested and
prepared into a single-cell suspension, seeded into 6-well plates
at a density of 1×106 cells/well, with 3 wells for each
group and cultured for 12 to 24 h. Cells were washed with
serum-free RPMI-1640 twice, then cultured with serum-free
antibiotic-free RPMI-1640 (1.5 ml/well). Cells were transfected
with siRNA (50 nM/well) for 4 h using Lipofectamine™ 2000 as a
vector and then cultured with RPMI-1640 containing 10% FBS. At 6 h
after transfection, cells were washed with PBS twice and observed
under a fluorescence microscope. Fluorescence-labeled siRNA
(FAM-siRNA) was used to assess transfection efficiency.
Nonsense-siRNA-transfected cells were used as the negative control;
untransfected cells were used as the blank control. At 48 to 72 h
after transfection, the expression of Wnt2, Wnt3, TCF4, β-catenin,
c-myc and cyclin D1 was detected by RT-PCR and western blotting;
the expression of Wnt2 and β-catenin was detected by
immunofluorescence microscopy.
 | Table IIsiRNA sequences. |
Table II
siRNA sequences.
| Gene | Target gene
sequences | siRNA sequences | Molecular weight |
|---|
| Wnt2 |
5′-GGATGCAAAGGAAAGGAAA-3′ |
5′-GGAUGCAAAGGAAAGGAAA-dTdT-3′ | 13,008.76 |
| |
3′-dTdT-CCUACGUUUCCUUUCCUUU-5′ | |
| β-catenin |
5′-GCCACAAGATTACAAGAAA-3′ |
5′-GCCACAAGAUUACAAGAAA-dTdT-3′ | 13,193.75 |
| |
3′-dTdT-CGGUGUUCUAAUGUUCUUU-5′ | |
Flow cytometry
At 48 to 72 h after siRNA transfection, cells were
cultured with 200 μl of RNase A (1 mg/ml) at 37°C for 30 min and
stained with propidium iodide (PI) at 4°C in the dark for 30 min.
Cell cycle distribution was assessed by FACScan flow cytometry with
an excitation wavelength of 488 nm. After Annexin V-FITC/PI double
staining, cell apoptosis was also assessed by FACScan flow
cytometry. Cells without Annexin V-FITC or PI staining were used as
the negative control.
MTT assay
Before siRNA transfection and at 24, 48, 72 h after
transfection, cells at a density of 5×105 cells/ml were
seeded into a 96-well plate (1×104 cells/well), with 8
wells for each group, and 5 mg/ml MTT (20 μl/well) was added and
cultured at 37°C for 4 h. After discarding the supernatant, cells
were mixed with DMSO (200 μl/well) for 10 min. The absorbance of
each well at 570 nm (A570) was measured by an
ultraviolet spectrophotometer, and the cell proliferation rate was
calculated as (A570 of siRNA-transfected cells -
A570 of blank control)/(A570 of negative
control - A570 of blank control) × 100%.
Statistical analyses
All experiments were repeated three times. The data
are presented as mean ± standard deviation (SD). SPSS13.0 software
was used for statistical analyses. Homogeneity of variance was
assessed by the Levene’s test. Intergroup comparison was performed
with the ANOVA test when the variance was homogeneous or with the
Wilcoxon test when the variance was heterogeneous. A value of
P<0.05 was considered to indicate a statistically significant
difference.
Results
mRNA expression of the Wnt
pathway-related factors and target genes as detected by RT-PCR
The integrity and purity of total RNA extracted from
the cells were confirmed by electrophoresis. The mRNA levels of
Wnt2, Wnt3, β-catenin, TCF4, c-myc and cyclin D1 varied in the 4
cell lines (Fig. 1). The mRNA
expression levels of Wnt2, Wnt3, β-catenin and c-myc were
detectable in all 4 cell lines, whereas TCF4 and cyclin D1 mRNA
were undetectable in RBE cells.
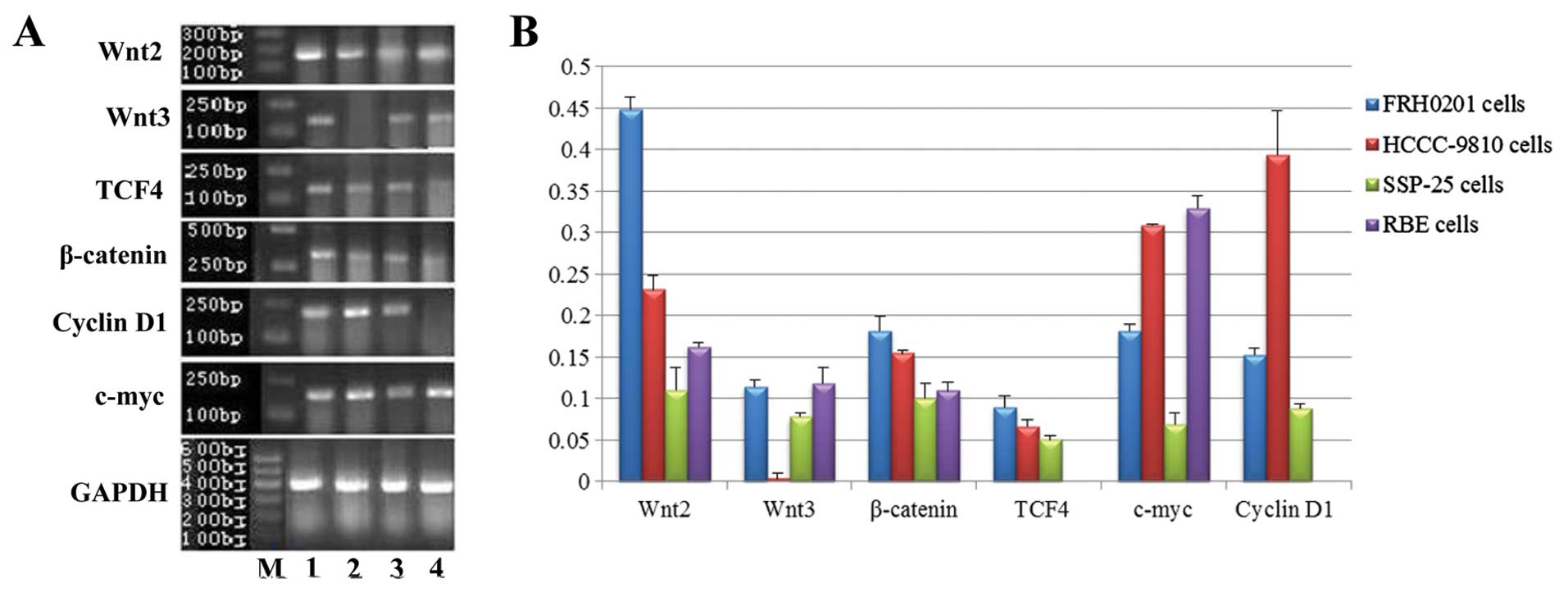 | Figure 1mRNA expression of Wnt2, Wnt3,
β-catenin, TCF4, c-myc and cyclin D1 in FRH0201, HCCC-9810, SSP-25
and RBE cells as detected by RT-PCR. (A) Lane M, marker; lane 1,
FRH0201 cells; lane 2, HCCC-9810 cells; lane 3, SSP-25 cells; lane
4, RBE cells. GAPDH was used as an internal reference. (B) Relative
mRNA levels of Wnt2, Wnt3, β-catenin, TCF4, c-myc and cyclin D1 as
compared with that of GAPDH. |
The mRNA levels (from high to low) in the cells were
FRH0201 > HCCC-9810 > RBE > SSP-25 for Wnt2 (F=199.499,
P<0.001); RBE ≈ FRH0201 > SSP-25 > HCCC-9810 for Wnt3
(F=68.927, P<0.001); FRH0201 ≈ HCCC-9810 > SSP-25 > RBE
for β-catenin (F=21.924, P<0.001); RBE ≈ HCCC-9810 > FRH0201
> SSP-25 for c-myc (F=179.284, P<0.001); FRH0201 ≈ HCCC-9810
≈ SSP-25 for TCF4 (P>0.05); and HCCC-9810 ≈ FRH0201 ≈ SSP-25 for
cyclin D1 (P>0.05).
Protein expression of the Wnt
pathway-related factors and target genes detected by western
blotting
The protein expression of Wnt2, Wnt3, β-catenin,
TCF4, c-myc and cyclin D1 was detected at various levels (Fig. 2). The protein expression levels of
Wnt2, Wnt3, β-catenin, TCF4 and c-myc were detectable in all 4 cell
lines, whereas cyclin D1 protein expression was undetectable in the
RBE cells.
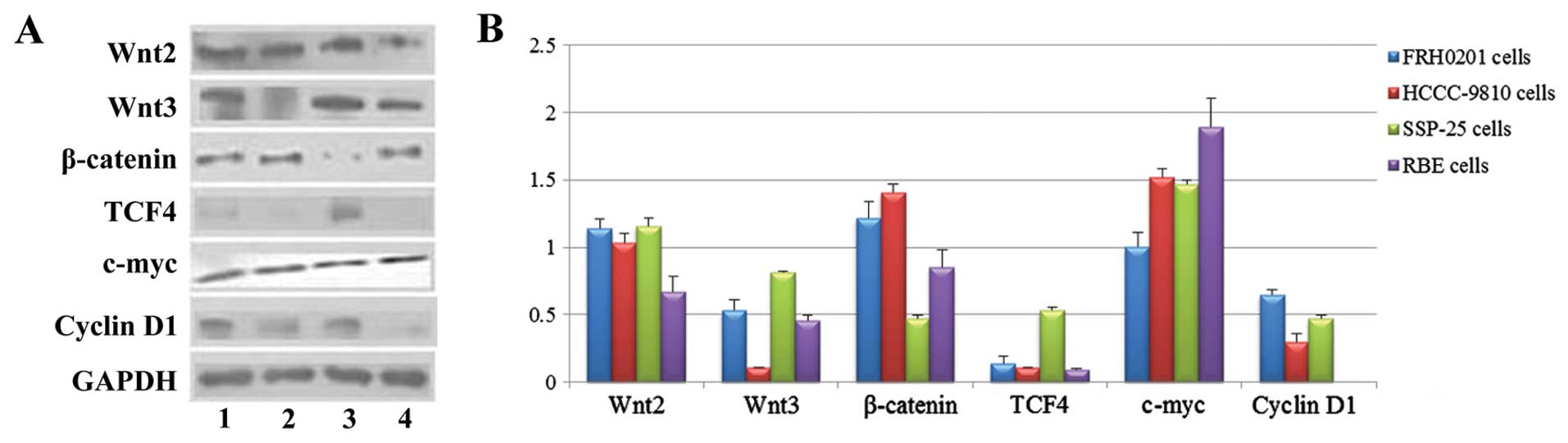 | Figure 2Protein expression of Wnt2, Wnt3,
β-catenin, TCF4, c-myc and cyclin D1 in FRH0201, HCCC-9810, SSP-25
and RBE cells as detected by western blotting. (A) Lane 1, FRH0201
cells; lane 2, HCCC-9810 cells; lane 3, SSP-25 cells; lane 4, RBE
cells. GAPDH was used as an internal reference. (B) Relative
protein levels of Wnt2, Wnt3, β-catenin, TCF4, c-myc and cyclin D1
as compared with that of GAPDH. |
The protein levels (from high to low) in the cells
were SSP-25 ≈ FRH0201 ≈ HCCC-9810 > RBE for Wnt2 (F=24.753,
P<0.001); SSP-25 ≈ FRH0201 ≈ RBE ≈ HCCC-9810 for Wnt3
(P>0.05); HCCC-9810 > FRH0201 > RBE > SSP-25 for
β-catenin (F=58.665, P<0.001); SSP-25 ≈ FRH0201 ≈ HCCC-9810 ≈
RBE for TCF4 (P>0.05); RBE > HCCC-9810 ≈ SSP-25 > FRH0201
for c-myc (F=25.208, P<0.001); and FRH0201 ≈ SSP-25 ≈ HCCC-9810
for cyclin D1 (P>0.05).
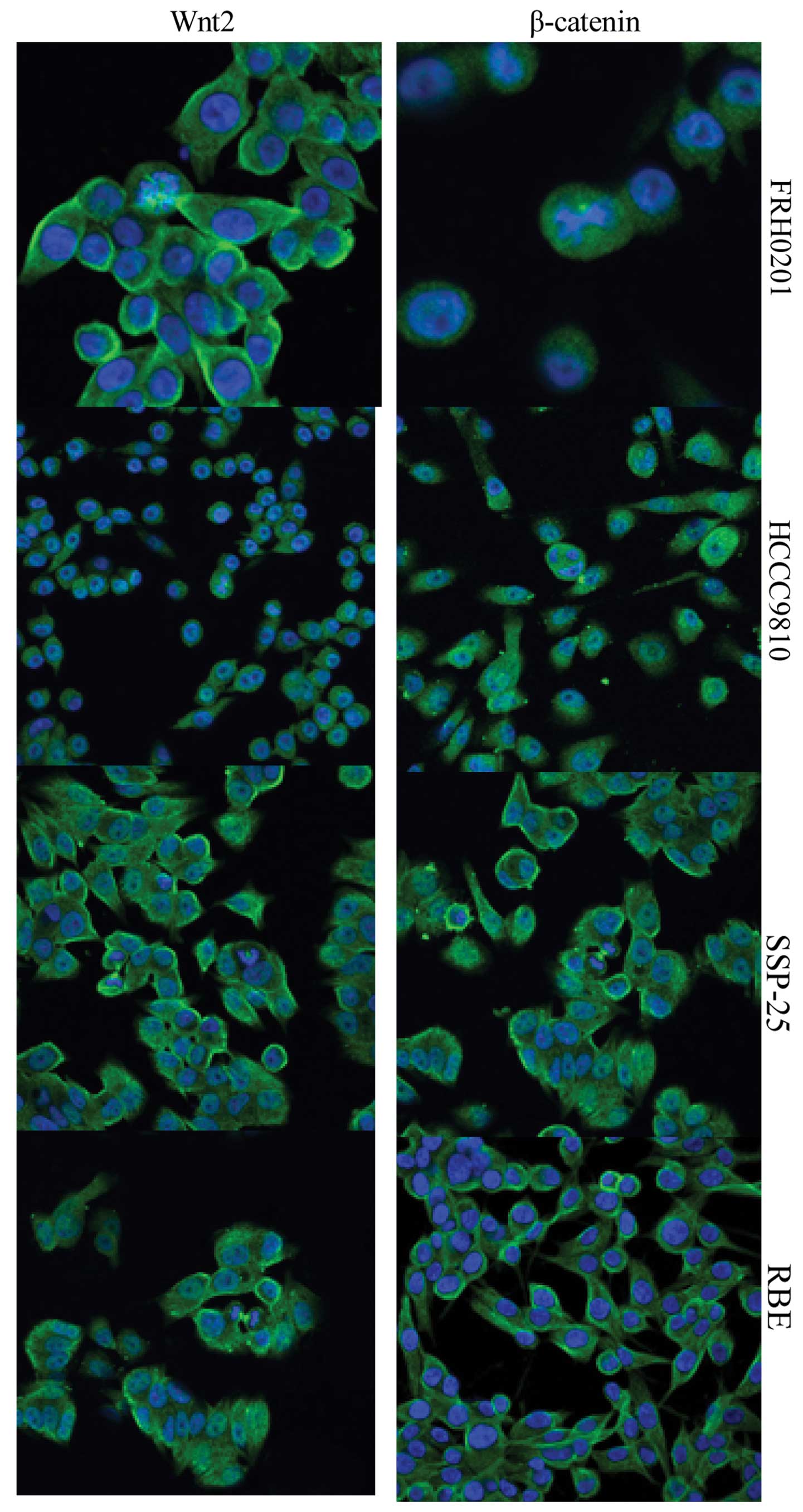
Expression of Wnt2 and β-catenin as
detected by immunofluorescence microscopy
Immunofluorescent staining showed that Wnt2 was
expressed both in the cytoplasm and on the cell membrane, whereas
β-catenin was expressed on the cell membrane and in cytoplasm and
nuclei (Fig. 3). Consistent with
the results of the western blot analysis, both Wnt2 and β-catenin
were highly expressed in the 4 cell lines. The expression of Wnt2
was similar in the FRH0201, HCCC-9810 and SSP-25 cells, and was
relatively weaker in the RBE cells. The expression of β-catenin was
stronger in the FRH0201 and HCCC-9810 cells, but was weaker in the
SSP-25 cells.
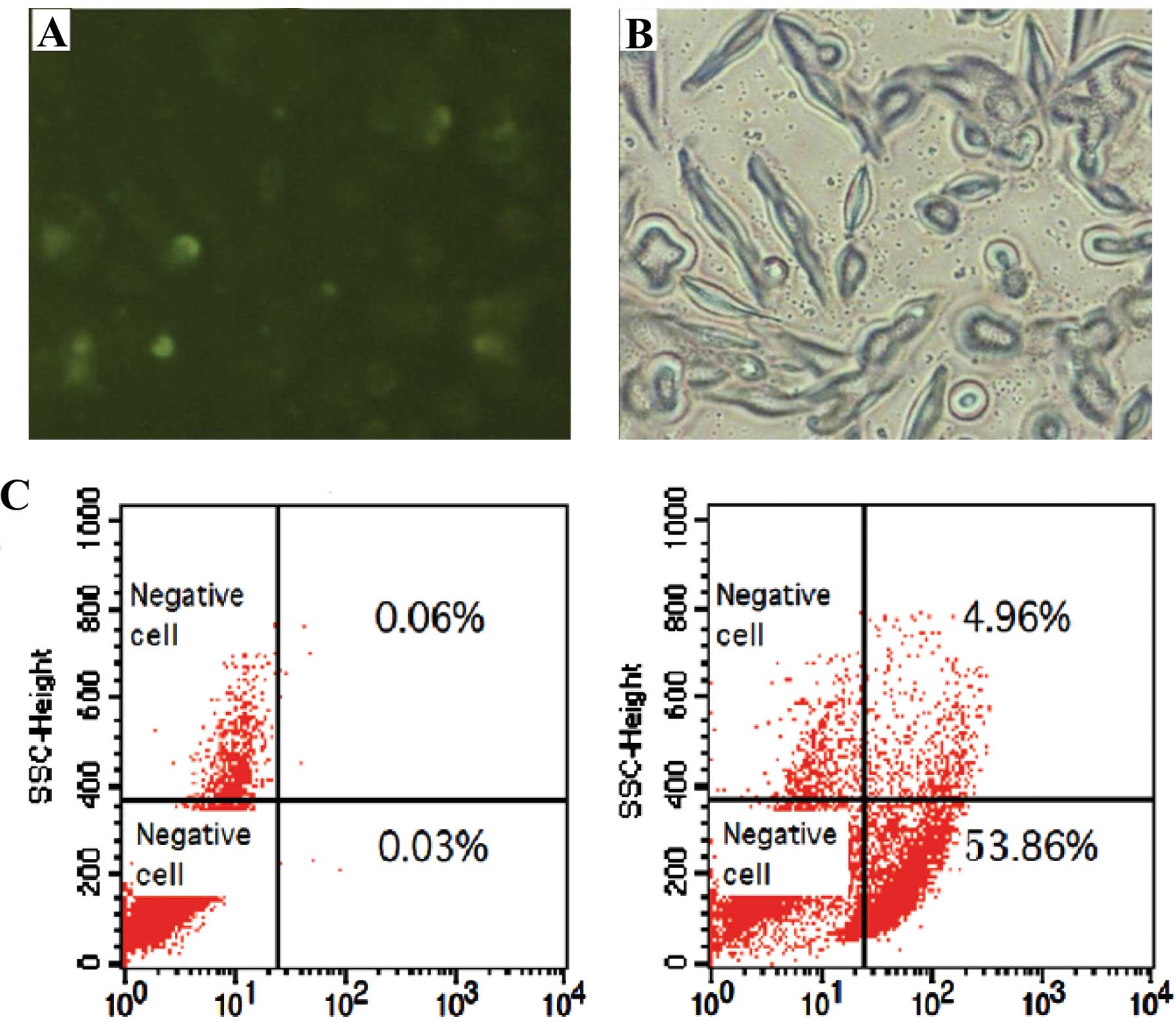
siRNA transfection efficiency
As detected by both fluorescence microscopy and flow
cytometry, siRNA was transfected into the FRH0201 cells, with a
transfection efficiency of 53.9% (Fig.
4).
mRNA expression of the Wnt
pathway-related factors and target genes after siRNA
transfection
After siRNA transfection, the mRNA expression of the
factors was obviously altered (Fig.
5). The expression of cyclin D1 was similar in the 4 groups. In
the Wnt2-siRNA group, the mRNA levels of Wnt2, Wnt3, TCF4 and c-myc
were downregulated, whereas that of β-catenin was upregulated. In
the β-catenin-siRNA group, the mRNA levels of β-catenin, Wnt3, TCF4
and c-myc were downregulated, whereas that of Wnt2 was upregulated.
The mRNA levels of Wnt3, β-catenin and TCF4 were lower in the
β-catenin-siRNA group than levels in the Wnt2-siRNA group, whereas
that of Wnt2 was lower in the Wnt2-siRNA group than that in the
β-catenin-siRNA group. No significant differences were observed
between the nonsense-siRNA and blank control groups.
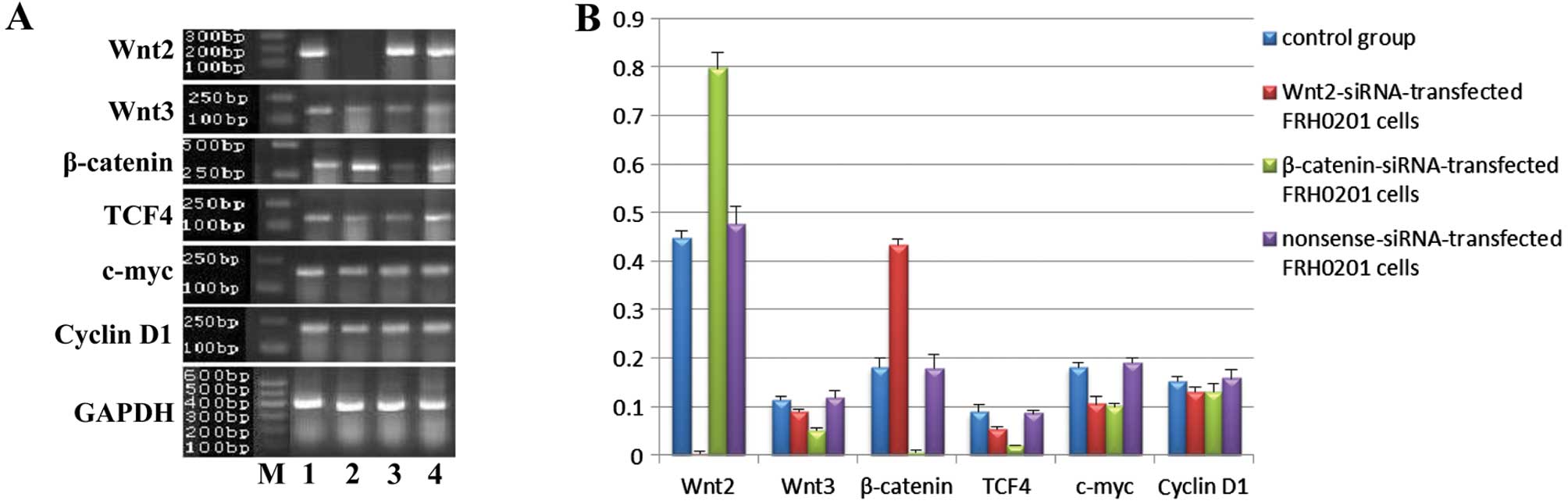 | Figure 5mRNA expression of Wnt2, Wnt3,
β-catenin, TCF4, c-myc and cyclin D1 in FRH0201 cells after siRNA
transfection. (A) Lane M, marker; lane 1, blank control; lane 2,
Wnt2-siRNA-transfected FRH0201 cells; lane 3,
β-catenin-siRNA-transfected FRH0201 cells; lane 4,
nonsense-siRNA-transfected FRH0201 cells. GAPDH was used as an
internal reference. (B) Relative mRNA levels of Wnt2, Wnt3,
β-catenin, TCF4, c-myc and cyclin D1 as compared with that of
GAPDH. |
Protein expression of the Wnt
pathway-related factors and target genes after siRNA
transfection
After siRNA transfection, the protein expression of
the factors was also obviously altered (Fig. 6). The expression of TCF4 and cyclin
D1 was similar in the 4 groups. In the Wnt2-siRNA group, the
protein levels of Wnt2 and Wnt3 were downregulated, whereas that of
c-myc was upregulated. In the β-catenin-siRNA group, the protein
level of β-catenin was downregulated, whereas that of c-myc was
upregulated. The protein levels of Wnt2 and Wnt3 were lower in the
Wnt2-siRNA group than levels in the β-catenin-siRNA group, whereas
that of β-catenin was lower in the β-catenin-siRNA group than that
in the Wnt2-siRNA group. No significant differences were observed
between the nonsense-siRNA and blank control groups.
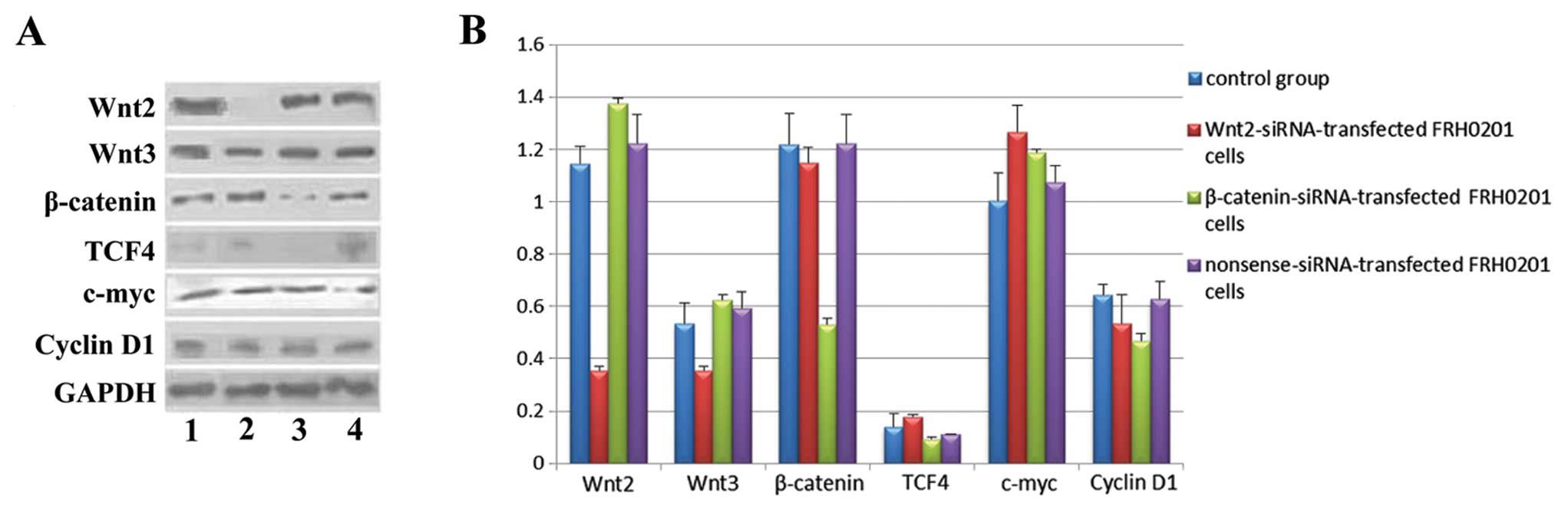 | Figure 6Protein expression of Wnt2, Wnt3,
β-catenin, TCF4, c-myc and cyclin D1 in FRH0201 cells after siRNA
transfection. (A) Lane 1, blank control; lane 2,
Wnt2-siRNA-transfected FRH0201 cells; lane 3,
β-catenin-siRNA-transfected FRH0201 cells; lane 4,
nonsense-siRNA-transfected FRH0201 cells. GAPDH was used as an
internal reference. (B) Relative protein levels of Wnt2, Wnt3,
β-catenin, TCF4, c-myc and cyclin D1 as compared with that of
GAPDH. |
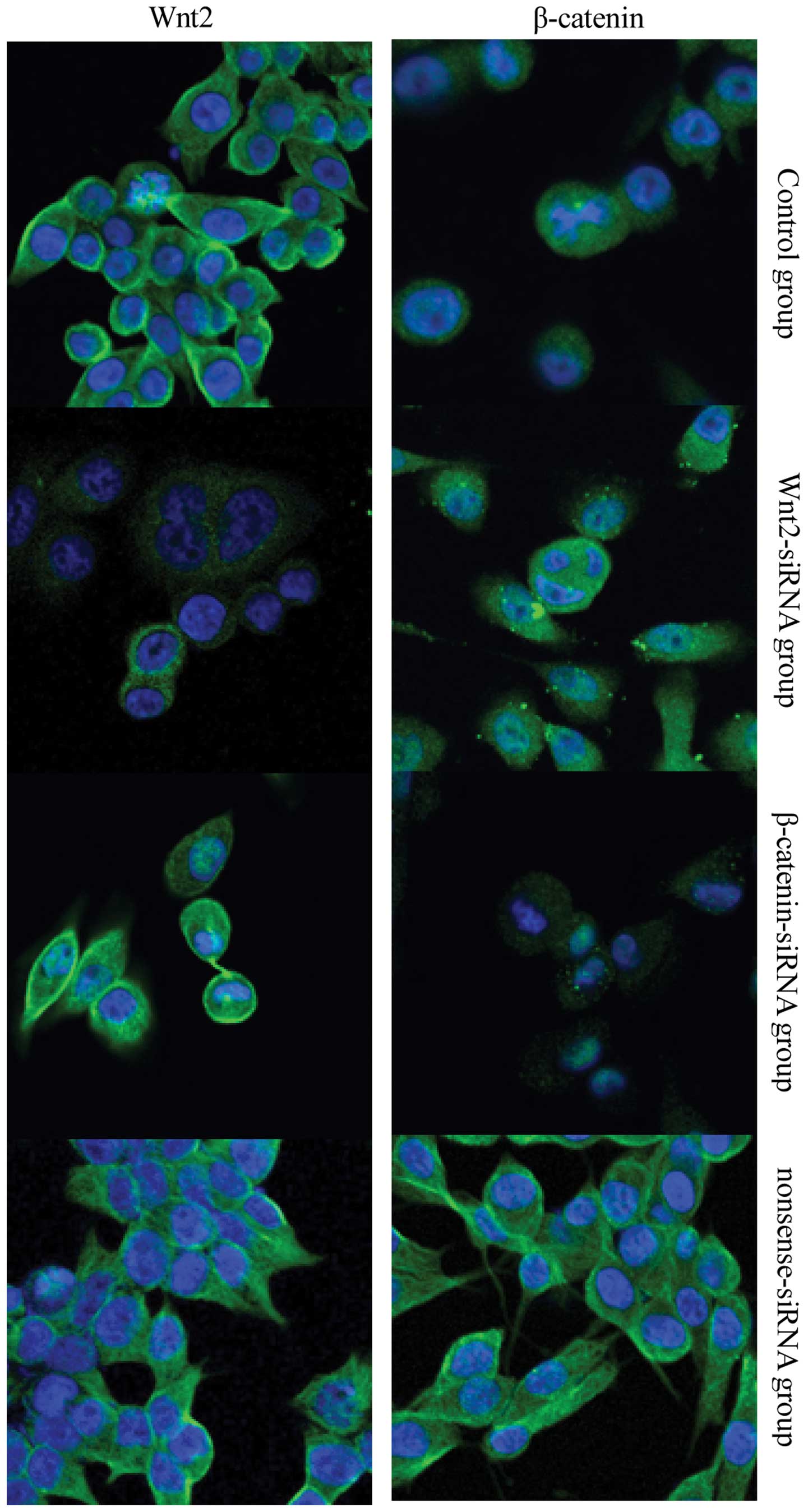
Protein expression of Wnt2 and β-catenin
after siRNA transfection
The results of fluorescence microscopy (Fig. 7) were consistent with the results of
the western blot analysis. In the Wnt2-siRNA group, the cytoplasm
and membrane staining was weaker for Wnt2, but the cytoplasmic
staining for β-catenin was unchanged. In the β-catenin-siRNA group,
the cytoplasm and membrane stained weaker for β-catenin, but the
staining for Wnt2 was unchanged. No significant differences were
observed between the nonsense-siRNA and blank control groups.
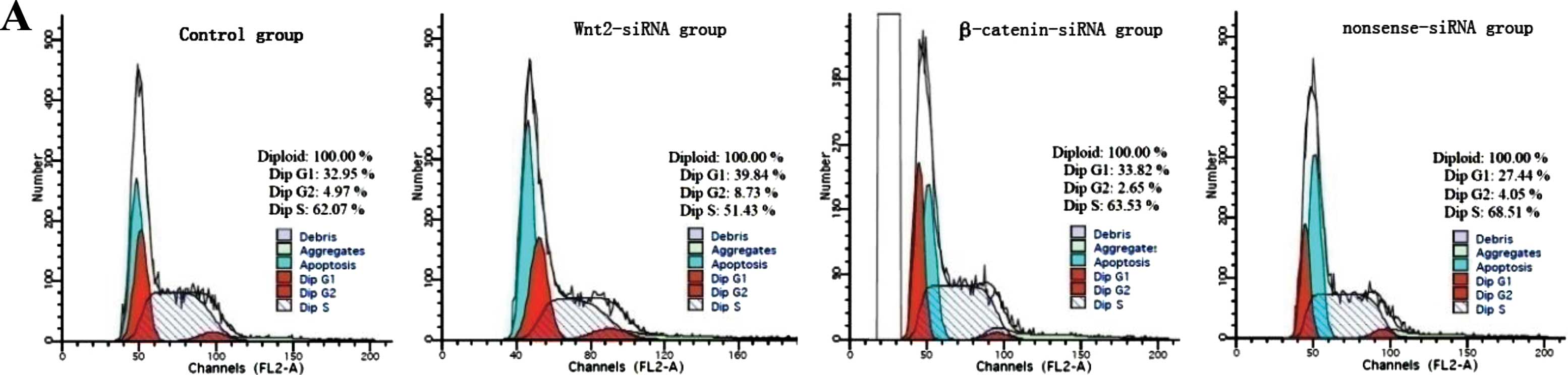
Cell cycle distribution, apoptosis, and
proliferation of FRH0201 cells after siRNA transfection
Flow cytometry showed that the proportion of S phase
cells was decreased after siRNA transfection;
G0/G1 phase arrest was observed in the
Wnt2-siRNA group (Fig. 8A). The
difference in cell apoptosis rate was significant between the 4
groups (45.1±2.9% for the blank control group; 50.1±2.8% for the
Wnt2-siRNA group; 50.5±3.1% for the β-catenin-siRNA group and
42.9±4.0% for the nonsense-siRNA group; F=6.589, P=0.015); the
apoptosis rate was significantly higher in the Wnt2-siRNA and
β-catenin-siRNA groups than in the blank control and nonsense-siRNA
groups (all P<0.05) (Fig. 8B).
The difference in cell proliferation rate was also significant
between the 4 groups (F=7.792, P=0.001); the proliferation rate was
significantly lower in the Wnt2-siRNA and β-catenin-siRNA groups
than in the blank control and nonsense-siRNA groups (all P<0.05)
(Fig. 8C).
Discussion
In the present study, we demonstrated that the Wnt
pathway is activated in cholangiocarcinoma cells. Blocking the Wnt
pathway by RNAi enhanced cell apoptosis and suppressed cell
proliferation.
Currently, only a few extrahepatic
cholangiocarcinoma cell lines have been established (17), and two of them were established in
China [QBC939 (18) and FRH0201
(19)]. These cell lines,
originating from either extrahepatic primary lesions or
intrahepatic metastases, provide ideal experimental models for the
study of hilar cholangiocarcinoma. In the present study, we
selected the FRH0201 cell line originating from primary hilar
cholangiocarcinoma lesions and 3 cell lines originating from
intrahepatic cholangiocarcinoma lesions as controls. We found high
expression of Wnt, β-catenin, TCF4 and target gene c-myc in all 4
cell lines, indicating activation of the Wnt pathway in both hilar
and intrahepatic cholangiocarcinoma cell lines. The expression
levels of the Wnt pathway-related factors varied in the 3
intrahepatic cholangiocarcinoma cell lines, but all were lower than
those in the hilar cholangiocarcinoma FRH0201 cells, suggesting
that the activation level of the Wnt pathway differs between hilar
cholangiocarcinoma and intrahepatic cholangiocarcinoma and that the
mechanisms of the Wnt pathway may be different in the two types of
cholangiocarcinomas.
Uematsu et al(20) and Davies et al(21) found that RNAi targeting the Wnt
pathway induced tumor cell apoptosis and inhibited cell
proliferation in renal cancer and non-small cell lung cancer.
Therefore, we hypothesized that blocking the Wnt pathway by RNAi
may also be a potential gene therapy for cholangiocarcinoma.
β-catenin, a key factor in the Wnt pathway, plays important roles
in the development and progression of hepatoblastoma, HCC and
cholangiocarcinoma. Sangkhathat et al(22) transfected β-catenin-siRNA into the
β-catenin-mutant pediatric hepatoblastoma cell line HuH-6 and the
HCC cell line HepG2. After transfection, the expression of
β-catenin, c-myc and cyclin D1 was downregulated, and cell
proliferation and invasion were suppressed (22). Wnt2, a member of the Wnt family, is
overexpressed in various digestive tract tumors and lung cancer. A
recent study with HCC cell lines found that Wnt3 and Frizzled7 also
activated the Wnt pathway and played important roles in the genesis
and progression of HCC (23).
Mazieres et al(24)
transfected Wnt2-siRNA into Wnt2-overexpressed malignant pleural
mesothelioma cell lines and found similar results. In the present
study, we found that the mRNA and protein levels of Wnt2 as well as
the mRNA level of β-catenin were downregulated after Wnt2-siRNA
transfection. Although the downregulation of β-catenin protein
level was not obvious, immunofluorescence microscopy revealed
decreased nuclear expression and increased cytoplasmic and membrane
expression of β-catenin, suggesting that Wnt2-siRNA inhibits the
nuclear accumulation of β-catenin, thus blocking the Wnt pathway.
We also found that the mRNA and protein levels of β-catenin as well
as the mRNA level of Wnt3 were downregulated after β-catenin-siRNA
transfection, confirming that Wnt3 was related to the cytoplasmic
expression of β-catenin. The ‘seesaw-like’ relationship between
Wnt2 and β-catenin at the translation level is considered to be
related with multiple factors that are involved in the activation
or inhibition of the Wnt pathway. Cyclin D1 was proven to play an
important role in hilar cholangiocarcinoma, but was not associated
with the activation of the Wnt pathway (25). Upregulation of cyclin D1 protein
level was noted after β-catenin-siRNA transfection, suggesting that
β-catenin regulates the expression of cyclin D1 via other pathways.
The detailed mechanisms need to be elucidated in our future
research.
Our results showed that using RNAi to knock down
Wnt2 or β-catenin and block the Wnt pathway obviously inhibited
cell proliferation, enhanced cell apoptosis, and arrested the cell
cycle at the G0/G1 phase. The regulation of
cell apoptosis and proliferation by c-myc and cyclin D1 was closely
related to alterations of many other genes (26). The mechanisms of the downregulation
of c-myc mRNA level and cyclin D1 protein level after
β-catenin-siRNA transfection warrant further study.
In conclusion, the Wnt pathway was activated in both
the hilar cholangiocarcinoma cell line FRH0201 and the intrahepatic
cholangiocarcinoma cell lines HCCC-9810, SSP-25 and RBE, but the
expression levels of its key factors Wnt2, Wnt3, TCF4 and β-catenin
and its target genes c-myc and cyclin D1 varied in the 4 cell
lines. RNAi targeting Wnt2 and β-catenin downregulated the
expression of these two genes in FRH0201 cells, inhibited the
activation of the Wnt pathway, downregulated the expression of
c-myc, promoted cell apoptosis and inhibited cell proliferation,
suggesting that Wnt2 and β-catenin could be key targets for the
gene therapy of hilar cholangiocarcinoma.
References
|
1
|
Saxena A, Chua TC, Chu FC and Morris DL:
Improved outcomes after aggressive surgical resection of hilar
cholangiocarcinoma: a critical analysis of recurrence and survival.
Am J Surg. 202:310–320. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lu D, Zhao Y, Tawatao R, et al: Activation
of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc
Natl Acad Sci USA. 101:3118–3123. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ilyas M: Wnt signalling and the
mechanistic basis of tumour development. J Pathol. 205:130–144.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Monga SP, Monga HK, Tan X, Mulé K,
Pediaditakis P and Michalopoulos GK: Beta-catenin antisense studies
in embryonic liver cultures: role in proliferation, apoptosis, and
lineage specification. Gastroenterology. 124:202–216. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Micsenyi A, Tan X, Sneddon T, Luo JH,
Michalopoulos GK and Monga SP: Beta-catenin is temporally regulated
during normal liver development. Gastroenterology. 126:1134–1146.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Monga SP, Pediaditakis P, Mule K, Stolz DB
and Michalopoulos GK: Changes in WNT/beta-catenin pathway during
regulated growth in rat liver regeneration. Hepatology.
33:1098–1109. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Benhamouche S, Decaens T, Godard C, et al:
Apc tumor suppressor gene is the ‘zonation-keeper’ of mouse liver.
Dev Cell. 10:759–770. 2006.
|
|
8
|
Funato Y, Michiue T, Asashima M and Miki
H: The thioredoxin-related redox-regulating protein nucleoredoxin
inhibits Wnt-beta-catenin signalling through dishevelled. Nat Cell
Biol. 8:501–508. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lian Z, Liu J, Li L, et al: Enhanced cell
survival of Hep3B cells by the hepatitis B x antigen effector,
URG11, is associated with upregulation of beta-catenin. Hepatology.
43:415–424. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Higashi N, Kojima N, Miura M, Imai K, Sato
M and Senoo H: Cell-cell junctions between mammalian (human and
rat) hepatic stellate cells. Cell Tissue Res. 317:35–43. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guan CN, Chen XM, Lou HQ, Liao XH, Chen BY
and Zhang PW: Clinical significance of axin and beta-catenin
protein expression in primary hepatocellular carcinomas. Asian Pac
J Cancer Prev. 13:677–681. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kaur P, Mani S, Cros MP, et al: Epigenetic
silencing of sFRP1 activates the canonical Wnt pathway and
contributes to increased cell growth and proliferation in
hepatocellular carcinoma. Tumour Biol. 33:325–336. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Loeppen S, Koehle C, Buchmann A and
Schwarz M: A beta-catenin-dependent pathway regulates expression of
cytochrome P450 isoforms in mouse liver tumors. Carcinogenesis.
26:239–248. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tien LT, Ito M, Nakao M, et al: Expression
of beta-catenin in hepatocellular carcinoma. World J Gastroenterol.
11:2398–2401. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tokumoto N, Ikeda S, Ishizaki Y, et al:
Immunohistochemical and mutational analyses of Wnt signaling
components and target genes in intrahepatic cholangiocarcinomas.
Int J Oncol. 27:973–980. 2005.PubMed/NCBI
|
|
16
|
Sugimachi K, Taguchi K, Aishima S, et al:
Altered expression of beta-catenin without genetic mutation in
intrahepatic cholangiocarcinoma. Mod Pathol. 14:900–905. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ku JL, Yoon KA, Kim IJ, et al:
Establishment and characterisation of six human biliary tract
cancer cell lines. Br J Cancer. 87:187–193. 2002.PubMed/NCBI
|
|
18
|
Takiyama I, Terashima M, Ikeda K, et al:
Establishment and characterization of a new human extrahepatic bile
duct carcinoma cell line (ICBD-1). Oncol Rep. 5:463–467.
1998.PubMed/NCBI
|
|
19
|
Tang WH, Yuan ST, Wang BS, Lu LJ, Ding J
and Yuan ZR: Establishment of a subcutaneous model of the human
extrahepatic bile duct carcinoma in nude mice via transplantation
of histologically intact tumor tissue. J Exp Clin Cancer Res.
23:661–667. 2004.PubMed/NCBI
|
|
20
|
Uematsu K, He B, You L, Xu Z, McCormick F
and Jablons DM: Activation of the Wnt pathway in non small cell
lung cancer: evidence of dishevelled overexpression. Oncogene.
22:7218–7221. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Davies JA, Ladomery M, Hohenstein P, et
al: Development of an siRNA-based method for repressing specific
genes in renal organ culture and its use to show that the Wt1
tumour suppressor is required for nephron differentiation. Hum Mol
Genet. 13:235–246. 2004. View Article : Google Scholar
|
|
22
|
Sangkhathat S, Kusafuka T, Miao J, et al:
In vitro RNA interference against β-catenin inhibits the
proliferation of pediatric hepatic tumors. Int J Oncol. 28:715–722.
2006.
|
|
23
|
Kim M, Lee HC, Tsedensodnom O, et al:
Functional interaction between Wnt3 and Frizzled-7 leads to
activation of the Wnt/beta-catenin signaling pathway in
hepatocellular carcinoma cells. J Hepatol. 48:780–791. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mazieres J, You L, He B, et al: Wnt2 as a
new therapeutic target in malignant pleural mesothelioma. Int J
Cancer. 117:326–332. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao P, Lu Y, Zhong M, Liu L and Li B:
Inverse correlation of aberrant expression of fragile histidine
triad (FHIT) protein with cyclin D1 protein and prognosis in
Chinese patients with cholangiocarcinoma. Acta Oncol. 47:1557–1563.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
El-Kady A, Sun Y, Li YX and Liao DJ:
Cyclin D1 inhibits whereas c-Myc enhances the cytotoxicity of
cisplatin in mouse pancreatic cancer cells via regulation of
several members of the NF-κB and Bcl-2 families. J Carcinog.
10:242011.PubMed/NCBI
|