Introduction
Hepatocellular carcinoma (HCC) is the most common
primary hepatic malignancy with the continually increasing
incidence, and the second major cause of cancer-related deaths
worldwide (1,2). In China, the most common cause of this
neoplasm is the chronic infection induced by the hepatitis B virus.
Other important etiological factor are cirrhosis, chronic viral
hepatitis (hepatitis C virus), alcohol abuse, obesity,
hemochromatosis, α1-antitripsin deficiency and toxins similar to
aflatoxin (3). Because recent
development of genomic technologies has empowered survey of
molecular aberrations and deregulations directly from patient
specimens in a comprehensive way, some presented
prognostic/predictive targets for HCC biomarkers improved the
prognosis of patients by guiding treatment decision and allocation
of medical resources (4–6). Cancer/testis genes are normally
expressed by gametes and trophoblasts, as well as aberrantly
expressed in a range of malignant tumors of unrelated histological
origin, including the liver. CT genes are a class of closely
cancer-related biomarkers. Since germline stem cells and their
trophoblastic derivatives share many characteristics with tumour
cells, the activation of portions of the germline gene-expression
in cancer cells could contribute to the malignant phenotype,
including proliferation, survival, invasiveness, immune evasion and
metastatic capacity (7).
A number of CT genes have been found expressed in
HCC and their products are promising targets for antigen-specific
immunotherapy of this tumor. For example, the mRNA expressions of
MAGE1, SSX-1, CTp11 and HCA587, were detectable in diverse
percentage of HCC samples examined (8,9). The
brother of the regulator of imprinted sites (BORIS) as a novel
member of the CT antigens may be an auxiliary diagnosis index and a
novel favorable prognostic indicator of HCC (10). However, whether their recurrent
expression contributes to tumorigenesis, is still under
investigation. Recent studies demonstrate the important role of CT
genes in malignant features of HCC cells. A known member of CT
antigens family, NY-ESO-1, whose expression is associated with
worse HCC outcome, increases tumor cell migration (11).
To identify novel CT genes, some genes with
testis-exclusive expression have been searched for through
published literature. In the present study, we paid attention to a
reported gene family, which included three members with
testis-exclusively expression, FAM9A, FAM9B and FAM9C (12) and investigated their expression
profiles in human hepatocellular carcinoma (HCC). Interestingly,
among the three members, only FMA9C was frequently upregulated in
HCC specimens compared with that of the corresponding non-cancerous
livers. Nevertheless, the physiological and the pathological
function of FAM9C is not understood. Therefore, we further
evaluated the role of FAM9C in HCC cells. Significantly, the
resulting data demonstrated that FAM9C overexpression promoted
proliferation, colony-forming growth and in vivo
tumorigenicity of HCC cells, and RNA interference against FAM9C
suppressed these phenotypes in HCC cells. We found that FAM9C
endowed HCC cells with resistance to apoptosis through PI3K-Akt
pathway. Our results reveal that upregulated FAM9C as a novel
cancer testis gene plays an anti-apoptotic role in human HCC.
Materials and methods
Tissue specimens and cell lines
HCC specimens used in the present study were
obtained from HCC patients who underwent liver resection and gave
their informed consent. The following liver tumor-derived cell
lines SSMC-7721, QGY-7703, BEL-7404, BEL-7405, YY-8103 and Huh-7,
were employed in the present study. These cell lines were grown in
Dulbecco’s modified Eagle’s medium (DMEM), supplemented with
heat-inactivated 10% fetal bovine serum (Gibco, Carlsbad, CA, USA)
at 37ºC in a 5% CO2 humidified incubator.
Antibodies and reagents
Antibodies against FAM9C, GAPDH, Lamin A and β-actin
were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA).
Antibodies against phospho-Akt (pAkt and Ser473) and Akt were from
Cell Signaling Technology (Beverly, MA, USA). Propidium iodide
(PI), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
(MTT), 5-Aza-2′-deoxycytidine (5-Aza-dC),
4′-6-diamidino-2-phenylindole (DAPI) and PI3K inhibitor LY294002
were purchased from Sigma Chemical Co. (St. Louis, MO, USA).
5-Aza-2′-deoxycytidine (5-Aza-dC)
treatment
SSMC-7721 and BEL-7405 cells were seeded at a
density of 5×105/100-mm dishes, cultured for 48 h, and
treated with 2 mM 5-Aza-dC (Sigma Chemical), a demethylating agent.
Forty-eight hours after treatment, cells are washed with PBS and
fresh medium was added. Cells are further incubated for another 48
h before isolation of total cellular RNA.
RNA extraction, reverse transcription-PCR
and real-time PCR
RNA from HCC specimens and cell lines were extracted
using TRIzol® Reagent (Invitrogen Life Technologies,
Carlsbad, CA, USA) according to the manufacturer’s instructions.
Total RNA was treated with DNase I to avoid DNA contamination, the
quality and quantity of extracted RNA was confirmed by NanoDrop
2000 (Thermo Fisher Scientific, Waltham, MA, USA). RNA reverse
transcription reaction was performed using M-MLV reverse
transcriptase (Promega), according to the manufacturer’s
instructions. The primers sequences used in this study were as
follows: upstream: 5′-TAG GAGCTGGAGGCTGAGAG-3′ and downstream:
5′-GGTTG AAGAGACAATGAAGTGCC-3′ for FAM9A gene amplification
(NM_174951.2); upstream: 5′-GCAGTGCAGTGGTG TGATCT-3′ and
downstream: 5′-CTTTCCTGCATGCTTCT TCC-3′ for FAM9B gene
amplification (NM_205849.1); upstream: 5′-CTGAGGGAGCAACATCAGGG-3′
and downstream: 5′-TTGGTATCAACCCCCGTGTG-3′ for FAM9C gene
amplification (NM_174901.4); and β-actin, a typical housekeeping
gene used as the internal control, upstream:
5′-AATCGTGCGTGACATTAAGGAG-3′ and downstream:
5′-ACTGTGTTGGCGTACAGGTCTT-3′. All reactions were performed in
triplicate in an Applied Biosystems 7300 Real-time PCR system. The
relative mRNA level of the target gene was calculated using the
comparative threshold cycle method (2−ΔΔCt) normalized
by β-actin expression (13).
Immunohistochemistry and
immunofluorescence assay
Formalin-fixed HCC samples were paraffin-embedded
and cut into 4-μm sections. The sections were deparaffinized and
dehydrated, and then treated with methanol containing 0.3%
H2O2 to inhibit endogenous peroxidase. The
slides were incubated with anti-FAM9C goat polyclonal antibody
(Santa Cruz Biotechnology) at 37ºC for 2 h and then at 4ºC
overnight, followed by incubation with a horseradish
peroxidase-conjugated anti-goat antibody (Dako Japan Ltd., Kyoto,
Japan) at 37ºC for 1 h. The signals were detected using
Diaminobenzidine Substrate kit (Vector Laboratories, Burlingame,
CA, USA). Counterstaining was performed with hematoxylin. In
addition, the slides with HCC specimens and the corresponding
adjacent non-HCC livers were simultaneously used in the
immunohistochemistry staining, and were then assessed by visual
inspection and the estimation of the percentage of immunopositive
cells. Immunofluorescence assay was performed to detect endogenous
FAM9C in BEL-7404 cells. These cells were plated on
polylysine-treated slides and then incubated at 37ºC for 1 h. The
fixed cells were blocked with phosphate-buffered saline buffer
containing 5% bovine serum albumin and then stained with anti-FAM9C
antibody (Santa Cruz Biotechnology) at 4ºC overnight, followed by
incubation with fluorescein isothiocyanate-conjugated goat anti-IgG
antibody (Gibco-BRL, Grand Island, NY, USA) at 4ºC for 2 h. After
rinsing, the slides were analyzed using immunofluorescence
microscopy.
RNA interference (RNAi)
Small interference RNAs (siRNAs) targeting FAM9C,
Akt and negative control siRNA (si-NC) were chemically synthesized
(Invitrogen-Life Technologies). Two siRNAs against FAM9C with high
efficiency were used in the functional experiment assays, and their
sequences were as follows: FAM9C-si1, sense, 5′-GGACCAGUUGGAGGUU
CAAdTdT-3′, antisense, 5′-UUGAACCUCCAACUGGUCC dTdT-3′; FAM9C-si2,
sense, 5′-CCAAGGAGCUAGAAGAU AUdTdT-3′, antisense,
5′-AUAUCUUCUAGCUCCUUGT dT-3′; si-NC, sense,
5′-UUCUCCGAACGUGUCACGUdT dT-3′, antisense,
5′-ACGUGACACGUUCGGAGAAdTdT-3′. As well, the sequences of a siRNA
against Akt were as follows: sense, 5′-GCACCUUCAUUGGCUACAAdTdT-3′,
antisense, 5′-UUGUAGCCAAUGAAGGUGCdTdT-3′.
Construction of recombinant plasmids
The full-length FAM9C ORF (501 bp, GenBank accession
number NM_174901.4) was amplified by RT-PCR from BEL-7404 cells
cDNA. The primers for FAM9C ORF were as follows: forward,
5′-TCAGAATTCATGGCTGCCAAGGACCAGT TGGAGG-3′; reverse,
5′-TCAGGTACCTCAATCTCTTTCA GTTCCTTTCCCA-3′. The PCR product was then
inserted into pcDNATM3.1B (Invitrogen) through EcoRI- and
KpnI- adapter. In addition, synthesized DNA nucleotide
fragments of short hairpin RNA (shRNA) for knocking down endogenous
FAM9C, was inserted into pSUPER (Oligoengine; Seattle, WA, USA).
The sequences of these synthesized oligonucleotides for FAM9C
silencing were as follows: forward, 5′-GAT CCCCGGACCAGTTGGAGGTTCAATTCAAGAGATTGA
ACCTCCAACTGGTCCTTTTTGGAAA-3′, reverse, 5′-AGCT
TTTCCAAAAAGGACCAGTTGGAGGTTCAATCTCTTGA
ATTGAACCTCCAACTGGTCCGGG-3′
(FAM9C-sh1); forward,
5′-GATCCCCCCAAGGAGCTAGAAGATATTTC AAGAGAATATCTTCTAGCTCCTTGGTTTTTGGAAA-3′,
reverse, 5′-AGCTTTTCCAAAAACCAAGGAGCTAGAAG
ATATTCTCTTGAAATATCTTCTAGCTCCTTGGGGG-3′
(FAM9C-sh2); pSUPER-shNC with irrelevant nucleotide sequence serves
as a negative control, and sequences of shNC were as follows:
forward, 5′-GATCCCCTTCTCCGAACGT
GTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATT
TTTGGAAA-3′, reverse, 5′-AGCTTTTCCAAAAATTCTCCG
AACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGA
GAAGGG-3′.
Cell transfection
Cell transfection was performed by Lipofectamine
2000 (Invitrogen) according to the manufacturer’s instructions.
Cell growth curve
HCC cells with FAM9C overexpression or knockdown
were seeded in triplicate into 96-well plates (3,000 cells/well)
respectively and then incubated with 5 mg/ml (10 μl) MTT for 4 h at
37ºC, then the medium was replaced with 100 μl dimethylsulfoxide
(DMSO) for 10 min every 24-h intervals for 5–6 days. Cell growth
curves were generated by calculating the mean value of the optical
density measurements at 492 nm using a microplate reader. All
experiments were independently repeated at least 3 times.
Colony formation assay
FAM9C was overexpressed in HCC cell lines Huh-7 and
YY-8103 through the transfection with recombinant pcDNA3.1-FAM9C
plasmid, additionally endogenous FAM9C was knocked down in
QGY-7703, BEL-7404 cells via pSUPER-shFAM9C plasmids. Transfected
cells were cultured on 100-mm dishes for colony formation, followed
by the addition of G418 (Life Technologies) to the medium at a
final concentration of 0.6–1 mg/ml. After 3–4 weeks, the remaining
colonies were washed twice with PBS and counted on crystal
violet-stained dishes. All experiments for observing colony
formation were independently repeated at least three times.
Establishment of stable cell lines
To establish a stably FAM9C-expressing HCC cell
line, HuH-7 cells were transfected with the recombinant plasmid
pcDNA3.1B-FMA9C, then cultured by G418 (Life Technologies). An
immunoblot assay was employed to examine the expression of FAM9C in
nine individual colonies. One colony with steady and strong FAM9C
expression was selected as study object, and another colony without
ectopic FAM9C expression was used as mock object. In addition,
BEL-7404 cell line with stable FAM9C knockdown was also established
by transfection with plasmid pSUPER-FAM9C-sh2, with the methods
mentioned above. A colony of BEL-7404 cells transfected with
plasmid pSUPER-shNC containing irrelevant sequence was used as a
negative control.
Western blot assay
Cultured HCC cells were harvested with trypsin-EDTA
and then centrifuged at 500 × g for 5 min. Cytoplasmic and nuclear
proteins were extracted using NE-PER® Nuclear and
Cytoplasmic Extraction reagents (Thermo Scientific Pierce),
according to the manufacturer’s instructions. Protein extracts were
separated by 12% SDS-PAGE and transferred onto Hybond-C
nitrocellulose membranes (Amersham Life Science, Buckinghamshire,
UK). After blocking with PBS containing 5% non-fat milk, the
membrane was incubated for immunoblot analysis with primary
antibodies, followed by incubation with an IRDye 800DX-conjugated,
affinity-purified secondary antibody. Signals were detected using
the Odyssey Infrared Imaging system (LI-COR Biosciences).
Flow cytometric analysis
Flow cytometry was used to analyze cell cycle and
cell apoptosis. For DNA content analysis, cells were fixed in 70%
ethanol, rehydrated in PBS, and treated for 30 min with RNase A (10
mg/ml) and for 5 min with propidium iodide (10 μg/ml). For
UV-induced apoptosis analysis, 3J/cm2, 5 min
UV-irradiated cells (1×106 cells) were harvested and
washed twice with cold PBS. Then, Annexin V-FITC staining was
performed according to the protocol provided by the manufacturer
(BD Biosciences Pharmingen, San Diego, CA, USA). Briefly, cells
were resuspended in binding buffer at a concentration of
1×106 cells/ml, and 5 μl of Annexin-FITC and 100 μl cell
suspension (1×105 cells) were gently mixed and incubated
for 15 min at room temperature in the dark. Cells were analyzed for
apoptosis by flow cytometry on a FACSCalibur (Becton-Dickinson,
Mountain View, CA, USA).
Tumorigenicity assay in nude mice
A total of 2×106 offspring Huh-7 cells
stably expressing ectopic FAM9C were subcutaneously injected into
the flank of nude mice, and the same amount of progeny Huh-7 cells
without ectopic FAM9C expression as mock control were injected into
the opposite flank of the same mice. Cells (2×106)
derived from BEL-7404 with stable knockdown of endogenous FAM9C
were injected subcutaneously into the flank of nude mice, and the
same amount of BEL-7404 cells from the colony transfected with
pSUPER-shNC were injected into the opposite flank of the same mice.
Tumor growth kinetics was estimated by measuring tumor size and
volume every 3–4 days. Tumor volume was determined using the
following formula: 1/2 × width2 × length.
Statistical analysis
Statistical analysis was performed by the Student’s
t-test using SSPS software (SPSS, Inc., Chicago, IL, USA). P-value
<0.05 was considered to indicate a statistically significant
result.
Results
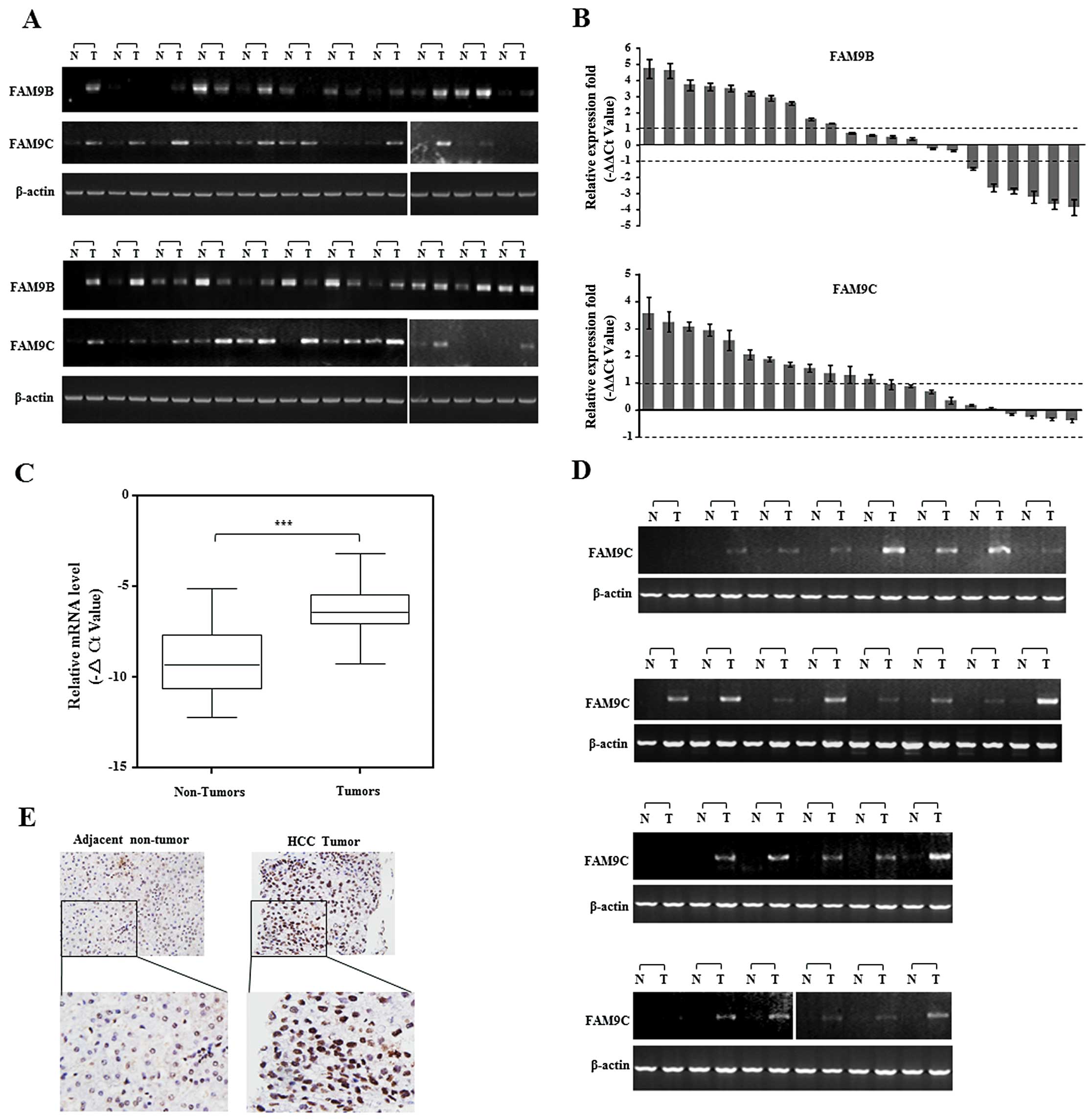
FAM9C was frequently upregulated in human
HCC
To evaluate the role of FAM9 members in HCC, we
first investigated the transcriptional levels of three FAM9 family
members, FAM9A, FAM9B and FAM9C, in 22 pairs of human HCC specimens
by semi-quantitative and real-time RT-PCR. The data showed that
only FAM9C mRNA was obviously elevated in more than a half of HCC
specimens examined, as compared with that of corresponding
non-cancerous livers, while FAM9A mRNA was not specifically
amplified and FAM9B exhibited disordered expression in these
specimens (Fig. 1A and B). To
confirm this result, FAM9C was further evaluated in additional 46
paired human HCC specimens by semi-quantitative and real-time PCR.
The result showed the relative mRNA level of FAM9C was
significantly upregulated in HCC tumor specimens compared with
corresponding adjacent non-tumor livers (P<0.001) (Fig. 1C), and the upregulation of FAM9C
mRNA was obvious in 25 of the 46 HCC specimens as shown by
semi-quantitative PCR (Fig. 1D).
Furthermore, we also examined the expression and location of FAM9C
protein in a pair of HCC specimens using immunohistochemical
staining with a specific antibody against FAM9C. The resulting data
showed that FAM9C protein was located in the nuclei, and presented
more expression in tumor tissue than adjacent non-tumor sample
(Fig. 1E). Collectively, these data
demonstrated that FAM9C was frequently elevated in human HCCs and
as a novel cancer testis gene.
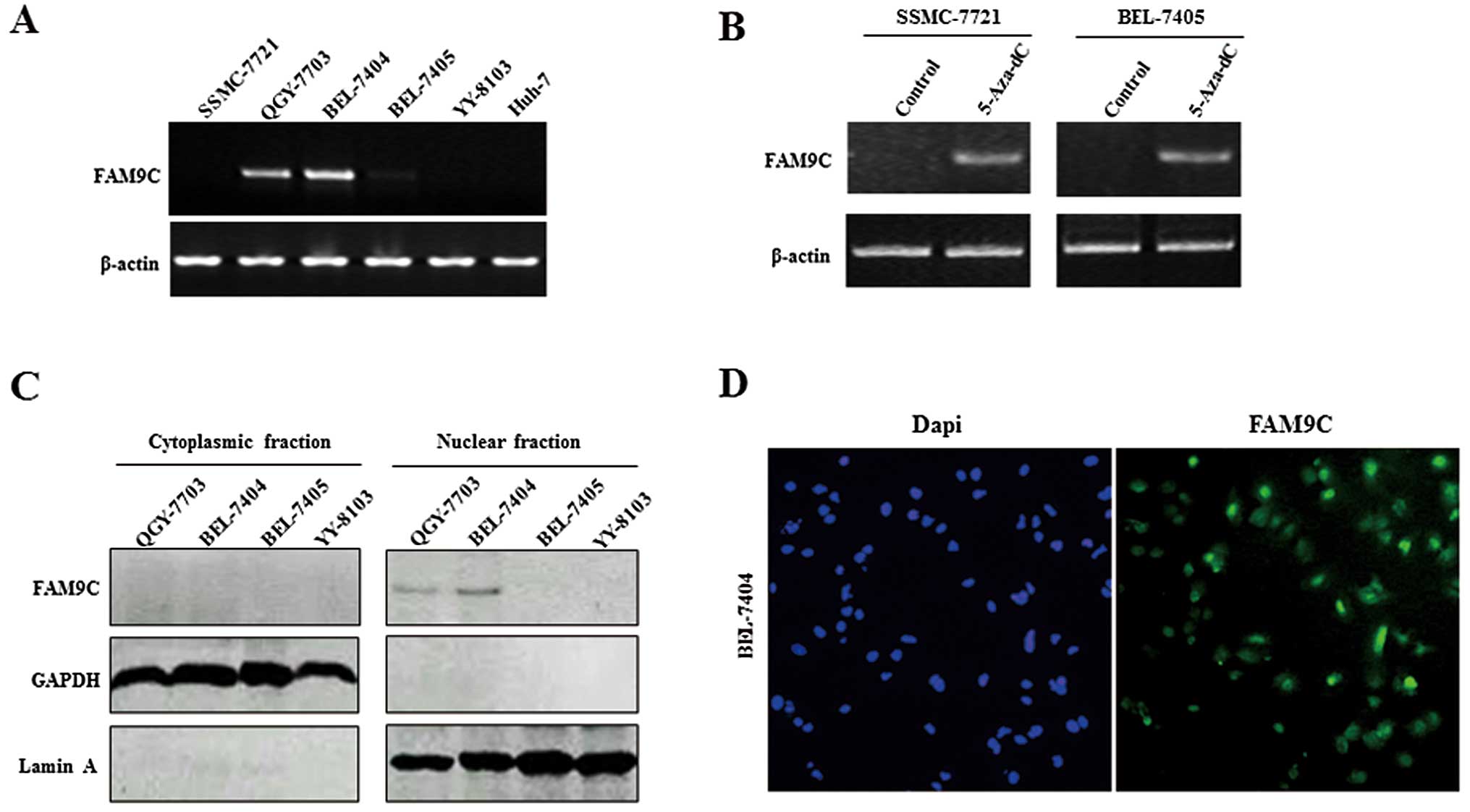
The expression and subcellular location
of FAM9C in HCC cell lines
The expression pattern of FAM9C was further
evaluated in some HCC cell lines by RT-PCR. The result showed that
FAM9C presented relatively high expression in QGY-7703 and BEL-7404
cells but low or no expression in other cell lines examined
(Fig. 2A). To investigate whether
DNA methylation regulates the FAM9C expression in HCC cells, we
treated two HCC cell lines SSMC-7721 and BEL-7405 with 5-Aza-dC, a
demethylating agent. RT-PCR analysis showed that FAM9C expression
was upregulated in these treated cells as compared with untreated
controls (Fig. 2B). In addition, we
determined the expression and subcellular location of FAM9C protein
in HCC cells by a western blot assay of cytoplasmic and nuclear
extracts, as well as a subcellular immunofluorescence assay. In
conformity with the mRNA expression levels of FAM9C, FAM9C protein
exhibited also relatively high expression in nuclear extracts of
QGY-7703 and BEL-7404 cells, but without expression in cytoplasmic
extracts of these cell lines (Fig.
2C). FAM9C was located in the nucleus in BEL-7404 cells, as
indicated by immunofluorescence assay (Fig. 2D).
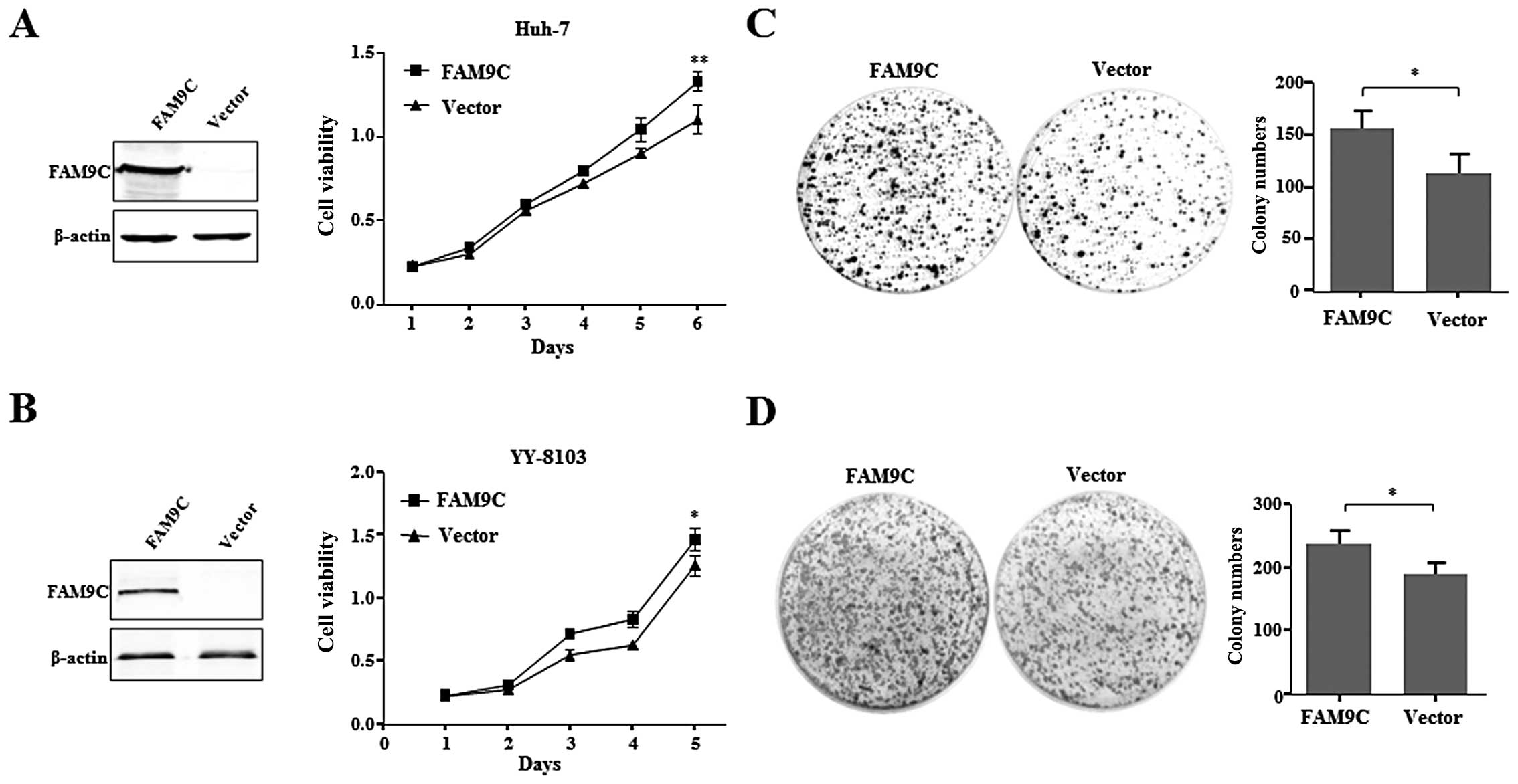
FAM9C overexpression promotes growth and
colony formation of HCC cells in vitro
To evaluate the role of FAM9C on HCC cells,
recombinant pcDNA3.1-FAM9C was transiently transfected into Huh-7
and YY-8103 cells with little FAM9C expression. The result showed
that overexpressed FAM9C, as demonstrated by western blot assay,
significantly promoted cell growth of the two HCC cell lines
compared with those transfected with empty vector (Fig. 3A and B). Furthermore, FAM9C also
promoted the anchorage-dependent colony formation of Huh-7 and
YY-8103 cells (Fig. 3C and D).
These data suggest that FAM9C overexpression plays an important
role in promoting HCC cell growth, colony formation in
vitro.
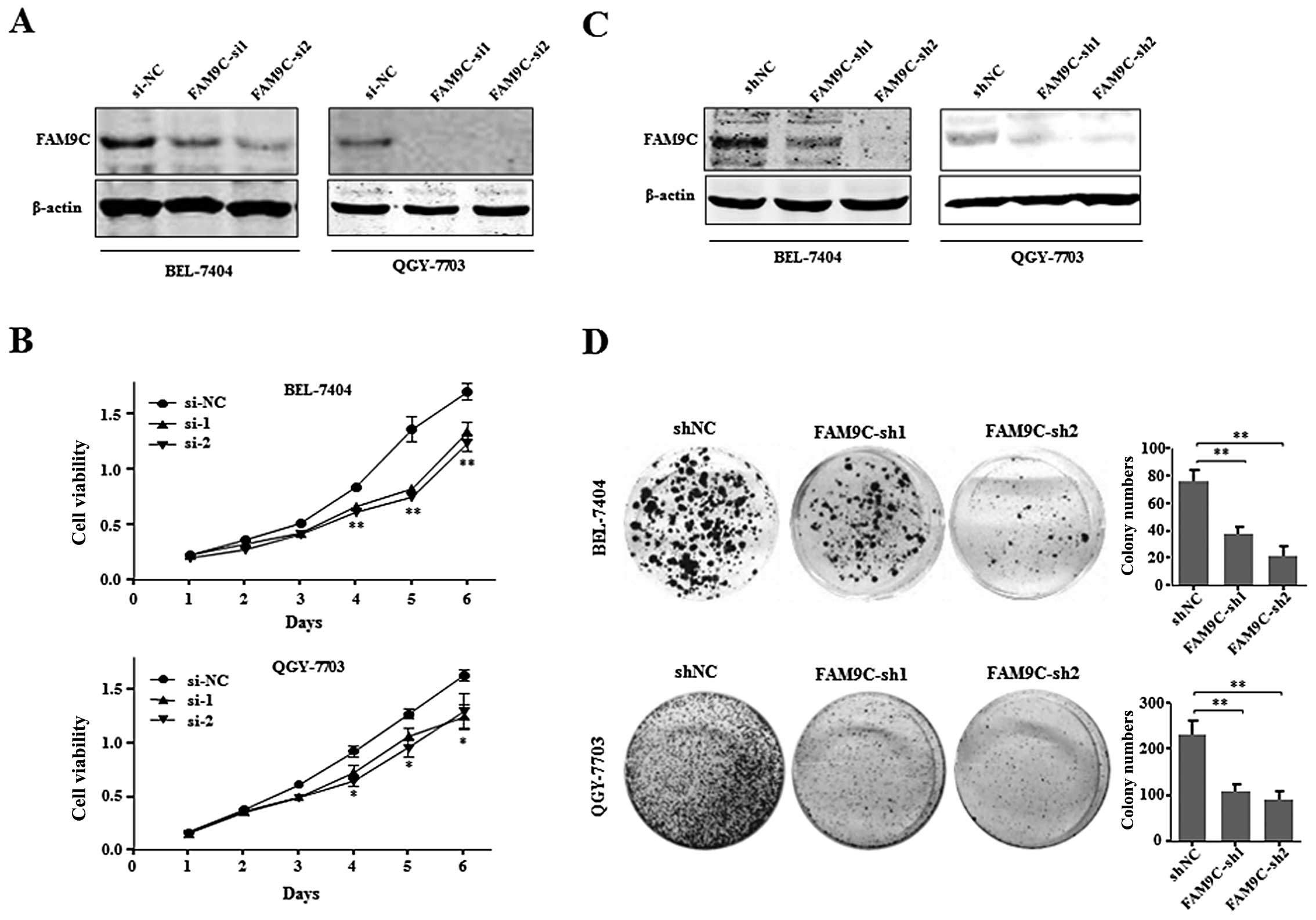
FAM9C knockdown inhibits cell
proliferation and colony formation of HCC cells in vitro
To further investigate the effect of FAM9C on HCC
cell proliferation and colony formation, we used chemically
synthesized siRNAs and shRNAs derived from recombinant pSUPER to
knock down endogenous FAM9C in BEL-7404 and QGY-7703 cells. Through
evaluating the knockdown efficiency of four siRNAs against FAM9C
(data not shown), the two efficient siRNAs (FAM9C-si1 and
FAM9C-si2) were used to silence endogenous FAM9C in BEL-7404 and
QGY-7703 cells (Fig. 4A). The
growth curve results showed that FAM9C knockdown significantly
inhibited the proliferation of BEL-7404 and QGY-7703 cells
(Fig. 4B). Moreover, pSUPER vectors
carrying shRNAs (FAM9C-sh1 and FAM9C-sh2) against FAM9C were
employed to FAM9C knockdown, where endogenous FAM9C was silenced in
the above two HCC cell lines respectively, as demonstrated by
western blot assay (Fig. 4C). The
colony formation assay demonstrated that FAM9C knockdown
significantly suppressed the anchorage-dependent clonogenicity of
BEL-7404 and QGY-7703 cells (Fig.
4D). These collective data implied that endogenous FAM9C could
be essential for maintaining the proliferation and
anchorage-dependent colony formation of HCC cells in
vitro.
The effect of FAM9C on the tumorigenicity
of HCC cells in nude mice
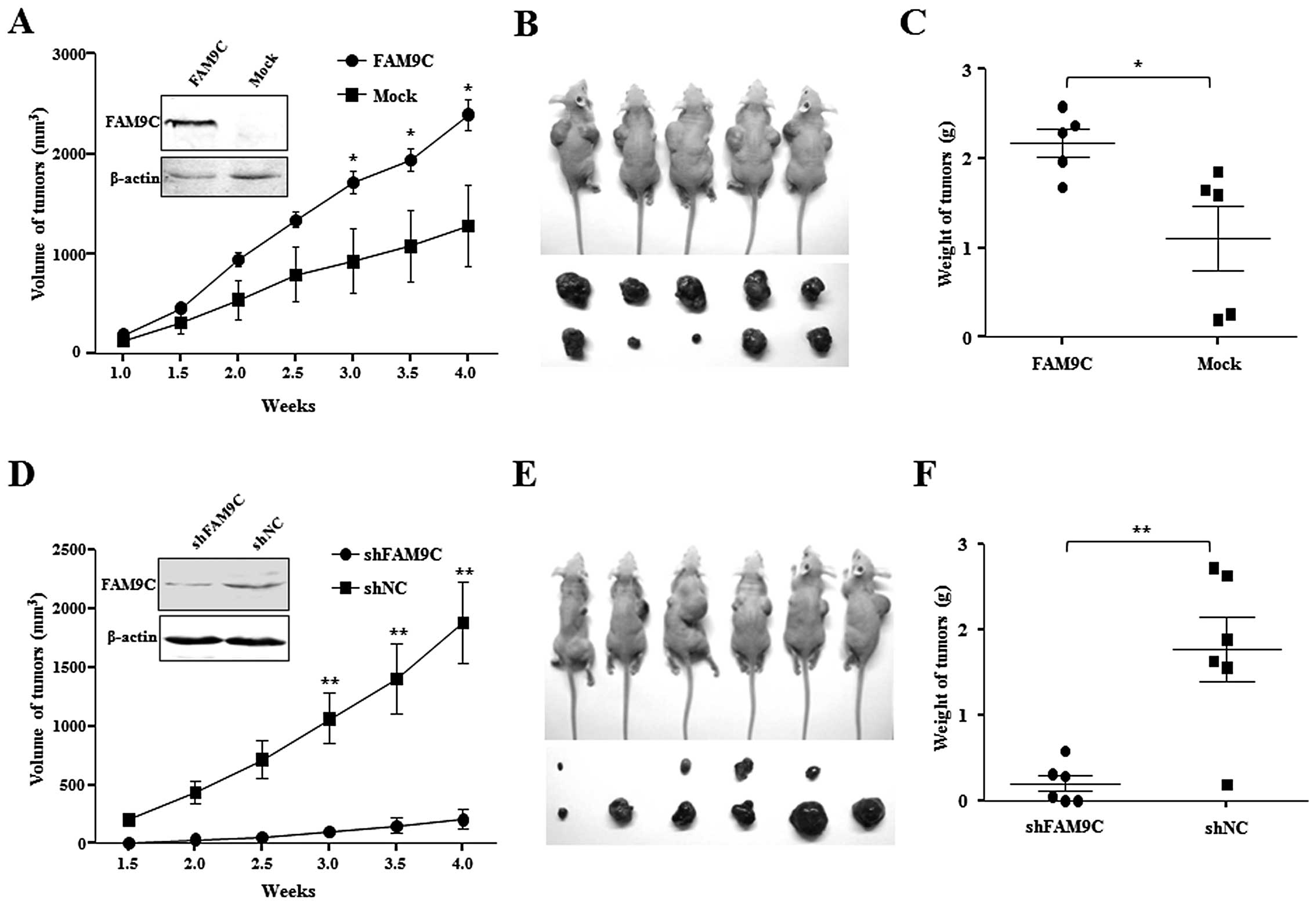
To determine whether the dysregulated FAM9C
contributes to hepatocarcinogenesis in vivo, Huh-7 cells
(2×106) stably expressing FAM9C were subcutaneously
injected into a flank of athymic nude mice, and the same amount of
subcloned Huh-7 cells transfected with pcDNA3.1-FAM9C but without
ectopic FAM9C expression as Mock control were subcutaneously
injected into the other flank of the same nude mice. As expected,
the stably FAM9C-expressing Huh-7 cells formed tumors faster than
the mock cells without FAM9C expression under the observation for
four weeks (P<0.05) (Fig. 5A).
The removed xenograft tumors from nude mice demonstrated that FAM9C
overexpression significantly promoted the tumor size and weight as
compared with that of mock objects (P<0.05) (Fig. 5B and C). These data indicated that
FAM9C overexpression significantly promoted tumorigenicity in
vivo of Huh-7 cells. Moreover, a total of 2×106
BEL-7404 cells, whose endogenous FAM9C was stably knocked down by
plasmid pSUPER-FAM9C-sh2, were subcutaneously injected into a flank
of athymic nude mice, and the same amount of cells transfected with
control plasmid (pSUPER-shNC) as negative control was injected into
the opposite flank of the same mice. Notably, FAM9C knockdown led
to significant tumor growth retardation under observation for four
weeks (P<0.01) (Fig. 5D), where
BEL-7404 cells with FAM9C silence had low tumorigenicity in
vivo in four of six mice tested, whereas cells used as controls
formed tumors in all six mice (Fig.
5E). The removed xenograft tumors from nude mice showed that
FAM9C silence significantly restrained the tumor size and weight as
compared with that of controls (P<0.01) (Fig. 5F). The result indicated that
silencing of FAM9C significantly inhibited tumorigenicity in
vivo of BEL-7404 cells.
FAM9C plays an anti-apoptotic role
through activating the PI3K-Akt signaling pathway in HCC cells
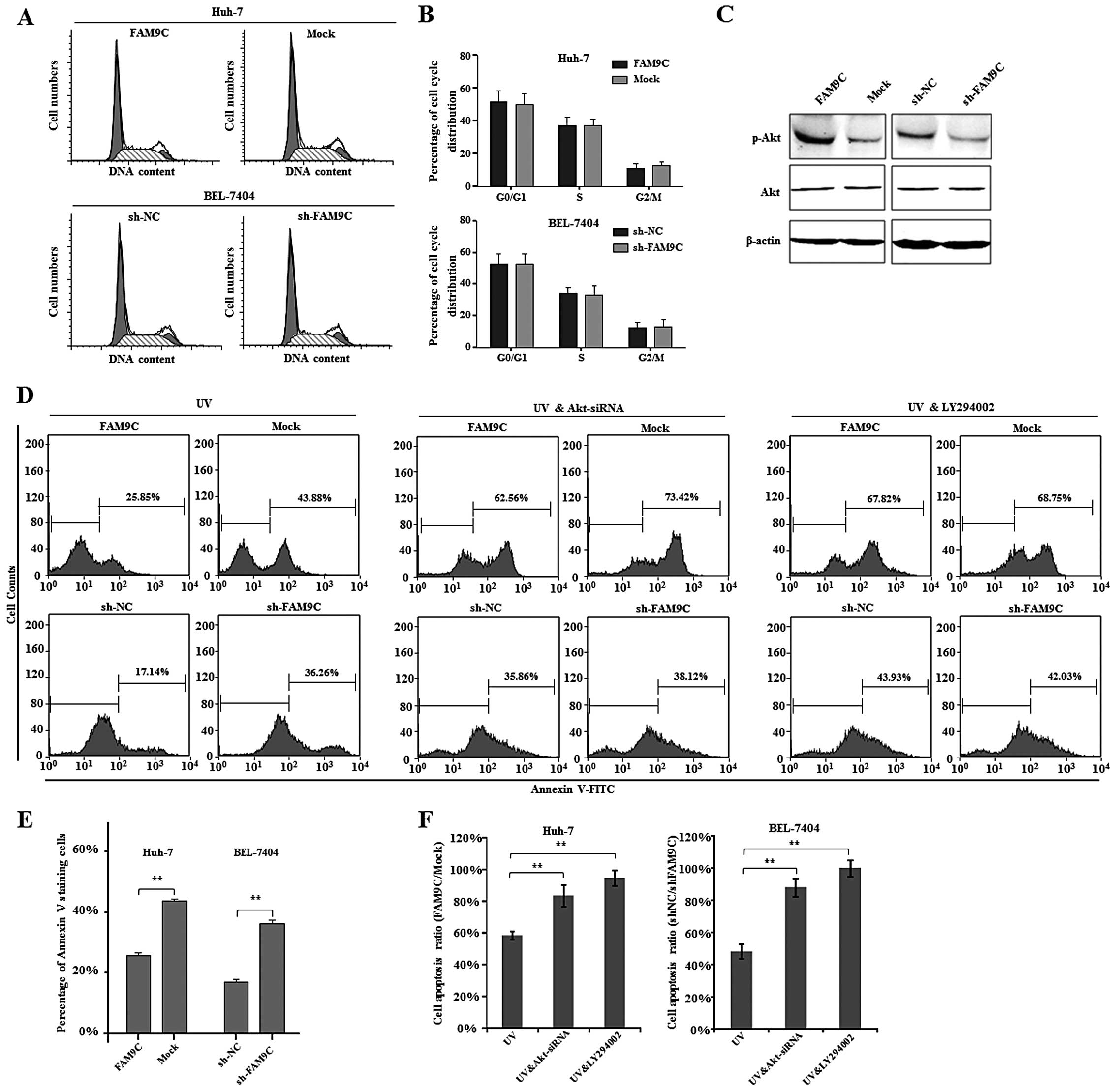
To explore the molecular mechanisms by which FAM9C
contributes to HCC cell proliferation, clonogenicity in
vitro and tumorigenicity in vivo, we first analyzed the
cell cycle of HCC cells with FAM9C overexpression or knockdown.
However, FAM9C overexpression and knockdown did not alter cell
cycle distribution of Huh-7 and BEL-7404 cells respectively
(Fig. 6A and B). As reported, FAM9C
protein exhibited similarity to SYCP3, a component of the
synaptonemal complex (12). More
importantly, SYCP3 led to activation of Akt, resulting in the
upregulation of anti-apoptotic proteins (14). To determine whether FAM9C as a
homologous gene of SYCP3 activates Akt signaling pathway, we
examined the activity of Akt pathway by western blot assay while
FAM9C overexpression or knockdown in HCC cells. We found that the
phosphorylation levels of Akt significantly increased in Huh-7
cells expressing FAM9C, whereas the phosphorylation levels of Akt
obviously decreased in BEL-7404 cells with FAM9C knockdown
(Fig. 6C). These data indicated
that FAM9C regulated activation of Akt.
To further address the role of FAM9C-induced
activation of Akt in HCC cells, we investigated cell apoptosis of
UV-irradiated HCC cells with FAM9C overexpression or knockdown by
flow cytometry method. The resulting data demonstrated that FAM9C
overexpression significantly reduced the proportion of apoptotic
Huh-7 cells (P<0.01), while FAM9C knockdown significantly
increased the apoptotic percentage of BEL-7404 cells (P<0.01),
indicating that FAM9C endowed HCC cells with resistance to
apoptosis (Fig. 6D and E). In
addition, to determine whether the activation of PI3K-Akt pathway
is directly responsible for the anti-apoptotic role of FAM9C in HCC
cells, we further performed cell apoptosis assays when Akt was
silenced by a validated siRNA (data not shown) or blocked by
LY294002, a known inhibitor of PI3K-Akt signaling pathway.
Interestingly, the results demonstrated that both siRNA against Akt
and LY294002 inhibitor significantly abolished or even prevented
the anti-apoptotic effect of FAM9C in Huh-7 and BEL-7404 cells with
UV exposure (P<0.01) (Fig. 6D and
F). Thus, these collective data suggest that FAM9C plays the
anti-apoptotic role through mediating PI3K-Akt signaling pathway in
HCC cells.
Discussion
FAM9 gene family mapped to Xp22.33-p22.31 includes
three members, FAM9A, FAM9B and FAM9C, which are expressed
exclusively in testis (12) and
have potential as cancer testis genes. In the present study, we
first investigated their expression profiling of the three FAM9
family members in human HCC specimens. Notably, only FAM9C was
frequently elevated in HCC specimens, suggesting that FAM9C could
be a novel cancer testis gene. FAM9C expression was reactivated in
HCC cells treated with the demethylating agent
5-aza-2′-deoxycitidine, implying that promoter demethylation was
related to FAM9C deregulation in HCC. Although global DNA
hypomethylation as a characteristic biomarker of liver cancer
(15) has been identified as a
cause of oncogenesis (16), some
specific mechanisms especially the involvement in reactivated
cancer testis genes are still obscure.
The crucial question is whether FAM9C upregulation
contributes to neoplastic phenotype of HCC. As previously reported,
restricted tissue-specific expression of FAM9C to testis,
subcellular localization of the encoded protein to nucleus, and
homology to SYCP3, encoding an essential structural component of
the synaptonemal complex which is involved in synapsis,
recombination and segregation of meiotic chromosomes, indicate
FAM9C protein may be involved in the meiotic process (12). SYCP3 has been reported to be
aberrantly expressed in tumors (17–19).
SYCP3 impairs mitotic recombination by interfering with BRCA2,
highlighting a new mechanism for chromosomal instability in cancers
(20). However, the function of
FAM9C gene is not yet determined by experiments. In the present
study, FAM9C overexpression promoted cell proliferation, in
vitro anchorage-dependent colony formation and in vivo
tumorigenicity of Huh-7 cells. On the contrary, FAM9C knockdown
induced by RNA interference inhibited cell proliferation, in
vitro clonogenicity and in vivo tumorigenicity of
BEL-7404 cells. These data showed that FAM9C as a novel cancer
testis gene contributed to HCC and thereby could be a potential
therapeutic target for HCC. However, further cell cycle analysis by
flow cytometry showed FAM9C had no significant effect on cell cycle
progression. Thus, whether FAM9C is involved in the formation of
the synaptonemal complex or DNA replication are worthy further
attention.
The molecular mechanism involved in these effects of
FAM9C on HCC cells in vitro and in vivo, indicated
that FAM9C was able to regulate the activation of Akt signaling
pathway. PI3K/Akt/mTOR signaling pathway is commonly deregulated in
a variety of cancers due to genetic alternations and promotes
cellular growth, proliferation, and survival (21–24).
Therefore, PI3K/Akt/mTOR signaling pathway represents a suitable
and promising therapeutic target in HCC, and many targeted agents
against this signaling pathway are in clinical trials (25–28).
In the present study, we revealed that upregulated FAM9C resulted
in activation of Akt in HCC cells, implying that mutually targeting
the FAM9C and PI3K/Akt/mTOR pathway may promote inhibitory effects
on tumors. We have noted that the demethylating reagent,
5-Aza-2′-deoxycytidine (5-Aza-dC) exerted antitumor effect on HCC
cells (29), but also caused the
upregulation of FAM9C in HCC cells. In light of the FAM9C-promoting
effects through the activation of Akt, we deduce that the
combination of 5-Aza-dC and inhibitors of PI3K-Akt signaling
pathway (e.g., LY294002) would more effectively inhibit
proliferation of HCC cells.
In conclusion, our findings demonstrated that FAM9C
as a novel cancer testis gene played an anti-apoptotic role through
regulating PI3K-Akt signaling pathway in human hepatocellular
carcinoma. We propose that FAM9C represents a novel target for
enhancing the response of HCC cells to demethylating reagents.
Acknowledgements
This research was supported in part by the Suzhou
Administration of Science and Technology (SYS201046 and
SZS201004).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar
|
|
3
|
Kew MC: Epidemiology of hepatocellular
carcinoma in sub-Saharan Africa. Ann Hepatol. 12:173–182.
2013.PubMed/NCBI
|
|
4
|
Hoshida Y, Moeini A, Alsinet C, Kojima K
and Villanueva A: Gene signatures in the management of
hepatocellular carcinoma. Semin Oncol. 39:473–485. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alsinet C and Villanueva A: Genomic
prognostic markers in hepatocellular carcinoma. Gastroenterol
Hepatol. 35:94–101. 2012.(In Spanish).
|
|
6
|
Sherman M: Modern approach to
hepatocellular carcinoma. Curr Gastroenterol Rep. 13:49–55. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Simpson AJ, Caballero OL, Jungbluth A,
Chen YT and Old LJ: Cancer/testis antigens, gametogenesis and
cancer. Nat Rev Cancer. 5:615–625. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao L, Mou DC, Leng XS, et al: Expression
of cancer-testis antigens in hepatocellular carcinoma. World J
Gastroenterol. 10:2034–2038. 2004.PubMed/NCBI
|
|
9
|
Peng JR, Chen HS, Mou DC, et al:
Expression of cancer/testis (CT) antigens in Chinese hepatocellular
carcinoma and its correlation with clinical parameters. Cancer
Lett. 219:223–232. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen K, Huang W, Huang B, et al: BORIS,
brother of the regulator of imprinted sites, is aberrantly
expressed in hepatocellular carcinoma. Genet Test Mol Biomarkers.
17:160–165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu H, Gu N, Liu ZB, et al: NY-ESO-1
expression in hepatocellular carcinoma: A potential new marker for
early recurrence after surgery. Oncol Lett. 3:39–44.
2012.PubMed/NCBI
|
|
12
|
Martinez-Garay I, Jablonka S, Sutajova M,
Steuernagel P, Gal A and Kutsche K: A new gene family (FAM9) of
low-copy repeats in Xp22.3 expressed exclusively in testis:
implications for recombinations in this region. Genomics.
80:259–267. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
|
|
14
|
Kang TH, Noh KH, Kim JH, et al: Ectopic
expression of X-linked lymphocyte-regulated protein pM1 renders
tumor cells resistant to antitumor immunity. Cancer Res.
70:3062–3070. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guerrero-Preston R, Santella RM, Blanco A,
Desai M, Berdasco M and Fraga M: Global DNA hypomethylation in
liver cancer cases and controls: a phase I preclinical biomarker
development study. Epigenetics. 2:223–226. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Akhavan-Niaki H and Samadani AA: DNA
methylation and cancer development: molecular mechanism. Cell
Biochem Biophys. Mar 19–2013.(Epub ahead of print).
|
|
17
|
Niemeyer P, Tureci O, Eberle T, Graf N,
Pfreundschuh M and Sahin U: Expression of serologically identified
tumor antigens in acute leukemias. Leuk Res. 27:655–660. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kalejs M, Ivanov A, Plakhins G, et al:
Upregulation of meiosis-specific genes in lymphoma cell lines
following genotoxic insult and induction of mitotic catastrophe.
BMC Cancer. 6:62006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mobasheri MB, Jahanzad I, Mohagheghi MA,
Aarabi M, Farzan S and Modarressi MH: Expression of two
testis-specific genes, TSGA10 and SYCP3, in different cancers
regarding to their pathological features. Cancer Detect Prev.
31:296–302. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hosoya N, Okajima M, Kinomura A, et al:
Synaptonemal complex protein SYCP3 impairs mitotic recombination by
interfering with BRCA2. EMBO Rep. 13:44–51. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
De Luca A, Maiello MR, D’Alessio A,
Pergameno M and Normanno N: The RAS/RAF/MEK/ERK and the PI3K/AKT
signalling pathways: role in cancer pathogenesis and implications
for therapeutic approaches. Expert Opin Ther Targets. 16(Suppl 2):
S17–S27. 2012.PubMed/NCBI
|
|
22
|
Zhou Q, Wong CH, Lau CP, et al: Enhanced
antitumor activity with combining effect of mTOR inhibition and
microtubule stabilization in hepatocellular carcinoma. Int J
Hepatol. 2013:1038302013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Touil Y, Zuliani T, Wolowczuk I, et al:
The PI3K/AKT signaling pathway controls the quiescence of the
low-Rhodamine123-retention cell compartment enriched for melanoma
stem cell activity. Stem Cells. 31:641–651. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Almhanna K, Strosberg J and Malafa M:
Targeting AKT protein kinase in gastric cancer. Anticancer Res.
31:4387–4392. 2011.PubMed/NCBI
|
|
25
|
Gedaly R, Angulo P, Chen C, et al: The
role of PI3K/mTOR inhibition in combination with sorafenib in
hepatocellular carcinoma treatment. Anticancer Res. 32:2531–2536.
2012.PubMed/NCBI
|
|
26
|
Muntane J, De la Rosa AJ, Docobo F,
Garcia-Carbonero R and Padillo FJ: Targeting tyrosine kinase
receptors in hepatocellular carcinoma. Curr Cancer Drug Targets.
13:300–312. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Temirak A, Abdulla M and Elhefnawi M:
Rational drug design for identifying novel multi-target inhibitors
for hepatocellular carcinoma. Anticancer Agents Med Chem.
12:1088–1097. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Grabinski N, Ewald F, Hofmann BT, et al:
Combined targeting of AKT and mTOR synergistically inhibits
proliferation of hepatocellular carcinoma cells. Mol Cancer.
11:852012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tao SF, Zhang CS, Guo XL, et al:
Anti-tumor effect of 5-aza-2′-deoxycytidine by inhibiting
telomerase activity in hepatocellular carcinoma cells. World J
Gastroenterol. 18:2334–2343. 2012.
|