Introduction
Lumican is a member of the class II small
leucine-rich proteoglycan (SLRP) family. Members of this family
have relatively small molecular sizes, with core proteins of
approximately 40 kDa, and possess 6–10 leucine-rich repeat units in
the core protein (1,2). Amino acid sequencing indicates that
lumican has 4 potential sites for N-linked keratan sulfate
(KS) or oligosaccharides (3,4).
Therefore, lumican includes a core protein, glycoprotein and
proteoglycan forms due to glycosylation (5). Lumican is a secreted collagen-binding
extracellular matrix protein of the cornea, dermis and tendon
stroma, arterial wall, and intestinal submucosa (6–9).
Corneal opacity, as well as skin and tendon fragility due to
disorganized and loosely packed collagen fibers in lumican-null
mice suggest that lumican plays an important role in collagen
fibrillogenesis (10,11).
Lumican was first reported as one of the major KS
proteoglycans in the chicken cornea (12). In addition to the cornea, lumican
expression has been reported in various human tissues, including
malignant tumor tissues (5,13–27).
Among the clinicopathological characteristics of pancreatic ductal
adenocarcinoma (PDAC), the localization of lumican in the stromal
tissue adjacent to cancer cells correlates with advanced cancer
stage, retroperitoneal and duodenal invasion, and residual tumor,
and tends to correlate with shorter survival (21). These reports suggest that lumican
localized in the stromal tissue is secreted from cancer cells and
affects cancer cells through an autocrine and paracrine mechanism.
We previously reported that PANC-1 cells, one of the PDAC cell
lines, secrete only 70-kDa glycosylated lumican into the
extracellular space. We also demonstrated that the secreted lumican
stimulated cell growth through ERK activation and inhibited cell
invasion and matrix metalloproteinase (MMP)-9 activation using
lumican-overexpressing PANC-1 cells and lumican-downregulated
PANC-1 cells (28). However, the
mechanism of how lumican affects cell growth and invasion remains
unclear.
In the present study, we performed shotgun liquid
chromatography (LC)/mass spectrometry (MS)-based global proteomic
analysis using protein from lumican-overexpressing PANC-1 cells and
lumican-downregulated PANC-1 cells to examine how lumican regulates
cell growth and invasion in PDAC cells. We identified 24 candidate
proteins that may play an important role in cell growth and
invasion and could be regulated by lumican.
Materials and methods
Materials
The following materials were purchased from Wako
Pure Chemical Industries (Osaka, Japan): urea,
3-(3-cholamidopropyl) dimethylammonio-1-propanesulphonate (CHAPS),
dithiothreitol (DTT), Tris (2-carboxyethyl) phosphine hydrochloride
(TCEP) and iodoacetamide (IAA); Amicon Ultra 0.5-ml 3K was from
Millipore (Tokyo, Japan), and thiourea from Nacalai Tesque, Inc.
(Kyoto, Japan). All other chemicals and reagents were purchased
from Sigma Chemical Corp. (St. Louis, MO, USA).
PDAC cell line
PANC-1 cells were obtained from the Cell Resource
Center for Biomedical Research, Institute of Development, Aging and
Cancer, Tohoku University (Sendai, Japan).
Protein extraction of lumican-regulated
PANC-1 cells
The lumican-overexpressing PANC-1 cells,
lumican-downregulated PANC-1 cells, and control cell lines (Mock
and NC, respectively) were prepared as previously described
(28). The lumican-regulated PANC-1
cells were cultured at a density of 5×105 cells in a
100-mm dish in RPMI-1640 medium with 10% fetal bovine serum (FBS)
for 72 h. Then, cells were solubilized in urea lysis buffer (7 M
urea, 2 M thiourea, 5% CHAPS, 1% Triton X-100). Protein
concentration was measured using the Bradford method.
In-solution trypsin digestion
A gel-free digestion approach was performed in
accordance with the protocol described by Bluemlein and Ralser
(29). In brief, 10 μg of protein
extract from each sample was reduced by addition of 45 mM DTT and
20 mM TCEP and was then alkylated using 100 mM IAA. Following
alkylation, samples were digested with Proteomics Grade Trypsin
(Agilent Technologies, Inc., Santa Clara, CA, USA) at 37°C for 24
h. Next, digests were evaporated in a vacuum concentrator
centrifuge and the residue was resuspended in 0.1% trifluoroacetic
acid/5% acetonitrile. The digests were filtered through Amicon
Ultra 0.5-ml 3K to remove undigested proteins and the flow-through
was used in the following analyses.
LC-MS/MS analysis of protein
identification
Approximately 2-μg peptide samples were injected
into a peptide L-trap column (Chemicals Evaluation and Research
Institute, Tokyo, Japan) using an HTC PAL autosampler (CTC
Analytics, Zwingen, Switzerland) and further separated through an
Advance-nano UHPLC using a Reverse-Phase C18-column (Zaplous column
α, 3-μm diameter gel particles and 100 Å pore size, 0.1×150 mm;
both from AMR, Inc., Tokyo, Japan). The mobile phase consisted of
solution A (0.1% formic acid in water) and solution B
(acetonitrile). The column was developed at a flow rate of 500
nl/min with a concentration gradient of acetonitrile from 5 to 45%
B over 120 min. Gradient-eluted peptides were analyzed using an
amaZon ETD ion-trap mass spectrometer (Bruker Daltonics, Billerica,
MA, USA). The results were acquired in a data-dependent manner in
which MS/MS fragmentation was performed on the 10 most intense
peaks of every full MS scan.
All MS/MS spectra data were searched against the
SwissProt Homo sapiens database using Mascot (v2.3.01;
Matrix Science, London, UK). The search criteria was set as
follows: enzyme, trypsin; allowance of up to two missed cleavage
peptides; mass tolerance ± 0.5 Da and MS/MS tolerance ± 0.5 Da; and
modifications of cysteine carbamidomethylation and methionine
oxidation.
Semi-quantitative analysis of identified
proteins
The fold-changes in expressed proteins on the base 2
logarithmic scale were calculated using Rsc based upon spectral
counting (30). Relative amounts of
identified proteins were also calculated using the normalized
spectral abundance factor (NSAF) (31). Differentially expressed proteins
were chosen so that their Rsc satisfy >1 or <−1, which
correspond to fold-changes of >2 or <0.5.
Bioinformatics
Functional annotations for the identified proteins
whose expression level was regulated by lumican were processed
using the Database for Annotation, Visualization and Integrated
Discovery (DAVID), v6.7 (http://david.abcc.ncifcrf.gov/home.jsp) (32–34).
Results
Protein identification and profile in
lumican-regulated PANC-1 cells
To examine the effect of lumican on cell growth and
invasion of PDAC cells, we created two types of PANC-1 cells whose
lumican expression level was regulated: lumican-overexpressing
PANC-1 cells and lumican-downregulated PANC-1 cells (28). We then investigated the molecular
profile of proteins whose expression level was regulated by lumican
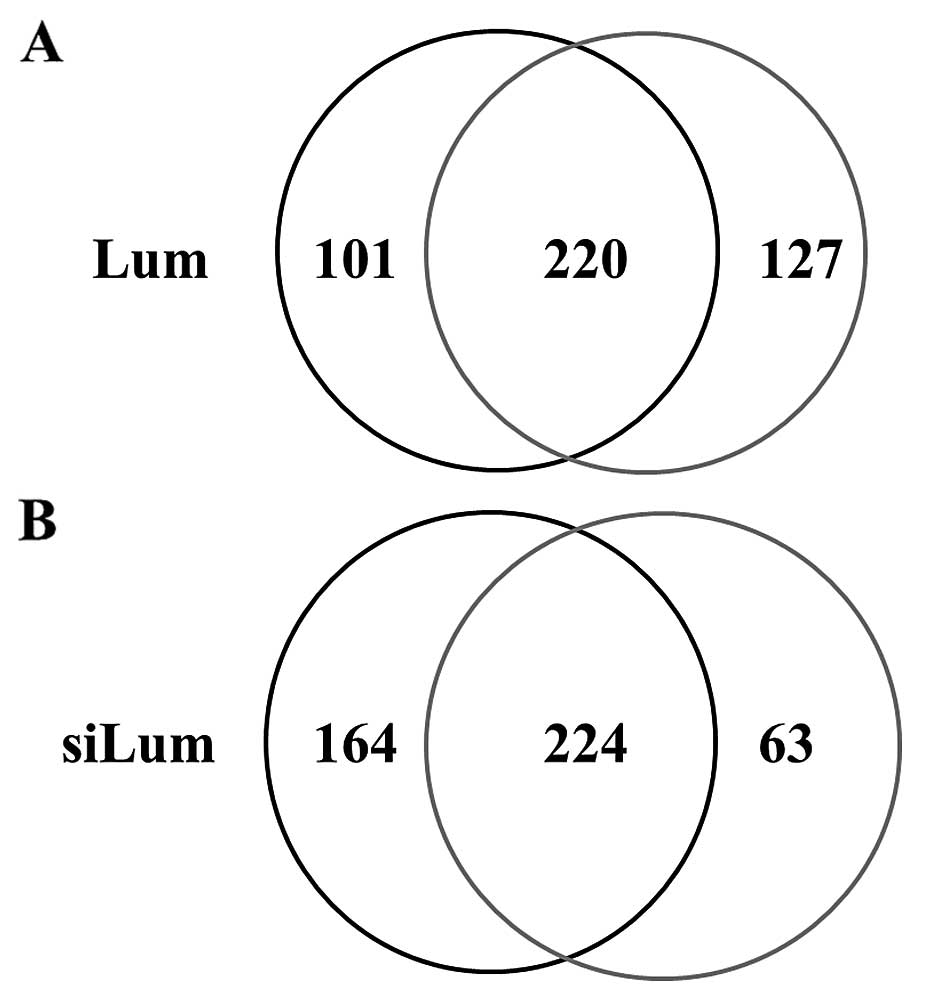
using shotgun proteomics. Fig. 1
shows the Venn map for the identified proteins in lumican-regulated
PANC-1 cells. In lumican-overexpressing PANC-1 cells (Lum), 321
proteins were identified, and 347 were identified in control cells
(Mock) under the search parameter settings used (Fig. 1A). On the other hand, 388 proteins
were identified in lumican-downregulated PANC-1 cells (siLum) and
287 in control cells (NC) (Fig.
1B). Among the 448 proteins identified from lumican upregulated
cells, 220 (49.1%) proteins were identified in both cell lines,
while 101 (22.6%) and 127 (28.3%) proteins were unique to Lum and
Mock, respectively (Fig. 1A). Of
the 451 total proteins identified from lumican downregulated cells,
224 (49.7%) proteins were identified in both cell lines, whereas
164 (36.3%) and 63 (14.0%) proteins were unique to siLum and NC,
respectively (Fig. 1B).
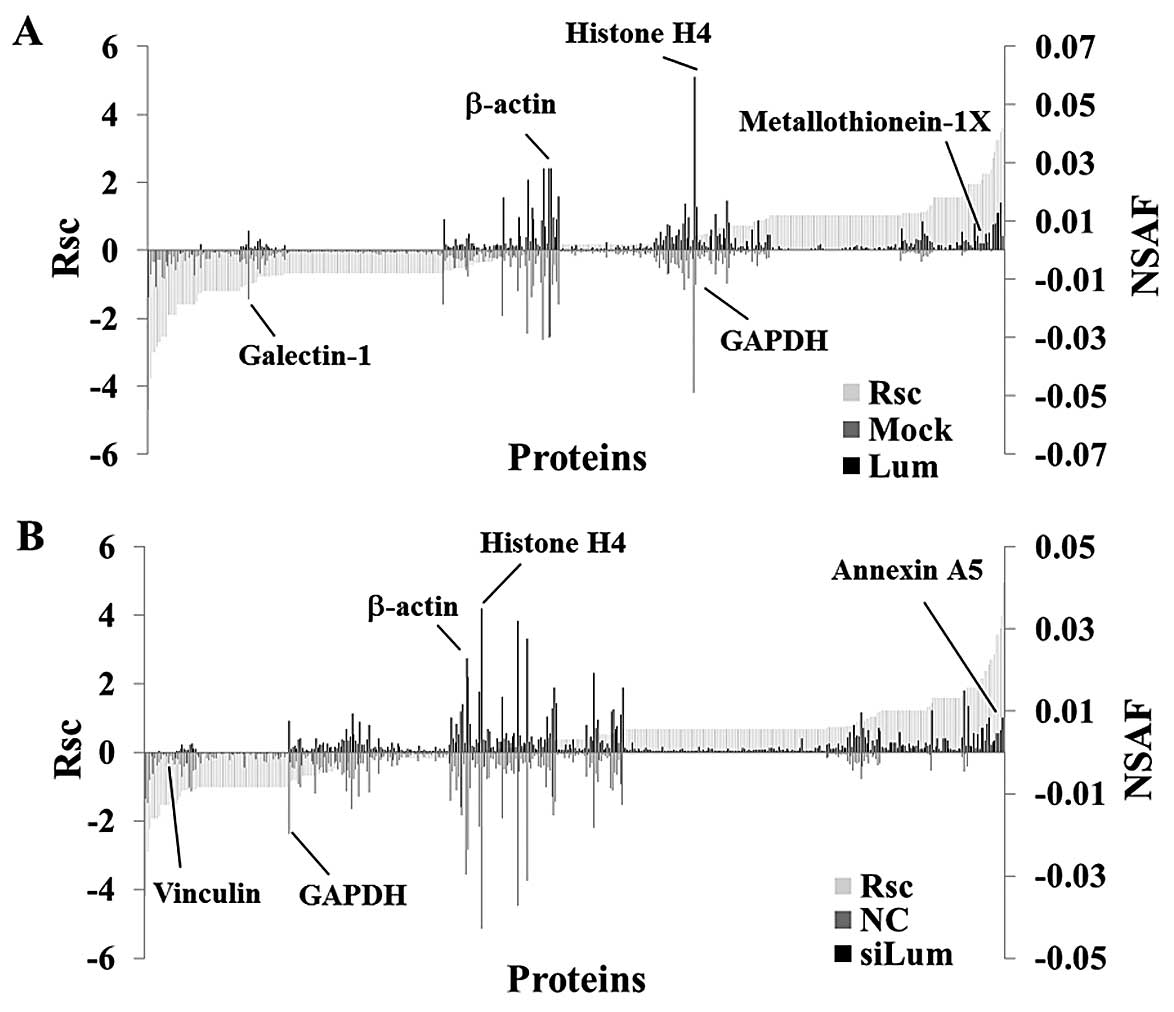
Semi-quantitative comparison of
identified proteins in lumican-regulated PANC-1 cells
Next, we performed a label-free semi-quantitative
method based on spectral counting, as described in the Materials
and methods section, to find proteins whose expression levels were
regulated by lumican. The Rsc value was plotted against the
corresponding protein (X-axis) from left to right for proteins
identified in the Lum and Mock groups (Fig. 2A). The positive and negative Rsc
values indicate increased and decreased expression, respectively,
in the Lum group. The NSAF value (bar) plotted against the
corresponding protein (X-axis), NSAF of Lum (black bar) and Mock
(gray bar) proteins are indicated above and below the X-axis,
respectively (Fig. 2A). Proteins
with either a high positive or negative Rsc value were considered
candidate proteins whose expression level was regulated by lumican.
Fig. 2B shows the Rsc and NSAF
values of the siLum and NC groups as described above. In the
lumican upregulated cells, the Rsc of metallothionein (MT)-1X was
positive, and the Rsc of galectin-1 was negative (Fig. 2A). In the lumican downregulated
cells, the Rsc of Annexin A5 was positive and the Rsc of vinculin
was negative (Fig. 2B).
Housekeeping proteins such as β-actin, histone H4 and GAPDH were
located near the center of the X-axis (Fig. 2).
As a result of semi-quantification, 174
differentially expressed proteins were identified in lumican
upregulated PANC-1 cells (Table I),
and a total of 143 differentially expressed proteins were
identified in lumican downregulated PANC-1 cells (Table II). However, the expression level
of housekeeping proteins such as β-actin, GAPDH and histone H4 was
not changed in lumican upregulated cells or lumican downregulated
cells. In order to identify proteins whose expression levels were
regulated by lumican, we selected proteins whose Rsc value was
inversely associated between lumican upregulated cells and lumican
downregulated cells. Twenty four proteins were identified as
candidates (Table III).
 | Table IDifferentially expressed proteins in
lumican upregulated PANC-1 cells. |
Table I
Differentially expressed proteins in
lumican upregulated PANC-1 cells.
| | | | Spectral
counting |
|---|
| | | |
|
|---|
| No. | ID | Accession no. and
description | No. of amino
acids | Mock | Lum | Fold-change
(Rsc) |
|---|
| 1 | ACTH_HUMAN | (P63267) Actin,
γ-enteric smooth muscle | 376 | 35 | 0 | −4.69652 |
| 2 | ACTC_HUMAN | (P68032) Actin, α
cardiac muscle 1 | 377 | 18 | 0 | −3.7706617 |
| 3 | TBB2A_HUMAN | (Q13885) Tubulin β-2A
chain | 445 | 10 | 0 | −2.9897736 |
| 4 | H2B1H_HUMAN | (Q93079) Histone H2B
type 1-H | 126 | 9 | 0 | −2.8547299 |
| 5 | HS904_HUMAN | (Q58FG1) Putative
heat shock protein HSP 90-α A4 | 418 | 8 | 0 | −2.7058891 |
| 6 | TBA4A_HUMAN | (P68366) Tubulin
α-4A chain | 448 | 8 | 0 | −2.7058891 |
| 7 | H2AJ_HUMAN | (Q9BTM1) Histone
H2A.J | 129 | 7 | 0 | −2.540088 |
| 8 | H2B3B_HUMAN | (Q8N257) Histone
H2B type 3-B | 126 | 7 | 0 | −2.540088 |
| 9 | RLA0_HUMAN | (P05388) 60S acidic
ribosomal protein P0 | 317 | 7 | 0 | −2.540088 |
| 10 | H2B1O_HUMAN | (P23527) Histone
H2B type 1-O | 126 | 4 | 0 | −1.885788 |
| 11 | K2C71_HUMAN | (Q3SY84) Keratin,
type II cytoskeletal 71 | 523 | 4 | 0 | −1.885788 |
| 12 | NDKB_HUMAN | (P22392) Nucleoside
diphosphate kinase B | 152 | 4 | 0 | −1.885788 |
| 13 | RA1L2_HUMAN | (Q32P51)
Heterogeneous nuclear ribonucleoprotein A1-like 2 | 320 | 4 | 0 | −1.885788 |
| 14 | CH10_HUMAN | (P61604) 10 kDa
heat shock protein, mitochondrial | 102 | 4 | 0 | −1.885788 |
| 15 | H2A1H_HUMAN | (Q96KK5) Histone
H2A type 1-H | 128 | 3 | 0 | −1.5801931 |
| 16 | HNRPK_HUMAN | (P61978)
Heterogeneous nuclear ribonucleoprotein K | 463 | 3 | 0 | −1.5801931 |
| 17 | K1C25_HUMAN | (Q7Z3Z0) Keratin,
type I cytoskeletal 25 | 450 | 3 | 0 | −1.5801931 |
| 18 | K22O_HUMAN | (Q01546) Keratin,
type II cytoskeletal 2 oral | 638 | 3 | 0 | −1.5801931 |
| 19 | MYL6_HUMAN | (P60660) Myosin
light polypeptide 6 | 151 | 3 | 0 | −1.5801931 |
| 20 | NFH_HUMAN | (P12036)
Neurofilament heavy polypeptide | 1026 | 3 | 0 | −1.5801931 |
| 21 | TAGL3_HUMAN | (Q9UI15)
Transgelin-3 | 199 | 3 | 0 | −1.5801931 |
| 22 | YI016_HUMAN | (A6NKZ8) Putative
tubulin β chain-like protein | 372 | 3 | 0 | −1.5801931 |
| 23 | ANXA5_HUMAN | (P08758) Annexin
A5 | 320 | 3 | 0 | −1.5801931 |
| 24 | MT1G_HUMAN | (P13640)
Metallothionein-1G | 62 | 3 | 0 | −1.5801931 |
| 25 | K1C14_HUMAN | (P02533) Keratin,
type I cytoskeletal 14 | 472 | 6 | 1 | −1.5040946 |
| 26 | K2C80_HUMAN | (Q6KB66) Keratin,
type II cytoskeletal 80 | 452 | 5 | 1 | −1.2892288 |
| 27 | ACTBL_HUMAN | (Q562R1)
β-actin-like protein 2 | 376 | 13 | 4 | −1.259281 |
| 28 | 1433B_HUMAN | (P31946) 14-3-3
protein β/α | 246 | 2 | 0 | −1.1924301 |
| 29 | AINX_HUMAN | (Q16352)
α-internexin | 499 | 2 | 0 | −1.1924301 |
| 30 | H2AZ_HUMAN | (H2AZ_HUMAN)
Histone H2A.Z | 128 | 2 | 0 | −1.1924301 |
| 31 | H2B1D_HUMAN | (P58876) Histone
H2B type 1-D | 126 | 2 | 0 | −1.1924301 |
| 32 | H2B1J_HUMAN | (P06899) Histone
H2B type 1-J | 126 | 2 | 0 | −1.1924301 |
| 33 | K1C13_HUMAN | (P13646) Keratin,
type I cytoskeletal 13 | 458 | 2 | 0 | −1.1924301 |
| 34 | K2C6C_HUMAN | (P48668) Keratin,
type II cytoskeletal 6C | 564 | 2 | 0 | −1.1924301 |
| 35 | KRT85_HUMAN | (P78386) Keratin,
type II cuticular Hb5 | 507 | 2 | 0 | −1.1924301 |
| 36 | MLL3_HUMAN | (Q8NEZ4)
Histone-lysine N-methyltransferase MLL3 | 4911 | 2 | 0 | −1.1924301 |
| 37 | NDK8_HUMAN | (O60361) Putative
nucleoside diphosphate kinase | 137 | 2 | 0 | −1.1924301 |
| 38 | NFM_HUMAN | (P07197)
Neurofilament medium polypeptide | 916 | 2 | 0 | −1.1924301 |
| 39 | SERPH_HUMAN | (P50454) Serpin
H1 | 418 | 2 | 0 | −1.1924301 |
| 40 | U17L5_HUMAN | (A8MUK1) Ubiquitin
carboxyl-terminal hydrolase 17-like protein 5 | 530 | 2 | 0 | −1.1924301 |
| 41 | URFB1_HUMAN | (Q6BDS2)
UHRF1-binding protein 1 | 1440 | 2 | 0 | −1.1924301 |
| 42 | MDHM_HUMAN | (P40926) Malate
dehydrogenase, mitochondrial | 338 | 2 | 0 | −1.1924301 |
| 43 | IF5A1_HUMAN | (P63241) Eukaryotic
translation initiation factor 5A-1 | 154 | 2 | 0 | −1.1924301 |
| 44 | SFPQ_HUMAN | (P23246) Splicing
factor, proline- and glutamine-rich | 707 | 2 | 0 | −1.1924301 |
| 45 | NACA_HUMAN | (Q13765) Nascent
polypeptide-associated complex subunit α | 215 | 2 | 0 | −1.1924301 |
| 46 | EF1B_HUMAN | (P24534) Elongation
factor 1-β | 225 | 2 | 0 | −1.1924301 |
| 47 | RS4X_HUMAN | (P62701) 40S
ribosomal protein S4, X isoform | 263 | 2 | 0 | −1.1924301 |
| 48 | IF4A2_HUMAN | (Q14240) Eukaryotic
initiation factor 4A-II | 407 | 9 | 3 | −1.0866682 |
| 49 | EF2_HUMAN | (P13639) Elongation
factor 2 | 858 | 16 | 6 | −1.0697973 |
| 50 | HSP72_HUMAN | (P54652) Heat
shock-related 70 kDa protein 2 | 639 | 11 | 4 | −1.0396137 |
| 51 | DESM_HUMAN | (P17661)
Desmin | 470 | 13 | 5 | −1.0068984 |
| 52 | LEG1_HUMAN | (P09382)
Galectin-1 | 135 | 13 | 5 | −1.0068984 |
| 53 | CC113_HUMAN | (Q9H0I3)
Coiled-coil domain-containing protein 113 | 377 | 0 | 1 | 1.03639903 |
| 54 | H11_HUMAN | (Q02539) Histone
H1.1 | 215 | 0 | 1 | 1.03639903 |
| 55 | UBA1_HUMAN | (P22314)
Ubiquitin-like modifier-activating enzyme 1 | 1058 | 0 | 1 | 1.03639903 |
| 56 | TCPH_HUMAN | (Q99832) T-complex
protein 1 subunit eta | 543 | 0 | 1 | 1.03639903 |
| 57 | RL31_HUMAN | (P62899) 60S
ribosomal protein L31 | 125 | 0 | 1 | 1.03639903 |
| 58 | ADT4_HUMAN | (Q9H0C2) ADP/ATP
translocase 4 | 315 | 0 | 1 | 1.03639903 |
| 59 | BRE1B_HUMAN | (O75150) E3
ubiquitin-protein ligase BRE1B | 1001 | 0 | 1 | 1.03639903 |
| 60 | CALX_HUMAN | (P27824)
Calnexin | 592 | 0 | 1 | 1.03639903 |
| 61 | CAP1_HUMAN | (Q01518) Adenylyl
cyclase-associated protein 1 | 475 | 0 | 1 | 1.03639903 |
| 62 | CAZA1_HUMAN | (P52907)
F-actin-capping protein subunit α-1 | 286 | 0 | 1 | 1.03639903 |
| 63 | CCNT1_HUMAN | (O60563)
Cyclin-T1 | 726 | 0 | 1 | 1.03639903 |
| 64 | CGNL1_HUMAN | (Q0VF96)
Cingulin-like protein 1 | 1302 | 0 | 1 | 1.03639903 |
| 65 | CISY_HUMAN | (O75390) Citrate
synthase, mitochondrial | 466 | 0 | 1 | 1.03639903 |
| 66 | CLIP1_HUMAN | (P30622) CAP-Gly
domain-containing linker protein 1 | 1438 | 0 | 1 | 1.03639903 |
| 67 | CN37_HUMAN | (P09543)
2′,3′-cyclic-nucleotide 3′-phosphodiesterase | 421 | 0 | 1 | 1.03639903 |
| 68 | CTSR1_HUMAN | (Q8NEC5) Cation
channel sperm-associated protein 1 | 780 | 0 | 1 | 1.03639903 |
| 69 | CX057_HUMAN | (Q6NSI4)
Uncharacterized protein CXorf57 | 855 | 0 | 1 | 1.03639903 |
| 70 | DDX21_HUMAN | (Q9NR30) Nucleolar
RNA helicase 2 | 783 | 0 | 1 | 1.03639903 |
| 71 | DYXC1_HUMAN | (Q8WXU2) Dyslexia
susceptibility 1 candidate gene 1 protein | 420 | 0 | 1 | 1.03639903 |
| 72 | EFTU_HUMAN | (P49411) Elongation
factor Tu, mitochondrial | 452 | 0 | 1 | 1.03639903 |
| 73 | EHD1_HUMAN | (Q9H4M9) EH
domain-containing protein 1 | 534 | 0 | 1 | 1.03639903 |
| 74 | EIF3H_HUMAN | (O15372) Eukaryotic
translation initiation factor 3 subunit H | 352 | 0 | 1 | 1.03639903 |
| 75 | EHD1_HUMAN | (Q9H4M9) EH
domain-containing protein 1 | 534 | 0 | 1 | 1.03639903 |
| 76 | FETUA_HUMAN | (P02765)
α-2-HS-glycoprotein | 367 | 0 | 1 | 1.03639903 |
| 77 | HIP1R_HUMAN | (O75146)
Huntingtin-interacting protein 1-related protein | 1068 | 0 | 1 | 1.03639903 |
| 78 | HMGN1_HUMAN | (P05114)
Non-histone chromosomal protein HMG-14 | 100 | 0 | 1 | 1.03639903 |
| 79 | HMGN4_HUMAN | (O00479) High
mobility group nucleosome-binding domain- containing protein 4 | 90 | 0 | 1 | 1.03639903 |
| 80 | HNRH3_HUMAN | (P31942)
Heterogeneous nuclear ribonucleoprotein H3 | 346 | 0 | 1 | 1.03639903 |
| 81 | IDE_HUMAN | (P14735)
Insulin-degrading enzyme | 1019 | 0 | 1 | 1.03639903 |
| 82 | IMA2_HUMAN | (P52292) Importin
subunit α-2 | 529 | 0 | 1 | 1.03639903 |
| 83 | IMDH2_HUMAN | (P12268)
Inosine-5′-monophosphate dehydrogenase 2 | 514 | 0 | 1 | 1.03639903 |
| 84 | K1C26_HUMAN | (Q7Z3Y9) Keratin,
type I cytoskeletal 26 | 468 | 0 | 1 | 1.03639903 |
| 85 | K6PL_HUMAN | (P17858)
6-phosphofructokinase, liver type | 780 | 0 | 1 | 1.03639903 |
| 86 | MYH11_HUMAN | (P35749)
Myosin-11 | 1972 | 0 | 1 | 1.03639903 |
| 87 | MYH14_HUMAN | (Q7Z406)
Myosin-14 | 1995 | 0 | 1 | 1.03639903 |
| 88 | PAR10_HUMAN | (Q53GL7) Poly
[ADP-ribose] polymerase 10 | 1025 | 0 | 1 | 1.03639903 |
| 89 | PI3R4_HUMAN | (Q99570)
Phosphoinositide 3-kinase regulatory subunit 4 | 1358 | 0 | 1 | 1.03639903 |
| 90 | PRDX4_HUMAN | (Q13162)
Peroxiredoxin-4 | 271 | 0 | 1 | 1.03639903 |
| 91 | PRS6A_HUMAN | (P17980) 26S
protease regulatory subunit 6A | 439 | 0 | 1 | 1.03639903 |
| 92 | PSB3_HUMAN | (P49720) Proteasome
subunit β type-3 | 205 | 0 | 1 | 1.03639903 |
| 93 | PTN22_HUMAN | (Q9Y2R2)
Tyrosine-protein phosphatase non-receptor type 22 | 807 | 0 | 1 | 1.03639903 |
| 94 | RAB10_HUMAN | (P61026)
Ras-related protein Rab-10 | 200 | 0 | 1 | 1.03639903 |
| 95 | RBBP7_HUMAN | (Q16576)
Histone-binding protein RBBP7 | 425 | 0 | 1 | 1.03639903 |
| 96 | RL18A_HUMAN | (Q02543) 60S
ribosomal protein L18a | 176 | 0 | 1 | 1.03639903 |
| 97 | RL27A_HUMAN | (P46776) 60S
ribosomal protein L27a | 148 | 0 | 1 | 1.03639903 |
| 98 | RL34_HUMAN | (P49207) 60S
ribosomal protein L34 | 117 | 0 | 1 | 1.03639903 |
| 99 | RL3L_HUMAN | (Q92901) 60S
ribosomal protein L3-like | 407 | 0 | 1 | 1.03639903 |
| 100 | RL40_HUMAN | (P62987)
Ubiquitin-60S ribosomal protein L40 | 128 | 0 | 1 | 1.03639903 |
| 101 | ROAA_HUMAN | (Q99729)
Heterogeneous nuclear ribonucleoprotein A/B | 332 | 0 | 1 | 1.03639903 |
| 102 | SET_HUMAN | (Q01105) Protein
SET | 290 | 0 | 1 | 1.03639903 |
| 103 | SODC_HUMAN | (P00441) Superoxide
dismutase [Cu-Zn] | 154 | 0 | 1 | 1.03639903 |
| 104 | SPSY_HUMAN | (P52788) Spermine
synthase | 366 | 0 | 1 | 1.03639903 |
| 105 | SYRC_HUMAN | (P54136)
Arginyl-tRNA synthetase, cytoplasmic | 660 | 0 | 1 | 1.03639903 |
| 106 | TADBP_HUMAN | (Q13148) TAR
DNA-binding protein 43 | 414 | 0 | 1 | 1.03639903 |
| 107 | TALDO_HUMAN | (P37837)
Transaldolase | 337 | 0 | 1 | 1.03639903 |
| 108 | TECT2_HUMAN | (Q96GX1)
Tectonic-2 | 697 | 0 | 1 | 1.03639903 |
| 109 | TEBP_HUMAN | (Q15185)
Prostaglandin E synthase 3 | 160 | 0 | 1 | 1.03639903 |
| 110 | TPD54_HUMAN | (O43399) Tumor
protein D54 | 206 | 0 | 1 | 1.03639903 |
| 111 | UGPA_HUMAN | (Q16851) UTP -
glucose-1-phosphate uridylyltransferase | 508 | 0 | 1 | 1.03639903 |
| 112 | WDR53_HUMAN | (Q7Z5U6) WD
repeat-containing protein 53 | 358 | 0 | 1 | 1.03639903 |
| 113 | ZC12A_HUMAN | (Q5D1E8)
Ribonuclease ZC3H12A | 599 | 0 | 1 | 1.03639903 |
| 114 | RL22_HUMAN | (P35268) 60S
ribosomal protein L22 | 128 | 0 | 1 | 1.03639903 |
| 115 | RL13_HUMAN | (P26373) 60S
ribosomal protein L13 | 211 | 0 | 1 | 1.03639903 |
| 116 | RL23_HUMAN | (P62829) 60S
ribosomal protein L23 | 140 | 0 | 1 | 1.03639903 |
| 117 | RS11_HUMAN | (P62280) 40S
ribosomal protein S11 | 158 | 0 | 1 | 1.03639903 |
| 118 | SUMO2_HUMAN | (P61956) Small
ubiquitin-related modifier 2 | 95 | 0 | 1 | 1.03639903 |
| 119 | TFR1_HUMAN | (P02786)
Transferrin receptor protein 1 | 760 | 0 | 1 | 1.03639903 |
| 120 | RL11_HUMAN | (P62913) 60S
ribosomal protein L11 | 178 | 0 | 1 | 1.03639903 |
| 121 | EF1A2_HUMAN | (Q05639) Elongation
factor 1-α 2 | 463 | 10 | 19 | 1.04422777 |
| 122 | CBX1_HUMAN | (P83916) Chromobox
protein homolog 1 | 185 | 1 | 3 | 1.10688593 |
| 123 | RL12_HUMAN | (P30050) 60S
ribosomal protein L12 | 165 | 1 | 3 | 1.10688593 |
| 124 | RAN_HUMAN | (P62826)
GTP-binding nuclear protein Ran | 216 | 1 | 3 | 1.10688593 |
| 125 | PDIA6_HUMAN | (Q15084) Protein
disulfide-isomerase A6 | 440 | 1 | 3 | 1.10688593 |
| 126 | RL10L_HUMAN | (Q96L21) 60S
ribosomal protein L10-like | 214 | 1 | 3 | 1.10688593 |
| 127 | SRC8_HUMAN | (Q14247) Src
substrate cortactin | 550 | 1 | 3 | 1.10688593 |
| 128 | RL8_HUMAN | (P62917) 60S
ribosomal protein L8 | 257 | 1 | 3 | 1.10688593 |
| 129 | RS23_HUMAN | (P62266) 40S
ribosomal protein S23 | 143 | 1 | 3 | 1.10688593 |
| 130 | RS13_HUMAN | (P62277) 40S
ribosomal protein S13 | 151 | 1 | 3 | 1.10688593 |
| 131 | RL24_HUMAN | (P83731) 60S
ribosomal protein L24 | 157 | 1 | 3 | 1.10688593 |
| 132 | RL37A_HUMAN | (P61513) 60S
ribosomal protein L37a | 92 | 2 | 5 | 1.13371212 |
| 133 | PAL4A_HUMAN | (Q9Y536)
Peptidylprolyl cis-trans isomerase A-like 4A/B/C | 164 | 2 | 5 | 1.13371212 |
| 134 | RL18_HUMAN | (Q07020) 60S
ribosomal protein L18 | 188 | 2 | 5 | 1.13371212 |
| 135 | K1C9_HUMAN | (P35527) Keratin,
type I cytoskeletal 9 | 623 | 8 | 17 | 1.17638585 |
| 136 | TBB1_HUMAN | (Q9H4B7) Tubulin
β-1 chain | 451 | 3 | 8 | 1.31408061 |
| 137 | PAIRB_HUMAN | (Q8NC51)
Plasminogen activator inhibitor 1 RNA-binding protein | 408 | 2 | 6 | 1.34868122 |
| 138 | CATD_HUMAN | (P07339) Cathepsin
D | 412 | 0 | 2 | 1.56775608 |
| 139 | FLNB_HUMAN | (O75369)
Filamin-B | 2602 | 0 | 2 | 1.56775608 |
| 140 | MYL6B_HUMAN | (P14649) Myosin
light chain 6B | 208 | 0 | 2 | 1.56775608 |
| 141 | PSB2_HUMAN | (P49721) Proteasome
subunit β type-2 | 201 | 0 | 2 | 1.56775608 |
| 142 | RS7_HUMAN | (P62081) 40S
ribosomal protein S7 | 194 | 0 | 2 | 1.56775608 |
| 143 | IQGA1_HUMAN | (P46940) Ras
GTPase-activating-like protein IQGAP1 | 1657 | 0 | 2 | 1.56775608 |
| 144 | DX39B_HUMAN | (Q13838)
Spliceosome RNA helicase DDX39B | 428 | 0 | 2 | 1.56775608 |
| 145 | GANAB_HUMAN | (Q9BS14)Neutral
α-glucosidase AB | 944 | 0 | 2 | 1.56775608 |
| 146 | H13_HUMAN | (P16402) Histone
H1.3 | 221 | 0 | 2 | 1.56775608 |
| 147 | H14_HUMAN | (P10412) Histone
H1.4 | 219 | 0 | 2 | 1.56775608 |
| 148 | HNRH2_HUMAN | (P55795)
Heterogeneous nuclear ribonucleoprotein H2 | 449 | 0 | 2 | 1.56775608 |
| 149 | ATPA_HUMAN | (P25705) ATP
synthase subunit α, mitochondrial | 553 | 0 | 2 | 1.56775608 |
| 150 | H31T_HUMAN | (Q16695) Histone
H3.1t | 136 | 0 | 2 | 1.56775608 |
| 151 | RL10A_HUMAN | (P62906) 60S
ribosomal protein L10a | 217 | 0 | 2 | 1.56775608 |
| 152 | H12_HUMAN | (P16403) Histone
H1.2 | 213 | 0 | 2 | 1.56775608 |
| 153 | TBB8_HUMAN | (Q3ZCM7) Tubulin
β-8 chain | 444 | 5 | 15 | 1.57505162 |
| 154 | EF1D_HUMAN | (P29692) Elongation
factor 1-δ | 281 | 1 | 5 | 1.6649664 |
| 155 | RLA0L_HUMAN | (Q8NHW5) 60S acidic
ribosomal protein P0-like | 317 | 1 | 6 | 1.8799355 |
| 156 | RS8_HUMAN | (P62241) 40S
ribosomal protein S8 | 208 | 0 | 3 | 1.95562202 |
| 157 | CCD50_HUMAN | (Q8IVM0)
Coiled-coil domain-containing protein 50 | 306 | 0 | 3 | 1.95562202 |
| 158 | K2C6A_HUMAN | (P02538) Keratin,
type II cytoskeletal 6A | 564 | 0 | 3 | 1.95562202 |
| 159 | MT1X_HUMAN | (P80297)
Metallothionein-1X | 61 | 0 | 3 | 1.95562202 |
| 160 | RL17_HUMAN | (P18621) 60S
ribosomal protein L17 | 184 | 0 | 3 | 1.95562202 |
| 161 | RS26_HUMAN | (P62854) 40S
ribosomal protein S26 | 115 | 0 | 3 | 1.95562202 |
| 162 | H15_HUMAN | (P16401) Histone
H1.5 | 226 | 0 | 3 | 1.95562202 |
| 163 | ATPB_HUMAN | (P06576) ATP
synthase subunit β, mitochondrial | 529 | 1 | 7 | 2.06719342 |
| 164 | ADT1_HUMAN | (P12235) ADP/ATP
translocase 1 | 298 | 0 | 4 | 2.26131991 |
| 165 | H2A1_HUMAN | (P0C0S8) Histone
H2A type 1 | 130 | 0 | 4 | 2.26131991 |
| 166 | RS6_HUMAN | (P62753) 40S
ribosomal protein S6 | 249 | 0 | 4 | 2.26131991 |
| 167 | H2B1B_HUMAN | (P33778) Histone
H2B type 1-B | 126 | 0 | 4 | 2.26131991 |
| 168 | FLNA_HUMAN | (P21333)
Filamin-A | 2647 | 1 | 9 | 2.38204237 |
| 169 | H2B1M_HUMAN | (Q99879) Histone
H2B type 1-M | 126 | 0 | 6 | 2.72867159 |
| 170 | H2AX_HUMAN | (P16104) Histone
H2A.x | 143 | 0 | 7 | 2.9159295 |
| 171 | H2A1B_HUMAN | (P04908) Histone
H2A type 1-B/E | 130 | 0 | 9 | 3.23077846 |
| 172 | H2A1A_HUMAN | (Q96QV6) Histone
H2A type 1-A | 131 | 0 | 9 | 3.23077846 |
| 173 | H2B1C_HUMAN | (P62807) Histone
H2B type 1-C/E/F/G/I | 126 | 0 | 11 | 3.48962935 |
| 174 | ACTA_HUMAN | (P62736) Actin,
aortic smooth muscle | 377 | 0 | 15 | 3.90067941 |
 | Table IIDifferentially expressed proteins in
lumican downregulated PANC-1 cells. |
Table II
Differentially expressed proteins in
lumican downregulated PANC-1 cells.
| | | | Spectral
counting |
|---|
| | | |
|
|---|
| No. | ID | Accession no. and
description | No. of amino
acids | NC | siLum | Fold-change
(Rsc) |
|---|
| 1 | ACTG_HUMAN | (P63261) Actin,
cytoplasmic 2 | 375 | 19 | 0 | −4.1933172 |
| 2 | H2B1D_HUMAN | (P58876) Histone
H2B type 1-D | 126 | 7 | 0 | −2.8944219 |
| 3 | H2A2A_HUMAN | (Q6FI13) Histone
H2A type 2-A | 130 | 4 | 0 | −2.2414896 |
| 4 | HNRPU_HUMAN | (Q00839)
Heterogeneous nuclear ribonucleoprotein U | 825 | 3 | 0 | −1.9363502 |
| 5 | H2B1M_HUMAN | (Q99879) Histone
H2B type 1-M | 126 | 3 | 0 | −1.9363502 |
| 6 | KRT81_HUMAN | (Q14533) Keratin,
type II cuticular Hb1 | 505 | 3 | 0 | −1.9363502 |
| 7 | RL10_HUMAN | (P27635) 60S
ribosomal protein L10 | 214 | 3 | 0 | −1.9363502 |
| 8 | K2C6B_HUMAN | (P04259) Keratin,
type II cytoskeletal 6B | 564 | 6 | 1 | −1.8595343 |
| 9 | CAPZB_HUMAN | (P47756)
F-actin-capping protein subunit β | 277 | 2 | 0 | −1.5490424 |
| 10 | FUBP2_HUMAN | (Q92945) Far
upstream element-binding protein 2 | 711 | 2 | 0 | −1.5490424 |
| 11 | IPO7_HUMAN | (O95373)
Importin-7 | 1038 | 2 | 0 | −1.5490424 |
| 12 | K2C6C_HUMAN | (P48668) Keratin,
type II cytoskeletal 6C | 564 | 2 | 0 | −1.5490424 |
| 13 | ML12A_HUMAN | (P19105) Myosin
regulatory light chain 12A | 171 | 2 | 0 | −1.5490424 |
| 14 | NUCB1_HUMAN | (Q02818)
Nucleobindin-1 | 461 | 2 | 0 | −1.5490424 |
| 15 | EF1B_HUMAN | (P24534) Elongation
factor 1-β | 225 | 2 | 0 | −1.5490424 |
| 16 | NDKA_HUMAN | (P15531) Nucleoside
diphosphate kinase A | 152 | 2 | 0 | −1.5490424 |
| 17 | VINC_HUMAN | (P18206)
Vinculin | 1134 | 7 | 2 | −1.5155258 |
| 18 | PDIA3_HUMAN | (P30101) Protein
disulfide-isomerase A3 | 505 | 4 | 1 | −1.3933005 |
| 19 | K2C5_HUMAN | (P13647) Keratin,
type II cytoskeletal 5 | 590 | 8 | 3 | −1.2936552 |
| 20 | K1C19_HUMAN | (P08727) Keratin,
type I cytoskeletal 19 | 400 | 11 | 5 | −1.1429912 |
| 21 | ECH1_HUMAN | (Q13011)
δ(3,5)-δ(2,4)-dienoyl-CoA isomerase, mitochondrial | 328 | 5 | 2 | −1.1144173 |
| 22 | PAIRB_HUMAN | (Q8NC51)
Plasminogen activator inhibitor 1 RNA-binding protein | 408 | 5 | 2 | −1.1144173 |
| 23 | RS3_HUMAN | (P23396) 40S
ribosomal protein S3 | 243 | 3 | 1 | −1.0881611 |
| 24 | CRIP1_HUMAN | (P50238)
Cysteine-rich protein 1 | 77 | 3 | 1 | −1.0881611 |
| 25 | RS28_HUMAN | (P62857) 40S
ribosomal protein S28 | 69 | 3 | 1 | −1.0881611 |
| 26 | RS13_HUMAN | (P62277) 40S
ribosomal protein S13 | 151 | 3 | 1 | −1.0881611 |
| 27 | RS27A_HUMAN | (P62979)
Ubiquitin-40S ribosomal protein S27a | 156 | 3 | 1 | −1.0881611 |
| 28 | CAPG_HUMAN | (P40121)
Macrophage-capping protein | 348 | 1 | 0 | −1.0182433 |
| 29 | CDC42_HUMAN | (P60953) Cell
division control protein 42 homolog | 191 | 1 | 0 | −1.0182433 |
| 30 | CX6B1_HUMAN | (P14854) Cytochrome
c oxidase subunit 6B1 | 86 | 1 | 0 | −1.0182433 |
| 31 | GDIA_HUMAN | (P31150) Rab GDP
dissociation inhibitor α | 447 | 1 | 0 | −1.0182433 |
| 32 | IQGA1_HUMAN | (P46940) Ras
GTPase-activating-like protein IQGAP1 | 1657 | 1 | 0 | −1.0182433 |
| 33 | RL12_HUMAN | (P30050) 60S
ribosomal protein L12 | 165 | 1 | 0 | −1.0182433 |
| 34 | RL35A_HUMAN | (P18077) 60S
ribosomal protein L35a | 110 | 1 | 0 | −1.0182433 |
| 35 | SQSTM_HUMAN | (Q13501)
Sequestosome-1 | 440 | 1 | 0 | −1.0182433 |
| 36 | RL10L_HUMAN | (Q96L21) 60S
ribosomal protein L10-like | 214 | 1 | 0 | −1.0182433 |
| 37 | RS14_HUMAN | (P62263) 40S
ribosomal protein S14 | 151 | 1 | 0 | −1.0182433 |
| 38 | AGR3_HUMAN | (Q8TD06) Anterior
gradient protein 3 homolog | 166 | 1 | 0 | −1.0182433 |
| 39 | ALMS1_HUMAN | (Q8TCU4) Alstrom
syndrome protein 1 | 4167 | 1 | 0 | −1.0182433 |
| 40 | AP3D1_HUMAN | (O14617) AP-3
complex subunit δ-1 | 1153 | 1 | 0 | −1.0182433 |
| 41 | BI2L2_HUMAN | (Q6UXY1)
Brain-specific angiogenesis inhibitor 1-associated protein 2-like
protein 2 | 529 | 1 | 0 | −1.0182433 |
| 42 | CH041_HUMAN | (Q6NXR4)
Uncharacterized protein C8orf41 | 508 | 1 | 0 | −1.0182433 |
| 43 | CORO7_HUMAN | (P57737)
Coronin-7 | 925 | 1 | 0 | −1.0182433 |
| 44 | CSPG4_HUMAN | (Q6UVK1)
Chondroitin sulfate proteoglycan 4 | 2322 | 1 | 0 | −1.0182433 |
| 45 | CXL14_HUMAN | (O95715) C-X-C
motif chemokine 14 | 111 | 1 | 0 | −1.0182433 |
| 46 | DDX17_HUMAN | (Q92841) Probable
ATP-dependent RNA helicase DDX17 | 650 | 1 | 0 | −1.0182433 |
| 47 | DNJB1_HUMAN | (P25685) DnaJ
homolog subfamily B member 1 | 340 | 1 | 0 | −1.0182433 |
| 48 | EIF3C_HUMAN | (Q99613) Eukaryotic
translation initiation factor 3 subunit C | 913 | 1 | 0 | −1.0182433 |
| 49 | FER_HUMAN | (P16591)
Tyrosine-protein kinase Fer | 822 | 1 | 0 | −1.0182433 |
| 50 | GELS_HUMAN | (P06396)
Gelsolin | 782 | 1 | 0 | −1.0182433 |
| 51 | INSRR_HUMAN | (P14616) Insulin
receptor-related protein | 1297 | 1 | 0 | −1.0182433 |
| 52 | KRT85_HUMAN | (P78386) Keratin,
type II cuticular Hb5 | 507 | 1 | 0 | −1.0182433 |
| 53 | MT1X_HUMAN | (P80297) MT-1X | 61 | 1 | 0 | −1.0182433 |
| 54 | NALP6_HUMAN | (P59044) NACHT, LRR
and PYD domains-containing protein 6 | 892 | 1 | 0 | −1.0182433 |
| 55 | PLEC_HUMAN | (Q15149)
Plectin | 4684 | 1 | 0 | −1.0182433 |
| 56 | PRS8_HUMAN | (P62195) 26S
protease regulatory subunit 8 | 406 | 1 | 0 | −1.0182433 |
| 57 | PUR2_HUMAN | (P22102)
Trifunctional purine biosynthetic protein adenosine-3 | 1010 | 1 | 0 | −1.0182433 |
| 58 | RFX6_HUMAN | (Q8HWS3)
DNA-binding protein RFX6 | 928 | 1 | 0 | −1.0182433 |
| 59 | RS17_HUMAN | (P08708) 40S
ribosomal protein S17 | 135 | 1 | 0 | −1.0182433 |
| 60 | RUNX1_HUMAN | (Q01196)
Runt-related transcription factor 1 | 453 | 1 | 0 | −1.0182433 |
| 61 | SEPT9_HUMAN | (Q9UHD8)
Septin-9 | 586 | 1 | 0 | −1.0182433 |
| 62 | SRSF4_HUMAN | (Q08170)
Serine/arginine-rich splicing factor 4 | 494 | 1 | 0 | −1.0182433 |
| 63 | SUMO3_HUMAN | (P55854) Small
ubiquitin-related modifier 3 | 103 | 1 | 0 | −1.0182433 |
| 64 | TIM13_HUMAN | (Q9Y5L4)
Mitochondrial import inner membrane translocase subunit Tim13 | 95 | 1 | 0 | −1.0182433 |
| 65 | UGDH_HUMAN | (O60701)
UDP-glucose 6-dehydrogenase | 494 | 1 | 0 | −1.0182433 |
| 66 | WDHD1_HUMAN | (O75717) WD repeat
and HMG-box DNA-binding protein 1 | 1129 | 1 | 0 | −1.0182433 |
| 67 | ZZEF1_HUMAN | (O43149) Zinc
finger ZZ-type and EF-hand domain-containing protein 1 | 2961 | 1 | 0 | −1.0182433 |
| 68 | RL22_HUMAN | (P35268) 60S
ribosomal protein L22 | 128 | 1 | 0 | −1.0182433 |
| 69 | BASP1_HUMAN | (P80723) Brain acid
soluble protein 1 | 227 | 1 | 0 | −1.0182433 |
| 70 | SPSY_HUMAN | (P52788) Spermine
synthase | 366 | 1 | 0 | −1.0182433 |
| 71 | RS25_HUMAN | (P62851) 40S
ribosomal protein S25 | 125 | 1 | 0 | −1.0182433 |
| 72 | RS9_HUMAN | (P46781) 40S
ribosomal protein S9 | 194 | 1 | 0 | −1.0182433 |
| 73 | HIP1R_HUMAN | (O75146)
Huntingtin-interacting protein 1-related protein | 1068 | 1 | 0 | −1.0182433 |
| 74 | RL15_HUMAN | (P61313) 60S
ribosomal protein L15 | 204 | 1 | 0 | −1.0182433 |
| 75 | G6PI_HUMAN | (P06744)
Glucose-6-phosphate isomerase | 558 | 1 | 0 | −1.0182433 |
| 76 | TERA_HUMAN | (P55072)
Transitional endoplasmic reticulum ATPase | 806 | 1 | 4 | 1.05291534 |
| 77 | RS5_HUMAN | (P46782) 40S
ribosomal protein S5 | 204 | 1 | 4 | 1.05291534 |
| 78 | PCBP3_HUMAN | (P57721)
Poly(rC)-binding protein 3 | 371 | 1 | 4 | 1.05291534 |
| 79 | K2C74_HUMAN | (Q7RTS7) Keratin,
type II cytoskeletal 74 | 529 | 4 | 11 | 1.0534109 |
| 80 | CH60_HUMAN | (P10809) 60 kDa
heat shock protein, mitochondrial | 573 | 7 | 20 | 1.19690092 |
| 81 | RAN_HUMAN | (P62826)
GTP-binding nuclear protein Ran | 216 | 0 | 2 | 1.20893401 |
| 82 | ADT2_HUMAN | (P05141) ADP/ATP
translocase 2 | 298 | 0 | 2 | 1.20893401 |
| 83 | CAZA1_HUMAN | (P52907)
F-actin-capping protein subunit α-1 | 286 | 0 | 2 | 1.20893401 |
| 84 | G6PD_HUMAN | (P11413)
Glucose-6-phosphate 1-dehydrogenase | 515 | 0 | 2 | 1.20893401 |
| 85 | H2AV_HUMAN | (Q71UI9) Histone
H2A.V | 128 | 0 | 2 | 1.20893401 |
| 86 | H2B1H_HUMAN | (Q93079) Histone
H2B type 1-H | 126 | 0 | 2 | 1.20893401 |
| 87 | H2B1J_HUMAN | (P06899) Histone
H2B type 1-J | 126 | 0 | 2 | 1.20893401 |
| 88 | H2B1K_HUMAN | (O60814) Histone
H2B type 1-K | 126 | 0 | 2 | 1.20893401 |
| 89 | HCD2_HUMAN | (Q99714)
3-hydroxyacyl-CoA dehydrogenase type-2 | 261 | 0 | 2 | 1.20893401 |
| 90 | MT1A_HUMAN | (P04731)
Metallothionein-1A | 61 | 0 | 2 | 1.20893401 |
| 91 | RL18A_HUMAN | (Q02543) 60S
ribosomal protein L18a | 176 | 0 | 2 | 1.20893401 |
| 92 | RL21_HUMAN | (P46778) 60S
ribosomal protein L21 | 160 | 0 | 2 | 1.20893401 |
| 93 | RS7_HUMAN | (P62081) 40S
ribosomal protein S7 | 194 | 0 | 2 | 1.20893401 |
| 94 | SPT6H_HUMAN | (Q7KZ85)
Transcription elongation factor SPT6 | 1726 | 0 | 2 | 1.20893401 |
| 95 | SRP14_HUMAN | (P37108) Signal
recognition particle 14 kDa protein | 136 | 0 | 2 | 1.20893401 |
| 96 | TBA4B_HUMAN | (Q9H853) Putative
tubulin-like protein α-4B | 241 | 0 | 2 | 1.20893401 |
| 97 | TCTP_HUMAN | (P13693)
Translationally-controlled tumor protein | 172 | 0 | 2 | 1.20893401 |
| 98 | UBB_HUMAN | (P0CG47)
Polyubiquitin-B | 229 | 0 | 2 | 1.20893401 |
| 99 | RL19_HUMAN | (P84098) 60S
ribosomal protein L19 | 196 | 0 | 2 | 1.20893401 |
| 100 | FKB1A_HUMAN | (P62942)
Peptidyl-prolyl cis-trans isomerase FKBP1A | 108 | 0 | 2 | 1.20893401 |
| 101 | MDHM_HUMAN | (P40926) Malate
dehydrogenase, mitochondrial | 338 | 0 | 2 | 1.20893401 |
| 102 | NDE1_HUMAN | (Q9NXR1) Nuclear
distribution protein nudE homolog 1 | 346 | 0 | 2 | 1.20893401 |
| 103 | PSA5_HUMAN | (P28066) Proteasome
subunit α type-5 | 241 | 0 | 2 | 1.20893401 |
| 104 | H2B1B_HUMAN | (P33778) Histone
H2B type 1-B | 126 | 0 | 2 | 1.20893401 |
| 105 | HNRH2_HUMAN | (P55795)
Heterogeneous nuclear ribonucleoprotein H2 | 449 | 1 | 5 | 1.30464663 |
| 106 | K2C1_HUMAN | (P04264) Keratin,
type II cytoskeletal 1 | 644 | 2 | 8 | 1.34002273 |
| 107 | K1C9_HUMAN | (P35527) Keratin,
type I cytoskeletal 9 | 623 | 0 | 3 | 1.59614949 |
| 108 | RLA0L_HUMAN | (Q8NHW5) 60S acidic
ribosomal protein P0-like | 317 | 0 | 3 | 1.59614949 |
| 109 | H2A1B_HUMAN | (P04908) Histone
H2A type 1-B/E | 130 | 0 | 3 | 1.59614949 |
| 110 | SRC8_HUMAN | (Q14247) Src
substrate cortactin | 550 | 0 | 3 | 1.59614949 |
| 111 | UBA1_HUMAN | (P22314)
Ubiquitin-like modifier-activating enzyme 1 | 1058 | 0 | 3 | 1.59614949 |
| 112 | H2A1H_HUMAN | (Q96KK5) Histone
H2A type 1-H | 128 | 0 | 3 | 1.59614949 |
| 113 | H2A2B_HUMAN | (Q8IUE6) Histone
H2A type 2-B | 130 | 0 | 3 | 1.59614949 |
| 114 | HN1_HUMAN | (Q9UK76)
Hematological and neurological expressed 1 protein | 154 | 0 | 3 | 1.59614949 |
| 115 | HNRPK_HUMAN | (P61978)
Heterogeneous nuclear ribonucleoprotein K | 463 | 0 | 3 | 1.59614949 |
| 116 | K1C14_HUMAN | (P02533) Keratin,
type I cytoskeletal 14 | 472 | 0 | 3 | 1.59614949 |
| 117 | K22O_HUMAN | (Q01546) Keratin,
type II cytoskeletal 2 oral | 638 | 0 | 3 | 1.59614949 |
| 118 | K6PP_HUMAN | (Q01813)
6-phosphofructokinase type C | 784 | 0 | 3 | 1.59614949 |
| 119 | POTEJ_HUMAN | (P0CG39) POTE
ankyrin domain family member J | 1038 | 0 | 3 | 1.59614949 |
| 120 | RADI_HUMAN | (P35241)
Radixin | 583 | 0 | 3 | 1.59614949 |
| 121 | HS904_HUMAN | (Q58FG1) Putative
heat shock protein HSP 90-α A4 | 418 | 2 | 10 | 1.62280828 |
| 122 | PROF1_HUMAN | (P07737)
Profilin-1 | 140 | 3 | 14 | 1.67515744 |
| 123 | H90B3_HUMAN | (Q58FF7) Putative
heat shock protein HSP 90-β-3 | 597 | 1 | 7 | 1.70556987 |
| 124 | TBB2A_HUMAN | (Q13885) Tubulin
β-2A chain | 445 | 7 | 33 | 1.88806959 |
| 125 | TIM50_HUMAN | (Q3ZCQ8)
Mitochondrial import inner membrane translocase subunit TIM50 | 353 | 0 | 4 | 1.90119651 |
| 126 | RL4_HUMAN | (P36578) 60S
ribosomal protein L4 | 427 | 0 | 4 | 1.90119651 |
| 127 | H2A1C_HUMAN | (Q93077) Histone
H2A type 1-C | 130 | 0 | 4 | 1.90119651 |
| 128 | K2C6A_HUMAN | (P02538) Keratin,
type II cytoskeletal 6A | 564 | 0 | 4 | 1.90119651 |
| 129 | NDK8_HUMAN | (O60361) Putative
nucleoside diphosphate kinase | 137 | 0 | 4 | 1.90119651 |
| 130 | TKT_HUMAN | (P29401)
Transketolase | 623 | 0 | 4 | 1.90119651 |
| 131 | H2B1O_HUMAN | (P23527) Histone
H2B type 1-O | 126 | 0 | 5 | 2.15292781 |
| 132 | H2B2F_HUMAN | (Q5QNW6) Histone
H2B type 2-F | 126 | 0 | 5 | 2.15292781 |
| 133 | ANXA5_HUMAN | (P08758) Annexin
A5 | 320 | 0 | 6 | 2.36724523 |
| 134 | H2A2C_HUMAN | (Q16777) Histone
H2A type 2-C | 129 | 0 | 6 | 2.36724523 |
| 135 | H2B1L_HUMAN | (Q99880) Histone
H2B type 1-L | 126 | 0 | 7 | 2.55385105 |
| 136 | HSP76_HUMAN | (P17066) Heat shock
70 kDa protein 6 | 643 | 0 | 8 | 2.71910308 |
| 137 | TBA3E_HUMAN | (Q6PEY2) Tubulin
α-3E chain | 450 | 0 | 8 | 2.71910308 |
| 138 | TBA3C_HUMAN | (Q13748) Tubulin
α-3C/D chain | 450 | 0 | 9 | 2.86739459 |
| 139 | TBA4A_HUMAN | (P68366) Tubulin
α-4A chain | 448 | 0 | 14 | 3.44154556 |
| 140 | TBA1A_HUMAN | (Q71U36) Tubulin
α-1A chain | 451 | 0 | 14 | 3.44154556 |
| 141 | TBB8_HUMAN | (Q3ZCM7) Tubulin
β-8 chain | 444 | 0 | 16 | 3.61971953 |
| 142 | ACTC_HUMAN | (P68032) Actin, α
cardiac muscle 1 | 377 | 0 | 21 | 3.98789716 |
| 143 | ACTS_HUMAN | (P68133) Actin, α
skeletal muscle | 377 | 0 | 42 | 4.95088405 |
 | Table IIICorrelation between lumican
expression level and identified protein expression level. |
Table III
Correlation between lumican
expression level and identified protein expression level.
| | Fold-change
(Rsc) |
|---|
| |
|
|---|
| ID | Accession no. and
description | Lumican
upregulation | Lumican
downregulation |
|---|
| ACTC_HUMAN | (P68032) Actin, α
cardiac muscle 1 | −3.770661694 | 3.987897158 |
| TBB2A_HUMAN | (Q13885) Tubulin
β-2A chain | −2.989773612 | 1.888069587 |
| H2B1H_HUMAN | (Q93079) Histone
H2B type 1-H | −2.854729916 | 1.208934009 |
| HS904_HUMAN | (Q58FG1) Putative
heat shock protein HSP 90-α A4 | −2.705889054 | 1.622808285 |
| TBA4A_HUMAN | (P68366) Tubulin
α-4A chain | −2.705889054 | 3.441545557 |
| H2B1O_HUMAN | (P23527) Histone
H2B type 1-O | −1.885788034 | 2.152927808 |
| ANXA5_HUMAN | (P08758) Annexin
A5 | −1.580193134 | 2.367245226 |
| H2A1H_HUMAN | (Q96KK5) Histone
H2A type 1-H | −1.580193134 | 1.596149489 |
| HNRPK_HUMAN | (P61978)
Heterogeneous nuclear ribonucleoprotein K | −1.580193134 | 1.596149489 |
| K22O_HUMAN | (Q01546) Keratin,
type II cytoskeletal 2 oral | −1.580193134 | 1.596149489 |
| K1C14_HUMAN | (P02533) Keratin,
type I cytoskeletal 14 | −1.504094641 | 1.596149489 |
| H2B1J_HUMAN | (P06899) Histone
H2B type 1-J | −1.192430072 | 1.208934009 |
| MDHM_HUMAN | (P40926) Malate
dehydrogenase, mitochondrial | −1.192430072 | 1.208934009 |
| NDK8_HUMAN | (O60361) Putative
nucleoside diphosphate kinase | −1.192430072 | 1.901196513 |
| HIP1R_HUMAN | (O75146)
Huntingtin-interacting protein 1-related protein | 1.036399032 | −1.018243251 |
| RL22_HUMAN | (P35268) 60S
ribosomal protein L22 | 1.036399032 | −1.018243251 |
| SPSY_HUMAN | (P52788) Spermine
synthase | 1.036399032 | −1.018243251 |
| RL10L_HUMAN | (Q96L21) 60S
ribosomal protein L10-like | 1.106885931 | −1.018243251 |
| RL12_HUMAN | (P30050) 60S
ribosomal protein L12 | 1.106885931 | −1.018243251 |
| RS13_HUMAN | (P62277) 40S
ribosomal protein S13 | 1.106885931 | −1.088161097 |
| PAIRB_HUMAN | (Q8NC51)
Plasminogen activator inhibitor 1 RNA-binding protein | 1.348681223 | −1.114417304 |
| IQGA1_HUMAN | (P46940) Ras
GTPase-activating-like protein IQGAP1 | 1.567756075 | −1.018243251 |
| MT1X_HUMAN | (P80297)
Metallothionein-1X | 1.955622017 | −1.018243251 |
| H2B1M_HUMAN | (Q99879) Histone
H2B type 1-M | 2.728671586 | −1.936350191 |
Functional annotation of proteins whose
expression level is regulated by lumican
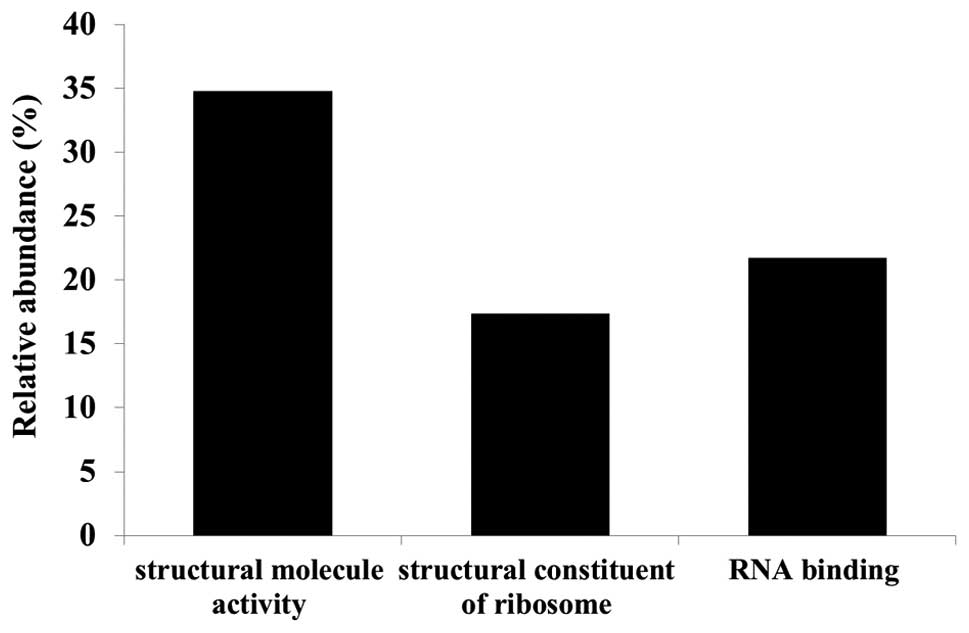
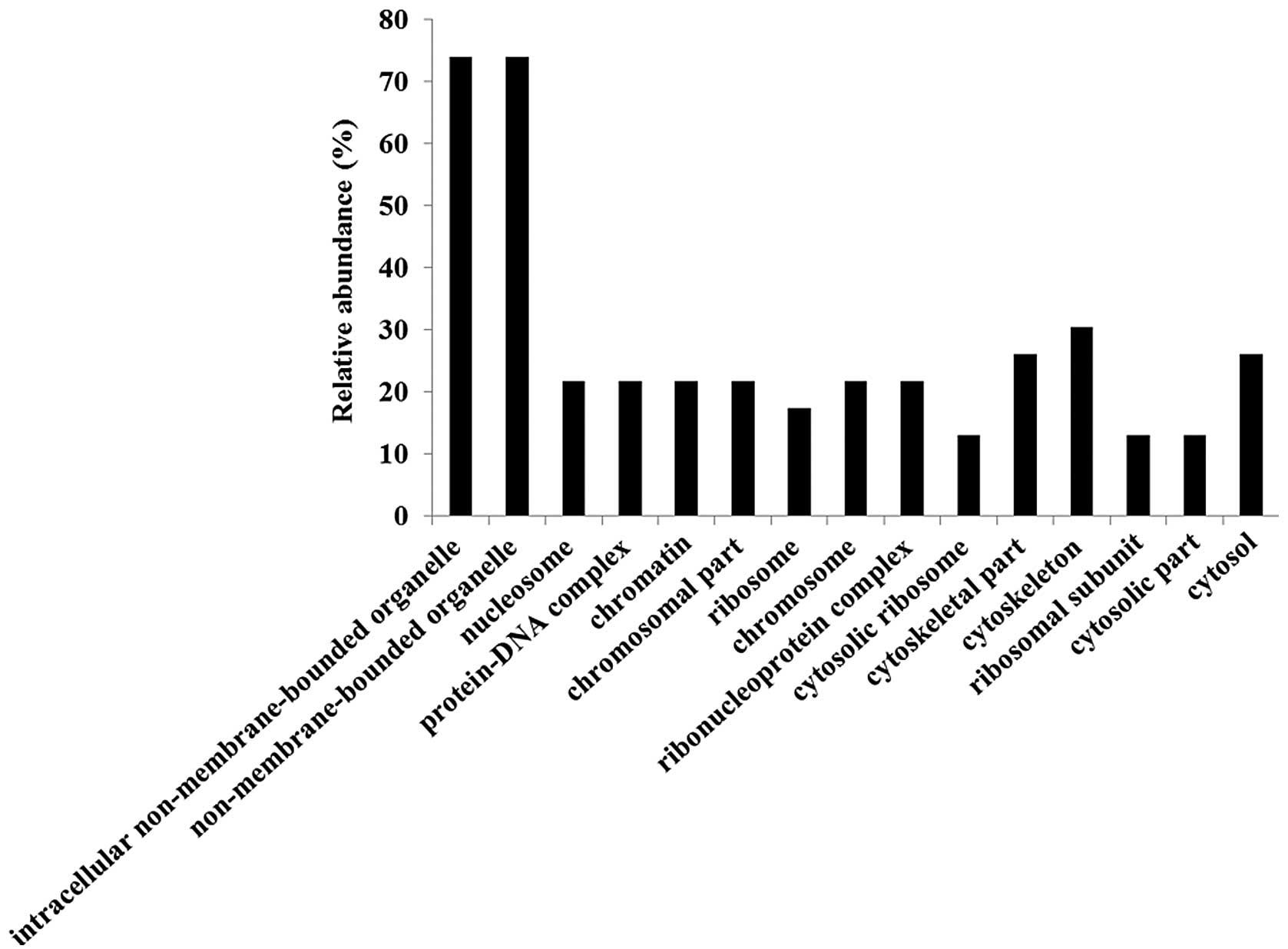
Gene ontology (GO) analyses were performed using the
identified candidate proteins for each molecular function (Fig. 3), biological process (Fig. 4) and cellular component (Fig. 5) using DAVID. We also analyzed
pathway terms, but no significant category was found. Functional
annotations were counted by normalizing to the total number of
proteins identified. Since a multifunctional protein yields more
than one annotation and some proteins are not defined by GO terms
yet, the total number of classified proteins resulted in more or
less than 100% (Fig. 3). Major GO
molecular function categories of the identified proteins were 34.8%
structural molecule activity, 17.4% structural constituents of
ribosomes, and 21.7% RNA binding proteins (Fig. 3).
Discussion
In the present study, we used a gel-free LC-MS-based
proteomics approach to examine the effect of lumican on cell growth
and invasion. Using semi-quantitative methods based on spectral
counting, we successfully identified several proteins whose
expression levels were altered more than 2-fold in both lumican
upregulated PANC-1 cells and lumican downregulated PANC-1 cells. A
limitation of spectral counting is in its accurate quantitative
capacity (35). Therefore, we
selected candidate proteins whose expression level was regulated by
lumican using the analysis results of two types of cells; lumican
upregulated cells and lumican downregulated cells. Thus, we
identified 24 proteins whose Rsc values were inversely correlated
among differentially expressed proteins in lumican upregulated and
downregulated cells as candidate proteins (Fig. 3).
To examine the role of these candidate proteins in
the effect of lumican on cell growth and invasion, we performed a
functional classification of the candidate proteins by GO analysis.
Although the GO terms for molecular function, biological processes,
and cellular components were examined, we focused on molecular
function. The molecular functions of candidate proteins were mainly
classified in the ‘structural molecule activity’ category. Since
structural molecule activity proteins contribute to the formation
of complexes within or outside of the cell, such candidate proteins
may be related to cell invasion regulated by lumican.
Annexin A5 expression may be regulated by lumican
since Annexin A5 expression was negatively correlated with the
lumican expression level. Annexin A5, also known as Annexin V, is
widely known as a marker of early stage apoptosis (36,37).
We previously demonstrated that the PDAC cell line secretes 70-kDa
glycosylated lumican and that this secreted lumican stimulates cell
growth (28). Thus, the induction
of cell growth by lumican may be related to an inhibition of
apoptosis through Annexin A5 expression. However, previous reports
suggest that lumican plays an important role in apoptosis induction
(22,38–40).
This discrepancy may be derived from the differences between
glycosylated lumican secreted from the PDAC cell line and other
cells.
Furthermore, MT-1X expression levels positively
correlated with the lumican expression level. MT family proteins
are encoded by 10 functional isoforms (MT-1A, MT-1B, MT-1E, MT-1F,
MT-1G, MT-1H, MT-1X, MT-2A, MT-3 and MT-4), and seven
non-functional isoforms (MT-1C, MT-1D, MT-1I, MT-1J, MT-1K, MT-L
and MT-2B). MT family proteins are involved in essential metal
homeostasis, cellular free radical scavenging, cell proliferation,
apoptosis, and metal detoxification. MT-1X and MT-2A transcripts
were significantly upregulated under hypoxia in human prostate
cancer cell lines, and siRNA-MT-2A treatment inhibited cell growth
and induction of apoptosis, but an effect of MT-1X on cell growth
and apoptosis was not demonstrated (41). Zinc is an abundant metal in the
human prostate, and zinc inhibits cell growth and induces apoptosis
in human prostate cancer cell lines (42,43).
These findings suggest that MT-2A may play an important role in
cell growth and apoptosis in prostate cancer through intracellular
zinc homeostasis. MT-1X function in cancer cells, particularly
PDAC, is not well understood. MT-1X is a known zinc-binding
protein, and MT-1X mRNA expression was induced, as well as MT-2A,
under hypoxic conditions (41).
Thus, MT-1X may inhibit apoptosis as well as MT-2A. Furthermore,
Ryu et al(44) suggested
that MT-1E could enhance the migration and invasion of human glioma
cells by inducing MMP-9 activation. MT-1X may have functions
resembling MT-1E, since MMP-9 is a zinc-requiring enzyme, and MT-1X
is classified into isoforms such as MT-1E. As mentioned above, it
may be postulated that MT-1X plays an important role in cell growth
and invasion by lumican. Further study is required to validate the
expression levels of these candidate proteins, and to clarify the
effect of candidate proteins, including MT-1X, on cell growth and
invasion that are affected by lumican.
In conclusion, we identified more than 400 proteins
from both lumican upregulated and lumican downregulated cells using
global shotgun proteomics. A label-free semi-quantitative method
based on spectral counting led to 24 candidate proteins whose
expression was regulated by lumican. These candidate proteins
included apoptosis-related and invasion-related proteins.
Therefore, lumican may be involved in cell growth and invasion by
altering the expression of these proteins.
Acknowledgements
The authors thank K. Teduka and T. Fujii for their
technical assistance (Department of Pathology, Integrative
Oncological Pathology). The present study was supported by a
Grant-in-Aid for Scientific Research from the Japan Society for the
Promotion of Science to T.Y. (C, no. 24591019) and Z.N. (C, no.
23590477).
References
|
1
|
Nikitovic D, Katonis P, Tsatsakis A,
Karamanos NK and Tzanakakis GN: Lumican, a small leucine-rich
proteoglycan. IUBMB Life. 60:818–823. 2008. View Article : Google Scholar
|
|
2
|
Iozzo RV: Matrix proteoglycans: from
molecular design to cellular function. Annu Rev Biochem.
67:609–652. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grover J, Chen XN, Korenberg JR and
Roughley PJ: The human lumican gene. Organization, chromosomal
location, and expression in articular cartilage. J Biol Chem.
270:21942–21949. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chakravarti S, Stallings RL, SundarRaj N,
Cornuet PK and Hassell JR: Primary structure of human lumican
(keratan sulfate proteoglycan) and localization of the gene (LUM)
to chromosome 12q21.3–q22. Genomics. 27:481–488. 1995.PubMed/NCBI
|
|
5
|
Naito Z: Role of the small leucine-rich
proteoglycan (SLRP) family in pathological lesions and cancer cell
growth. J Nippon Med Sch. 72:137–145. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rada JA, Cornuet PK and Hassell JR:
Regulation of corneal collagen fibrillogenesis in vitro by corneal
proteoglycan (lumican and decorin) core proteins. Exp Eye Res.
56:635–648. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Svensson L, Närlid I and Oldberg A:
Fibromodulin and lumican bind to the same region on collagen type I
fibrils. FEBS Lett. 470:178–182. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chakravarti S: Functions of lumican and
fibromodulin: lessons from knockout mice. Glycoconj J. 19:287–293.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vogel KG, Paulsson M and Heinegård D:
Specific inhibition of type I and type II collagen fibrillogenesis
by the small proteoglycan of tendon. Biochem J. 223:587–597.
1984.PubMed/NCBI
|
|
10
|
Chakravarti S, Magnuson T, Lass JH, Jepsen
KJ, LaMantia C and Carroll H: Lumican regulates collagen fibril
assembly: skin fragility and corneal opacity in the absence of
lumican. J Cell Biol. 141:1277–1286. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jepsen KJ, Wu F, Peragallo JH, et al: A
syndrome of joint laxity and impaired tendon integrity in lumican-
and fibromodulin-deficient mice. J Biol Chem. 277:35532–35540.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Blochberger TC, Cornuet PK and Hassell JR:
Isolation and partial characterization of lumican and decorin from
adult chicken corneas. A keratan sulfate-containing isoform of
decorin is developmentally regulated. J Biol Chem. 267:20613–20619.
1992.
|
|
13
|
Dolhnikoff M, Morin J, Roughley PJ and
Ludwig MS: Expression of lumican in human lungs. Am J Respir Cell
Mol Biol. 19:582–587. 1998. View Article : Google Scholar
|
|
14
|
Qin H, Ishiwata T and Asano G: Effects of
the extracellular matrix on lumican expression in rat aortic smooth
muscle cells in vitro. J Pathol. 195:604–608. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Baba H, Ishiwata T, Takashi E, Xu G and
Asano G: Expression and localization of lumican in the ischemic and
reperfused rat heart. Jpn Circ J. 65:445–450. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Onda M, Ishiwata T, Kawahara K, Wang R,
Naito Z and Sugisaki Y: Expression of lumican in thickened intima
and smooth muscle cells in human coronary atherosclerosis. Exp Mol
Pathol. 72:142–149. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ping Lu Y, Ishiwata T and Asano G: Lumican
expression in alpha cells of islets in pancreas and pancreatic
cancer cells. J Pathol. 196:324–330. 2002.PubMed/NCBI
|
|
18
|
Lu YP, Ishiwata T, Kawahara K, et al:
Expression of lumican in human colorectal cancer cells. Pathol Int.
52:519–526. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kelemen LE, Couch FJ, Ahmed S, et al:
Genetic variation in stromal proteins decorin and lumican with
breast cancer: investigations in two case-control studies. Breast
Cancer Res. 10:R982008. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Troup S, Njue C, Kliewer EV, et al:
Reduced expression of the small leucine-rich proteoglycans,
lumican, and decorin is associated with poor outcome in
node-negative invasive breast cancer. Clin Cancer Res. 9:207–214.
2003.PubMed/NCBI
|
|
21
|
Ishiwata T, Cho K, Kawahara K, et al: Role
of lumican in cancer cells and adjacent stromal tissues in human
pancreatic cancer. Oncol Rep. 18:537–543. 2007.PubMed/NCBI
|
|
22
|
Vuillermoz B, Khoruzhenko A, D’Onofrio MF,
et al: The small leucine-rich proteoglycan lumican inhibits
melanoma progression. Exp Cell Res. 296:294–306. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shinji S, Tajiri T, Ishiwata T, Seya T,
Tanaka N and Naito Z: Different expression levels of lumican in
human carcinoid tumor and neuroendocrine cell carcinoma. Int J
Oncol. 26:873–880. 2005.PubMed/NCBI
|
|
24
|
Seya T, Tanaka N, Shinji S, et al: Lumican
expression in advanced colorectal cancer with nodal metastasis
correlates with poor prognosis. Oncol Rep. 16:1225–1230.
2006.PubMed/NCBI
|
|
25
|
Matsuda Y, Yamamoto T, Kudo M, et al:
Expression and roles of lumican in lung adenocarcinoma and squamous
cell carcinoma. Int J Oncol. 33:1177–1185. 2008.PubMed/NCBI
|
|
26
|
Leygue E, Snell L, Dotzlaw H, et al:
Expression of lumican in human breast carcinoma. Cancer Res.
58:1348–1352. 1998.
|
|
27
|
Naito Z, Ishiwata T, Kurban G, et al:
Expression and accumulation of lumican protein in uterine cervical
cancer cells at the periphery of cancer nests. Int J Oncol.
20:943–948. 2002.PubMed/NCBI
|
|
28
|
Yamamoto T, Matsuda Y, Kawahara K,
Ishiwata T and Naito Z: Secreted 70kDa lumican stimulates growth
and inhibits invasion of human pancreatic cancer. Cancer Lett.
320:31–39. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bluemlein K and Ralser M: Monitoring
protein expression in whole-cell extracts by targeted label- and
standard-free LC-MS/MS. Nat Protoc. 6:859–869. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Old WM, Meyer-Arendt K, Aveline-Wolf L, et
al: Comparison of label-free methods for quantifying human proteins
by shotgun proteomics. Mol Cell Proteomics. 4:1487–1502. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zybailov B, Coleman MK, Florens L and
Washburn MP: Correlation of relative abundance ratios derived from
peptide ion chromatograms and spectrum counting for quantitative
proteomic analysis using stable isotope labeling. Analytical
chemistry. 77:6218–6224. 2005. View Article : Google Scholar
|
|
32
|
Dennis G Jr, Sherman BT, Hosack DA, et al:
DAVID: Database for Annotation, Visualization, and Integrated
Discovery. Genome Biol. 4:P32003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
34
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009.PubMed/NCBI
|
|
35
|
Lundgren DH, Hwang SI, Wu L and Han DK:
Role of spectral counting in quantitative proteomics. Expert Rev
Proteomics. 7:39–53. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vermes I, Haanen C, Steffens-Nakken H and
Reutelingsperger C: A novel assay for apoptosis. Flow cytometric
detection of phosphatidylserine expression on early apoptotic cells
using fluorescein labelled Annexin V. J Immunol Methods. 184:39–51.
1995. View Article : Google Scholar
|
|
37
|
Aubry JP, Blaecke A, Lecoanet-Henchoz S,
et al: Annexin V used for measuring apoptosis in the early events
of cellular cytotoxicity. Cytometry. 37:197–204. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vij N, Roberts L, Joyce S and Chakravarti
S: Lumican suppresses cell proliferation and aids Fas-Fas ligand
mediated apoptosis: implications in the cornea. Exp Eye Res.
78:957–971. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Brezillon S, Venteo L, Ramont L, et al:
Expression of lumican, a small leucine-rich proteoglycan with
antitumour activity, in human malignant melanoma. Clin Exp
Dermatol. 32:405–416. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Williams KE, Fulford LA and Albig AR:
Lumican reduces tumor growth via induction of fas-mediated
endothelial cell apoptosis. Cancer Microenviron. 4:115–126. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yamasaki M, Nomura T, Sato F and Mimata H:
Metallothionein is up-regulated under hypoxia and promotes the
survival of human prostate cancer cells. Oncol Rep. 18:1145–1153.
2007.PubMed/NCBI
|
|
42
|
Liang JY, Liu YY, Zou J, Franklin RB,
Costello LC and Feng P: Inhibitory effect of zinc on human
prostatic carcinoma cell growth. Prostate. 40:200–207. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Feng P, Li TL, Guan ZX, Franklin RB and
Costello LC: Direct effect of zinc on mitochondrial apoptogenesis
in prostate cells. Prostate. 52:311–318. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ryu HH, Jung S, Jung TY, et al: Role of
metallothionein 1E in the migration and invasion of human glioma
cell lines. Int J Oncol. 41:1305–1313. 2012.PubMed/NCBI
|