Introduction
The novel proliferation-related gene transforming
growth factor (TGF)-β-inducible nuclear protein 1 (TINP1) was
recently identified and cloned by using the gene-function screening
platform of the fluorescein reporter gene-phenotype system
(1). TINP1 (GenBank accession
number: NM_014886) is localized at chromosome 5q13, which shows
frequent abnormalities in hairy cell leukemia (HCL), indicating
that this chromosomal region harbors a gene involved in the
transformational events of HCL (2).
Indeed, the cell viability and the cell cycle analysis have shown
that TINP1 promotes HeLa cell proliferation, which provides
a basis for the further study of this gene’s function. The
TINP1 gene covers 1,105 bp with 6 exons and 5 introns for
coding a protein containing 260 amino acids (1,3,4). The
PSORT II program predicted that TINP1 may be a nuclear
protein (91.3%) and that the TINP1 gene contains a
Ribosomal_S8e conserved domain at amino acids 1–237 after searching
the Pfam database. Ribosomal_S8e is a very highly conserved domain
in eukaryotes, and its function is involved in RNA-protein
interactions that are associated with mRNA translation and protein
synthesis (3,5). In addition, a previous study indicated
that the human TINP1 gene is homologous to the NSA2
gene in Saccharomyces cerevisiae(6). The NSA2 gene has been
demonstrated to play an important role in Saccharomyces
cerevisiae growth, and it has been shown that the TINP1
gene exhibits similar functions to those of NSA2, which
significantly promotes the growth of gene-deficient
Saccharomyces cerevisiae(2).
However, the role of TINP1 in human carcinogenesis and cancer
progression remains largely unknown. It is clear that cell
proliferation is an essential characteristics of tumor initiation
and progression. Cell proliferation is due to cell cycle
progression, which is specifically regulated in a time- and
space-dependent manner, and abnormal cell proliferation induces the
development of multiple diseases (4,7,8),
including human cancer. Thus, the present study investigated the
signaling transduction pathway involved in human TINP1 gene
promotion of cell proliferation and the association between
TINP1 expression and multiple proliferation-related
signaling pathways. Furthermore, the molecular mechanisms
responsible for the TINP1-mediated gene pathway were
explored to provide a potential drug target for cancer
treatment.
Materials and methods
Cell lines and culture
A cervical cancer HeLa cell line, an osteosarcoma
U2OS cell line, a colorectal cancer HCT-16 cell line, a lung cancer
H1299 cell line, and a hepatocellular carcinoma HepG2 cell line
were maintained in our laboratory. A lung cancer A549 cell line, a
human embryonic kidney HEK293T cell line, a Jurkat cell line and a
THP-1 cell line were purchased from the Institute of Biochemistry
and Cell Biology, Shanghai Institutes of Biological Sciences,
Chinese Academy of Sciences (Shanghai, China). These cell lines
were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10%
fetal bovine serum (FBS) at 37°C in an incubator with 5%
CO2 and 95% air.
Design of siTINP1 and gene
transfection
TINP1 small interfering RNA (si-TINP1) and
non-silencing control siRNA were designed and purchased from Santa
Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). The non-silencing
siRNA was confirmed to have no matches with the complete human
genome by a BLAST search in NCBI (www.nibi.nlm.nih.gov). These siRNAs were used to
transfect tumor cells, i.e., tumor cells were seeded into 6-well
plates at a concentration of 2.5×105 cells/ml and grown
in DMEM containing 10% FBS for 24 h. siTINP1 and TINP1 cDNA
containing plasmid or non-silencing siRNA (negative control) were
transfected with Lipofectamine 2000 transfection reagent
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s
instructions.
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total cellular RNA from cultured tumor cells was
isolated using the TRIzol™ reagent (Invitrogen), according to the
manufacturer’s instructions, and reversely transcribed into
single-strand cDNA using the Reverse transcript™ kit (Invitrogen).
Next, PCR amplification was performed using specific primers for
the TINP1 gene: 5′-CTG CTG GAC AGA GAG GGA CAA-3′ and 5′-TTT
CTC CCT GGG CAC GTA CTT-3′. The housekeeping gene GAPDH was used as
a loading control of PCR amplification using the following primers:
5′-TGA AGG TCG GAG TCA ACG GAT TTG GT-3′ and 5′-CAT GTG GGC CAT GAG
GTC CAC CAC-3′. The RT-PCR amplification was performed with the
ThermoScript RT-PCR system (Invitrogen), according to the
manufacturer’s protocol, with an initial cycle of 94°C for 5 min
and 35 cycles of 94°C for 30 sec, 58°C for 30 sec, and 72°C for 30
sec, and a final extension at 72°C for 10 min. The PCR products
were separated in 2% agarose gel and semi-quantified according to
the levels of GAPDH mRNA. Then, the TINP1 expression was compared
with that of the negative control siRNA or vector only-transfected
cells.
Protein extraction and western blot
analysis
Total cellular protein was extracted from the
cultured cells using a RIPA lysis buffer (50 mM Tris-HCl, pH 7.4;
150 mM NaCl; 1% deoxycholate Na; 1% NP-40; 0.1% sodium dodecyl
sulfate, with freshly added protease inhibitor cocktail). Protein
concentrations were determined using the BCA protein assay reagent
(Pierce, Biotechnology, Inc., Rockford, IL, USA). Following
quantification, 20 μg of total protein samples were separated using
12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) and transferred onto a nitrocellulose membrane
(Hybond™, ECLTM; Amersham Pharmacia, Little Chalfont,
UK). After blocking for 2 h in 5% non-fat milk in Tris-buffer
supplemented with 0.05% Tween-20 (TBS-T), the membrane was
incubated with the corresponding primary antibody at 4°C overnight.
Polyclonal antibodies against p53, Erk 1/2, JNK/SPAK, p38, and
β-actin were obtained from Cell Signaling Technology (Boston, MA,
USA). The membrane was then rinsed in TBS-T three times for 10 min
each and incubated with the secondary antibody at room temperature
for 2 h. The membrane was rinsed again in TBS-T three times for 10
min each and visualized using an ECL chemiluminescence system
(Bio-Rad Laboratories, Hercules, CA, USA).
Cell viability cell counting kit-8
(CCK-8) assay
The cells were seeded into 96-well plates at a
density of 2×103 cells/ml in a volume of 100 μl/well and
placed in an incubator for 18–24 h. After reaching 40–60%
confluency, the cells were transfected with 20 ng of blank vector
as a negative control, TINP1 plasmid, and H-Ras (as a positive
control), respectively, using the VigoFect transfection reagent in
triplicate and then cultured for 5 days. At the end of the
experiments, 10 μl of CCK-8 reagent was added to each well, and the
cells were further incubated for 2 h. Subsequently, the absorbance
value was determined using a multifunctional plate reader (GENios
ProTM). Cell viability was summarized as the percentage
of control.
Flow cytometry
The cells were seeded into 6-well plates at a
density of 3×105 cells/ml and transfected with different
plasmids for up to 3 days. At the end of the experiments, the cells
were digested with 2.5% pancreatin for 24 h, harvested and prepared
into single-cell suspensions. After washing with pre-cooled
phosphate-buffered saline (PBS) twice, the cell suspension was
rotated on a Vortex rotator; cooled 75% ethanol was added dropwise
to the suspension and the cells were fixed at −20°C overnight. The
next day, the cell suspension was washed with PBS twice, incubated
with RNase (10 μg/ml) at 37°C for 30 min and stained with propidium
iodide. The cells were then analyzed by a FACSCalibur flow
cytometer (Becton-Dickinson, Franklin Lakes, NJ, USA).
Luciferase reporter assay
The cells were seeded into 96-well plates at a
density of 1.1×106 cells/ml in a volume of 100 μl/well
and placed in an incubator for 18–24 h. After reaching 40–60%
confluency, the cells were co-transfected with the cell
proliferation-related pathway reporters: 50 ng of NF-κB-LUC,
pWNT-LUC, pP53-LUC, pC/EBP-LUC, or pT-bet-LUC plus 50 ng of
pAP-1-LUC, 5 ng of pRL-TK-LUC, and 50 ng of TINP1 or the control
plasmid using the VigoFect transfection reagent (Beijing Vigorous,
Beijing, China). Cells transfected with blank vector served as
blank controls, while cells transfected with TNF-α, β-catenin, p53,
PKA, aj41 and MEKK served as positive controls. These transfections
were performed in triplicate and repeated at least once. Following
incubation for 24 h, the luciferase activity was determined using a
multifunctional plate reader (GENios Pro™). The relative
fluorescence value of the cells transfected with blank vector was
defined as 100%, and the results were expressed as the mean value
of three replicate wells.
Statistical analyses
The data are summarized as means ± standard
deviation (SD) as appropriate. The Student’s t-test was used to
generate a P-value using SPSS software (SPSS, Inc., Chicago, IL,
USA). A P-value <0.05 was considered to indicate a statistically
significant difference.
Results
Expression of TINP1 and knockdown of
TINP1 expression using siTINP1
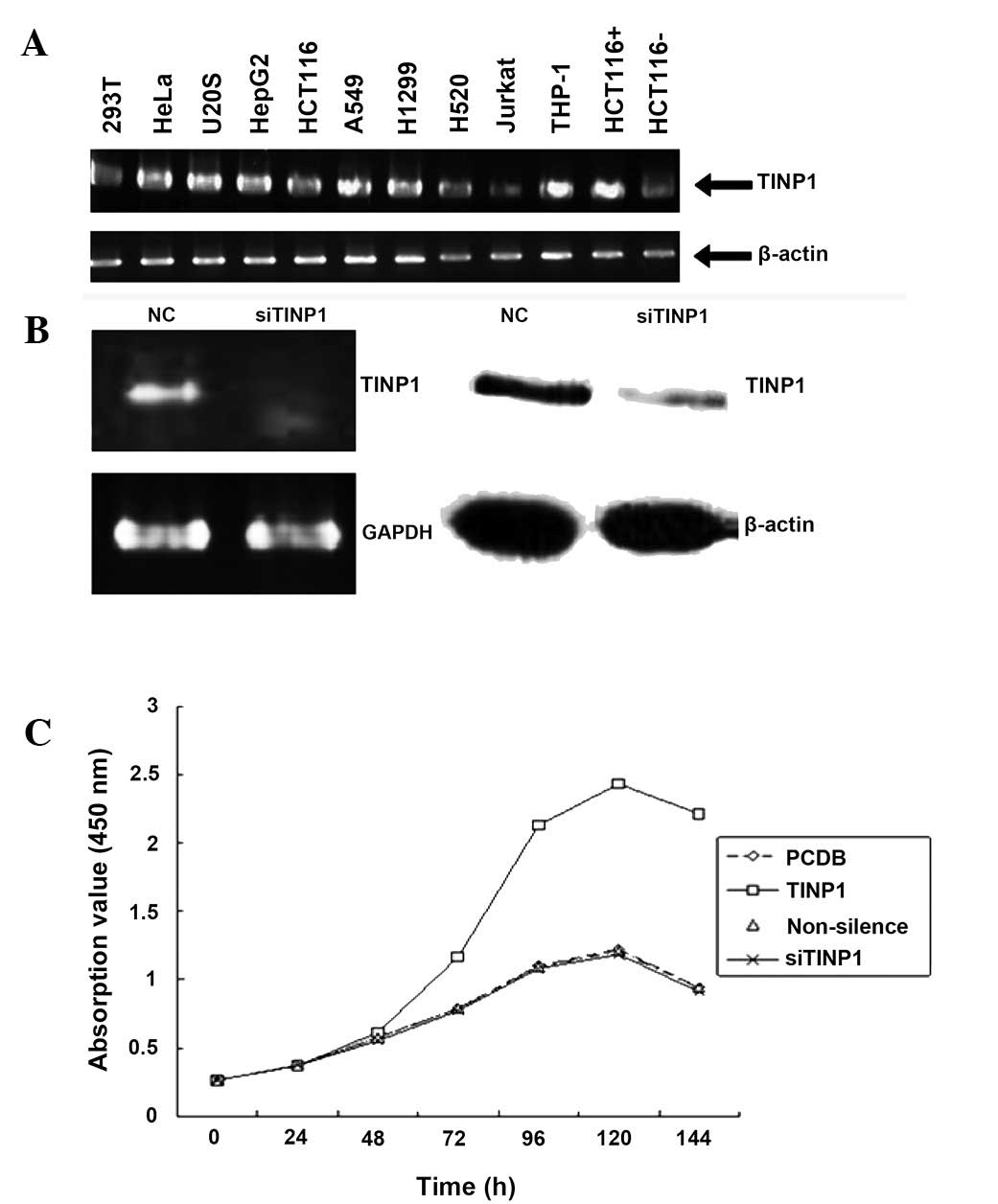
In the present study, we first detected TINP1
expression in multiple cell lines and found that TINP1 was
expressed in most of these cells using RT-PCR (Fig. 1A). Next, we chose HeLa cells for
knockdown of TINP1 expression using TINP1 siRNA nucleotides. After
24 and 48 h of gene transfection, the endogenous TINP1 mRNA
expression in siTINP1-transfected cells was significantly reduced
compared to that in negative control cells (Fig. 1B). Next, we assessed the effects of
TINP1 knockdown on the regulation of HeLa cell viability and found
that siTINP1 significantly reduced tumor cell viability (Fig. 1C).
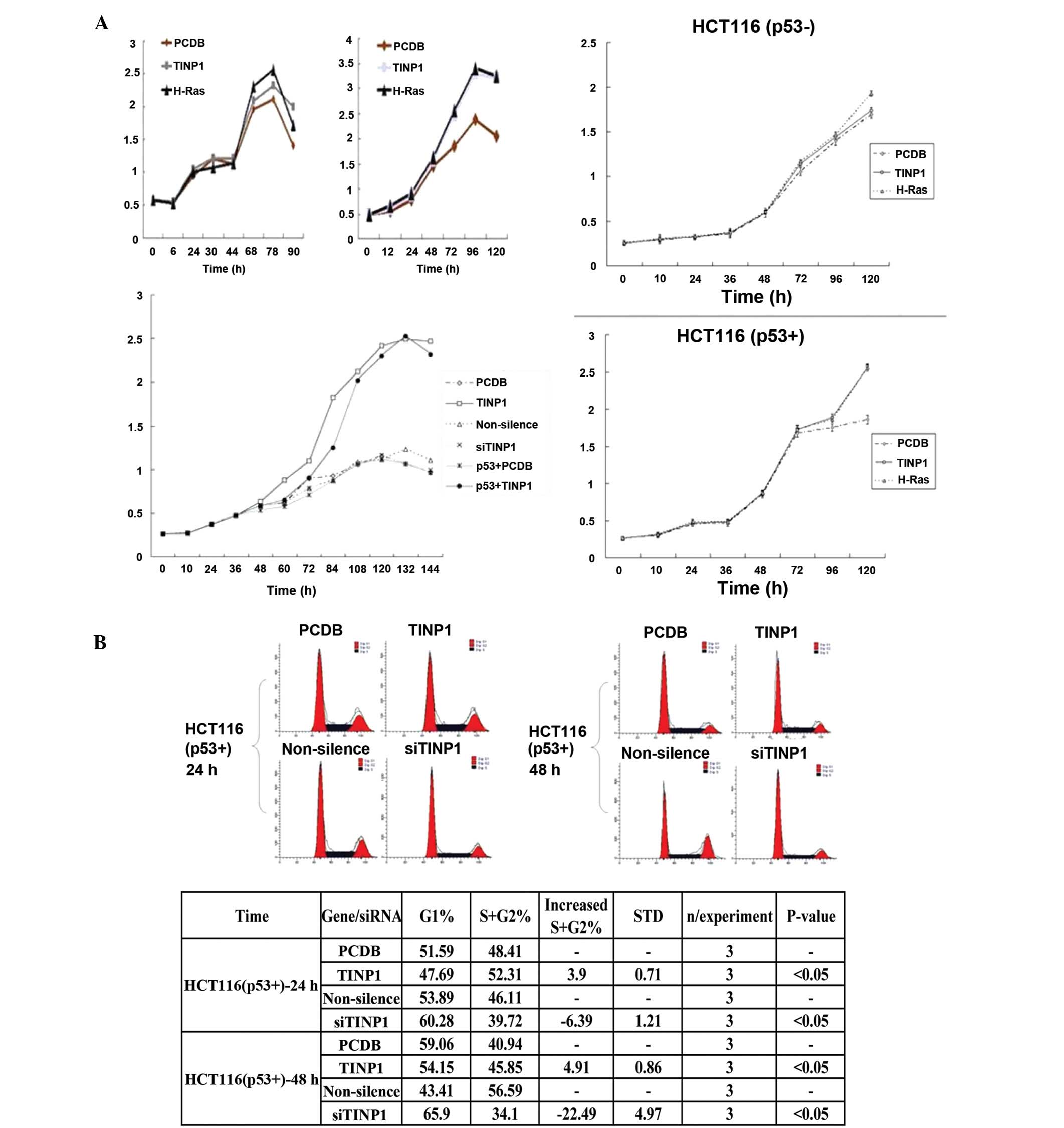
Effect of TINP1 overexpression on tumor
cell viability and cell cycle progression
We also assessed TINP1 overexpression on the
regulation of cell proliferation by transfecting TINP1 cDNA into
HeLa cells and found that TINP1 promoted cell growth (Fig. 2A). In addition, flow cytometry data
showed that TINP1 induced more cells at the S phase of the cell
cycle, further indicating cell proliferation (Fig. 2B).
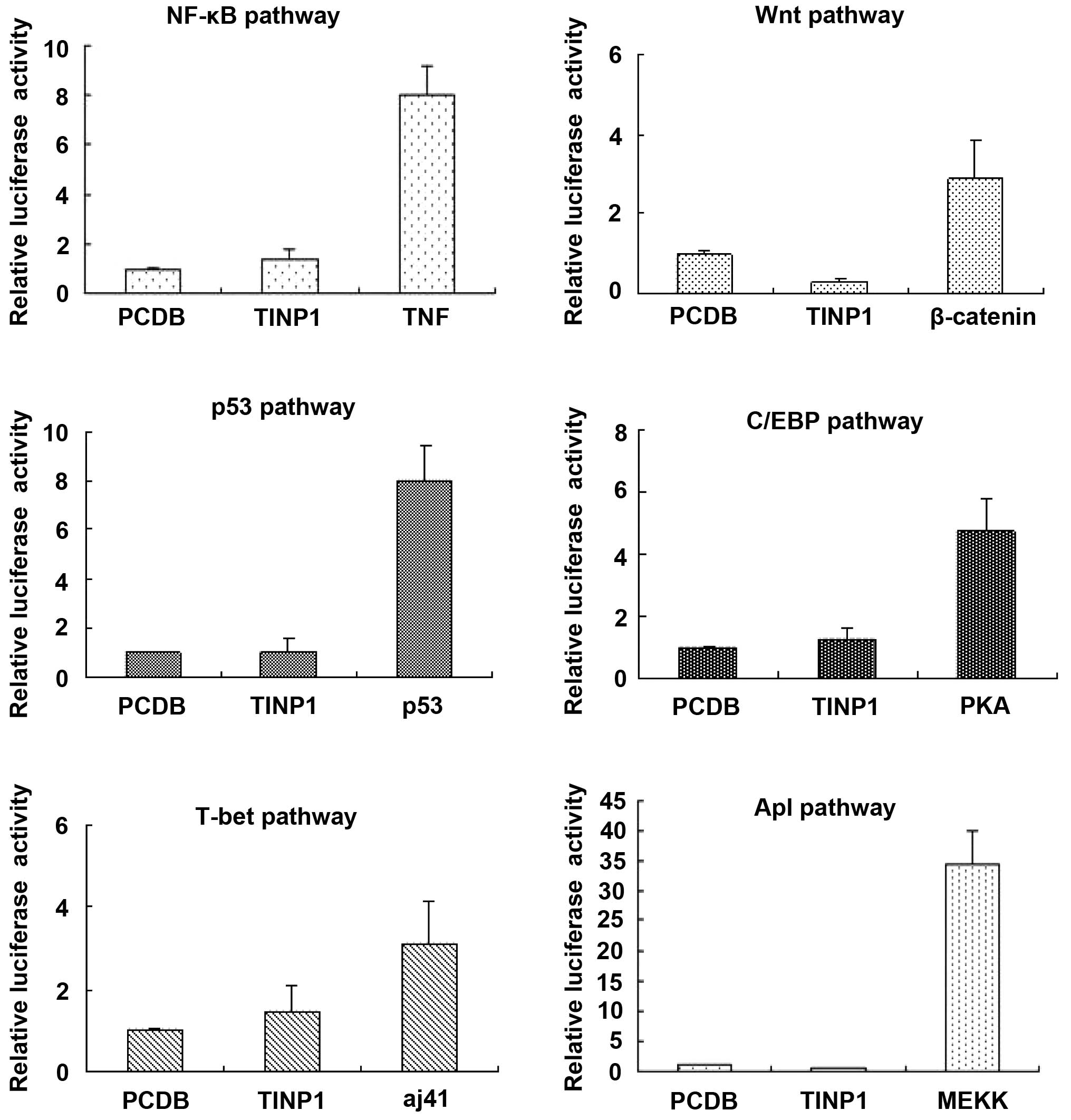
Effect of TINP1 overexpression on
signaling pathways
To explore the role of TINP1 in signaling pathway
regulation, we performed a dual-luciferase reporter assay to
investigate six signaling pathways (including NF-κB, WNT, p53,
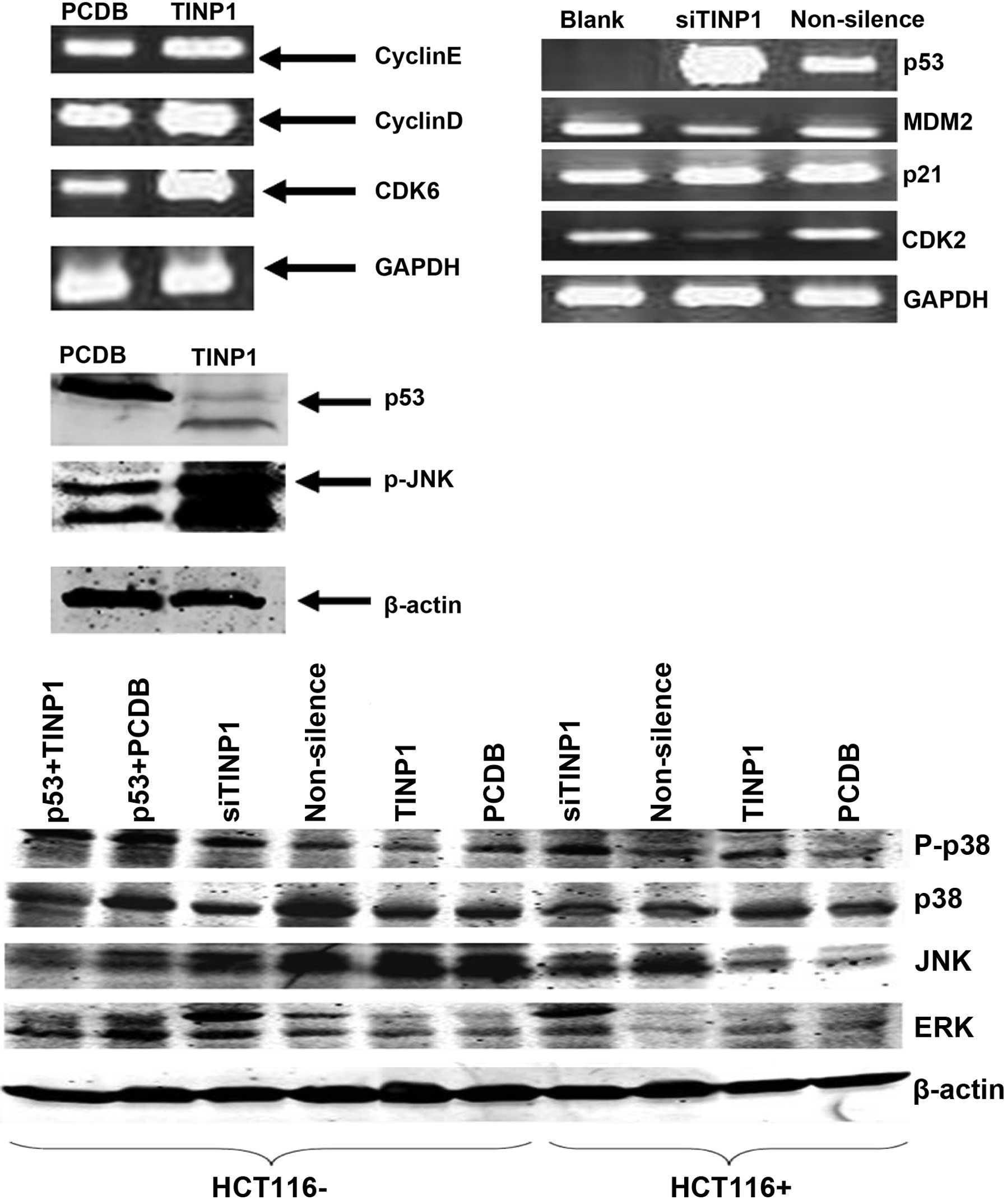
C/EBP, T-bet and AP1). The data showed that TINP1 did not
significantly alter the activities of these six signaling pathways
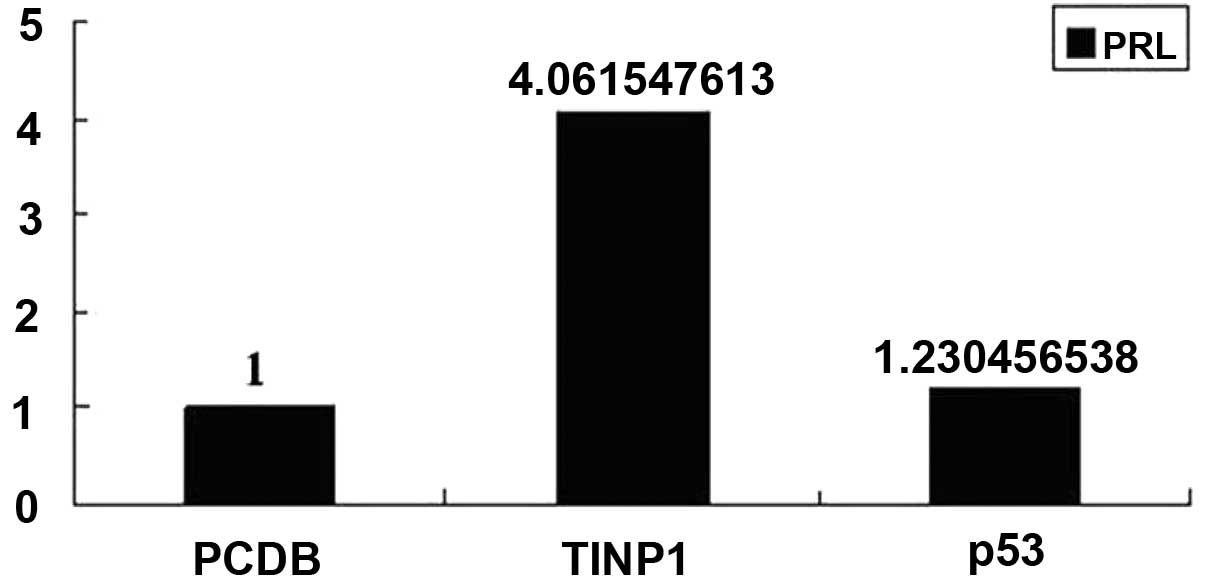
(Fig. 3). However, TINP1
significantly promoted pRL-TK-LUC activity in the p53 pathway
(Fig. 4). Next, we determined the
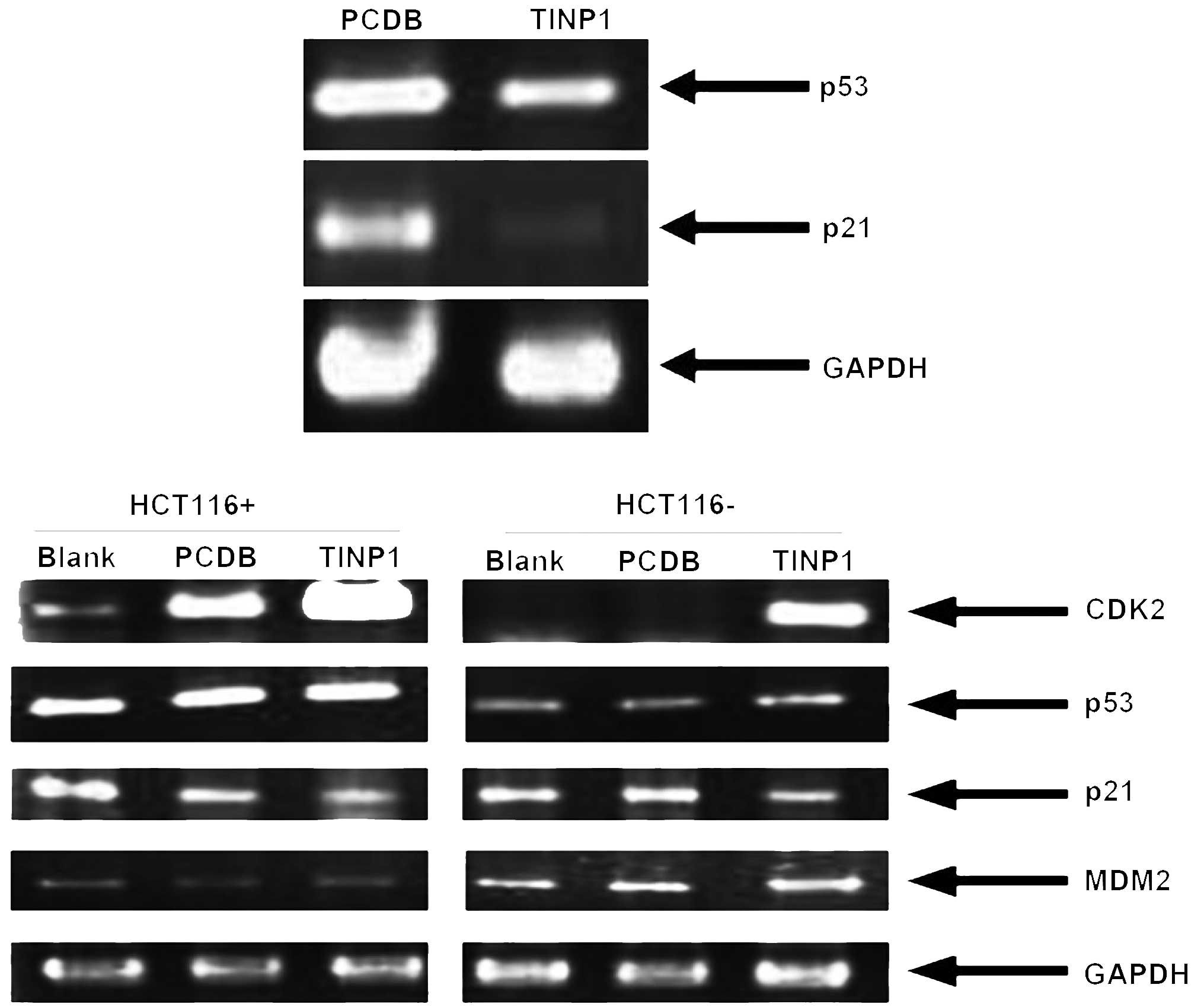
expression of these p53-related genes and found that TINP1
significantly inhibited p53 mRNA levels, silenced p21 mRNA
expression in HCT-116 cells, and induced cyclin D1 and CDK6
expression (Figs. 5 and 6). By contrast, TINP1 siRNA induced p53
and p21 expression but reduced the expression of MDM2 and CDK2 mRNA
(Fig. 5).
Discussion
In the present study, we investigated the effects of
TINP1 knockdown and overexpression in the regulation of cell growth
and gene expression in various cell lines. We found that TINP1 was
expressed in multiple cell lines and that knockdown of TINP1
expression reduced HeLa cell proliferation. By contrast, the
overexpression of TINP1 promoted cell proliferation in various cell
lines, which was associated with the inhibition of p21 and p53
expression to promote the S phase of the cell cycle. However, TINP1
overexpression did not affect the activities of five different
signaling pathways (NF-κB, WNT, C/EBP, AP1 and T-bet). These data
suggest that TINP1 plays a role in cell growth by regulating the
expression of p53 and p21-related genes. Future studies will
further investigate how TINP1 affects p53 and p21 gene expression
so that targeting TINP1 expression in tumor cells may be used in
clinical settings as a novel strategy to treat cancer.
The data from the present study clearly show that
TINP1 overexpression significantly promoted cell proliferation and
advanced the cells to the S phase of the cell cycle, whereas TINP1
knockdown reduced HeLa cell viability. These data confirmed a
previous study that indicated a proliferative role of TINP1 in
cells, since the human TINP1 gene is a homolog of the
NSA2 gene in Saccharomyces cerevisiae. The
NSA2 gene plays a role in the growth of Saccharomyces
cerevisiae(2). Furthermore, the
SymAtlas online application revealed that the human TINP1
gene is expressed in multiple tissues (2). Indeed, our present study demonstrated
that TINP1 was expressed in multiple cell lines. Since cell
proliferation (i.e., growth) is involved in multiple gene signaling
pathways, we assessed the role of TINP1 in the activation of
six different gene pathways. Our data showed that TINP1 does not
alter the luciferase activities of NF-κB, WNT, C/EBP, AP1 or T-bet
genes. However, further study showed that the overexpression of
TINP1 inhibited the expression of p53 and p21 mRNA but induced
cyclin D1 and CDK6 expression, whereas knockdown of TINP1 induced
p53 and p21 expression. Our present study linked TINP1 and the p53
gene pathway as promoting cell cycle progression. Further studies
will investigate how TINP1 inhibits p53 and p21 expression and will
also examine TINP1 as a novel target in cancer therapy.
Acknowledgements
This study was supported in part by grants from the
National Natural Science Foundation (no. 30900730) and the Shandong
Province Natural Science Foundation (Q2007D01).
References
|
1
|
Wu X, Ivanova G, Merup M, et al: Molecular
analysis of the human chromosome 5q13.3 region in patients with
hairy cell leukemia and identification of tumor suppressor gene
candidates. Genomics. 60:161–171. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ohnishi Y, Saika S, Yamanaka O, et al:
Investigation of mechanism of cell proliferation regulation and its
clinical application. Nihon Ganka Gakkai Zasshi. 109:865–884.
2005.(In Japanese).
|
|
3
|
Stanchi F, Bertocco E, Toppo S, et al:
Characterization of 16 novel human genes showing high similarity to
yeast sequences. Yeast. 18:69–80. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Strausberg RL, Feingold EA, Grouse LH, et
al: Generation and initial analysis of more than 15,000 full-length
human and mouse cDNA sequences. Proc Natl Acad Sci USA.
99:16899–16903. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Saveanu C, Namane A, Gleizes PE, et al:
Sequential protein association with nascent 60S ribosomal
particles. Mol Cell Biol. 23:4449–4460. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang H, Ma X, Shi T, Song Q, Zhao H and
Ma D: NSA2, a novel nucleolus protein regulates cell proliferation
and cell cycle. Biochem Biophys Res Commun. 391:651–658. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang SS and Huang JS: TGF-β control of
cell proliferation. J Cell Biochem. 96:447–462. 2005.
|
|
8
|
Chittaranjan S, McConechy M, Hou YC,
Freeman JD, Devorkin L and Gorski SM: Steroid hormone control of
cell death and cell survival: molecular insights using RNAi. PLoS
Genet. 5:e10003792009. View Article : Google Scholar : PubMed/NCBI
|