Introduction
Breast carcinogenesis is a multi-step process
characterized by tumor initiation and progression (1). There are well understood genetic and
epigenetic alterations associated with breast carcinogenesis.
Epigenetics is a heritable and reversible change in gene
expression, and epigenetic alterations include DNA methylation and
chromatin remodeling (2). DNA
methylation occurs when methyl groups are added to cytosines in CpG
dinucleotides resulting in the formation of methylcytosine
(5-methylcytosine) and it leads to changes in chromatin structure
and gene silencing (1–3). Several tumor suppressor genes contain
CpG islands in their promoters, and a number of them show evidence
of methylation silencing (3).
Hypermethylation of regulatory regions of several tumor suppressor
genes has been correlated with decreased gene expression, whereas
hypomethylation of normally methylated tumor suppressor genes plays
an important role in cancer development (3,4). Gene
specific epigenetic changes for breast cancer are likely to occur
early in tumorigenesis and have the potential to be used for early
detection and prevention (5). In
particular, abnormal promoter region methylation in candidate tumor
suppressor genes may be a useful biomarker by permitting early
diagnosis and predicting the clinical behavior of the breast
cancer.
The fragile histidine triad (FHIT) gene,
encompassing the FRA3B fragile site at chromosome 3p14.2, is a
tumor suppressor gene in several different types of cancer
(6). The FHIT gene is a
member of the histidine triad gene family, encoding a protein
similar to the yeast diadenosine tetraphosphates hydrolase, which
are intracellular and extracellular signaling molecules involved in
cellular differentiation and apoptosis (6). In breast cancer, abnormalities at the
FHIT locus have been demonstrated in considerably high
frequency (7). These include loss
of heterozygosity (LOH) (8,9), homozygous deletions (9,10),
hypermethylation of the promoter region (11), abnormally sized transcripts
(12) and reduced RNA and protein
expression (13).
Previous studies have shown that methylation is a
mechanism of the FHIT gene inactivation in breast cancer
(11,14), and the FHIT gene promoter
hypermethylation has been correlated with loss of gene expression
in several different types of cancer, including breast cancer
(15–19). Several studies (20–22)
have evaluated the association between the FHIT gene
hypermethylation and expression of Fhit protein encoded by the
FHIT gene with clinicopathological characteristics in breast
cancer, but with dissimilar results. There have been suggestions
that DNA methylation profiles are associated with human epidermal
growth factor receptor 2 (HER2) status of breast cancer (23) and Fhit cooperates with HER2 in
breast carcinogenesis (24).
However, limited information is available on the methylaton status
of the FHIT gene and Fhit expression associated with HER2
status in breast cancer.
To further clarify the role of Fhit expression in
breast cancer and its relation to gene hypermethylation, we
evaluated the association between methylation of the FHIT
gene and its expression in Korean breast cancer patients. We also
investigated whether the FHIT gene methylation and
expressions of Fhit correlate with clinicopathological
characteristics in the same patients, specifically, according to
HER2 status.
Materials and methods
Patients and materials
Formalin-fixed and paraffin-embedded primary breast
tumor tissue blocks from patients with breast cancer who underwent
surgery at Daegu Catholic University Hospital (Daegu, South Korea)
were examined. All specimens were reviewed by an experienced
pathologist and a total of 60 sporadic invasive ductal carcinoma
(IDC) tissue samples were included in the present study. The
clinicopathological characteristics such as age, menopausal state,
tumor size, nodal status, histologic grade, lymphovascular
invasion, and prognostic factors including estrogen receptor (ER),
progesterone receptor (PR), HER2, Bcl-2, Ki-67 and p53 expression
were evaluated based on pathological reports and medical records.
Pathological staging was assessed according to the seventh edition
of the American Joint Committee on Cancer (AJCC) staging manual for
breast cancer. We subclassified the breast cancer sample molecular
subtypes into basal-like, HER2, luminal A, and luminal B subtypes
according to immunohistochemical findings for the ER, PR, HER2 and
Ki-67 proliferation index (25).
Ethics approval for the study was obtained from the Institutional
Review Board at the Daegu Catholic University Hospital.
Construction of tissue microarrays
(TMA)
Representative paraffin tumor blocks were selected
according to the primary evaluation of hematoxylin and eosin
(H&E)-stained slides before they were prepared for TMA. Two
tumor tissue cores (2 mm in diameter) were obtained from each of
the donor breast cancer tissue blocks using a manual punch arrayer
(Quick-Ray™; Uni-Tech Science, Seoul, South Korea). The cores were
placed in a new recipient paraffin block that ultimately contained
50–60 tissue cores. Each array block contained both tumor and
control tissue samples. Multiple sections (5 μm thick) were cut
from the TMA blocks and then mounted onto microscope slides. The
TMA H&E-stained sections were reviewed by light microscopy to
confirm the presence of representative tumor areas.
Immunohistochemical staining and
interpretation
Immunohistochemical analysis was performed on
5-μm-thick TMA tissue sections using the Bond Polymer Intense
Detection System (Leica Microsystems, Victoria, Australia)
according to the manufacturer’s instructions with minor
modifications. Briefly, the 5-μm-thick sections of formalin-fixed
and paraffin-embedded TMA tissues were deparaffinized with Bond
Dewax Solution (Leica Microsystems), and an antigen retrieval
procedure was performed using Bond ER Solution (Leica Microsystems)
for 30 min at 100°C. The endogenous peroxidase was quenched by a
5-min incubation with hydrogen peroxide. Sections were incubated
for 15 min at ambient temperature with a rabbit polyclonal
anti-Fhit antibody (ab53074, 1:150; Abcam, Cambridge, UK), and
commercially available primary monoclonal antibodies for ER (1:100,
clone 6F11; Novocastra), PR (1:100, clone 16; Novocastra), HER2
(1:250, A0485; Dako), Ki-67 (1:200, MM1-L; Novocastra), Bcl-2 (1:4,
clone 124; Dako), p53 (1:200, BP53.12; Zymed Laboratories), p16
(1:200; Dako, Denmark) and epidermal growth factor receptor (EGFR)
(1:100, clone EGFR.25; Novocastra) using a biotin-free polymeric
horseradish peroxidase-linker antibody conjugate system in a
Bond-Max automatic slide stainer (Leica Microsystems).
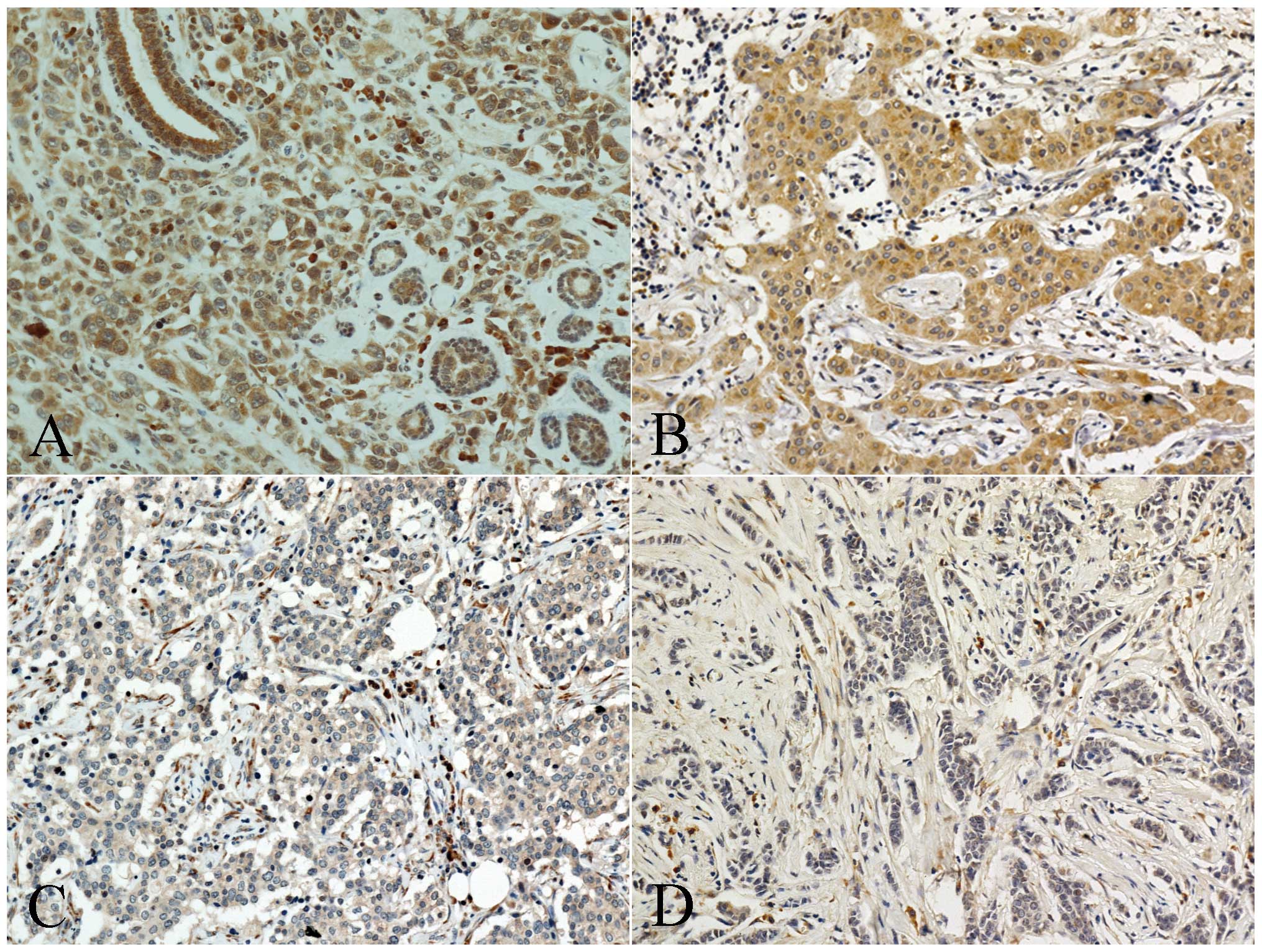
Fhit expression levels were graded on a scale of 0
to 3+ based on staining intensity and proportion of positive tumor
cells by an expert pathologist who was blinded to the patient
clinical records. The extent of positivity was scored as 0,
negative; 1+, weak intensity, <30% of cancer cells staining; 2+,
moderate intensity, 31–60%; and 3+, strong intensity, >60%
(Fig. 1). For statistical analysis,
diffuse absence of staining was regarded as negative expression,
whereas any level of staining, regardless of percentage of cancer
cell staining, was considered positive for Fhit expression.
A cut-off value of 10% for the stained nuclei was
used to define ER and PR positivity. Cytoplasmic staining of any
intensity in >10% of the tumor cells was scored as positive for
Bcl-2. Membranous staining for HER-2 with strong complete staining
in 10% of the tumor cells was regarded as HER-2 overexpression. p53
and p16 staining was scored positive if >10% of the cells were
stained with a strong intensity. The Ki-67 labeling index was
expressed as a percentage and was graded as ‘high’ if the number of
positive cells was ≥14%. Inflammation was assessed by scoring
infiltration of mononuclear cells in the tumor cell nests and
stroma (intratumoral) and adjacent stroma (peritumoral). The extent
of lymphocyte infiltration was scored as 0, no mononuclear cell
infiltration; 1+, focal scattered infiltration; 2+, focal and
clustered infiltration; and 3+, diffuse infiltration and formation
of lymphoid follicle. For statistical analysis, absence of
mononuclear cell infiltration was defined as negative, and any
level of mononuclear cell infiltration was considered positive for
intratumoral or peritumoral inflammation.
DNA extraction and sodium bisulfate
treatment
For DNA extraction, eight 5–10-μm thick tissue
sections were obtained from paraffin-embedded primary breast
cancer. Genomic DNA was isolated using QIAamp DNA FFPE Tissue kit
(Qiagen, Hilden, Germany) by following the manufacturer’s protocol.
The purified DNA was quantified using an ND-1000 spectrophotometer
(NanoDrop Technologies, Inc., Wilmington, DE, USA). The quality of
the DNA was verified by performing gel electrophoresis. Sodium
bisulfate modification of 200–500 ng genomic DNA was performed
using the EZ DNA Methylation-Gold kit (Zymo Research, Orange, CA,
USA) according to the manufacturer’s protocol.
Pyrosequencing
Methylation was analyzed using pyrosequencing.
Primer was designed using the PyroMark Assay Design program ver.
2.0.1.15 (Qiagen). For polymerase chain reaction (PCR), the forward
primer was 5′-GGGAGGTAAGTT TAAGTGGAATATTG-3′ and the reverse primer
was 5′-CCACTAAACTCCCAAATAATAACCTAAC-3′. PCR was performed using
bisulfate-treated DNA under the following conditions: 95°C for 5
min; 45 cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C for 30
sec; and final extension of 5 min at 72°C. PCR was conducted using
a PCR PreMix (Enzynomics, Daejeon, Korea) and the quality and
quantity of the PCR product was confirmed by performing agarose gel
(2%) electrophoresis by loading 4 μl of 20 PCR products.
Pyrosequencing was performed using the Pyro Gold kit and PSQ 96 MA
instrument (Qiagen) as instructed by the manufacturer. The Primer
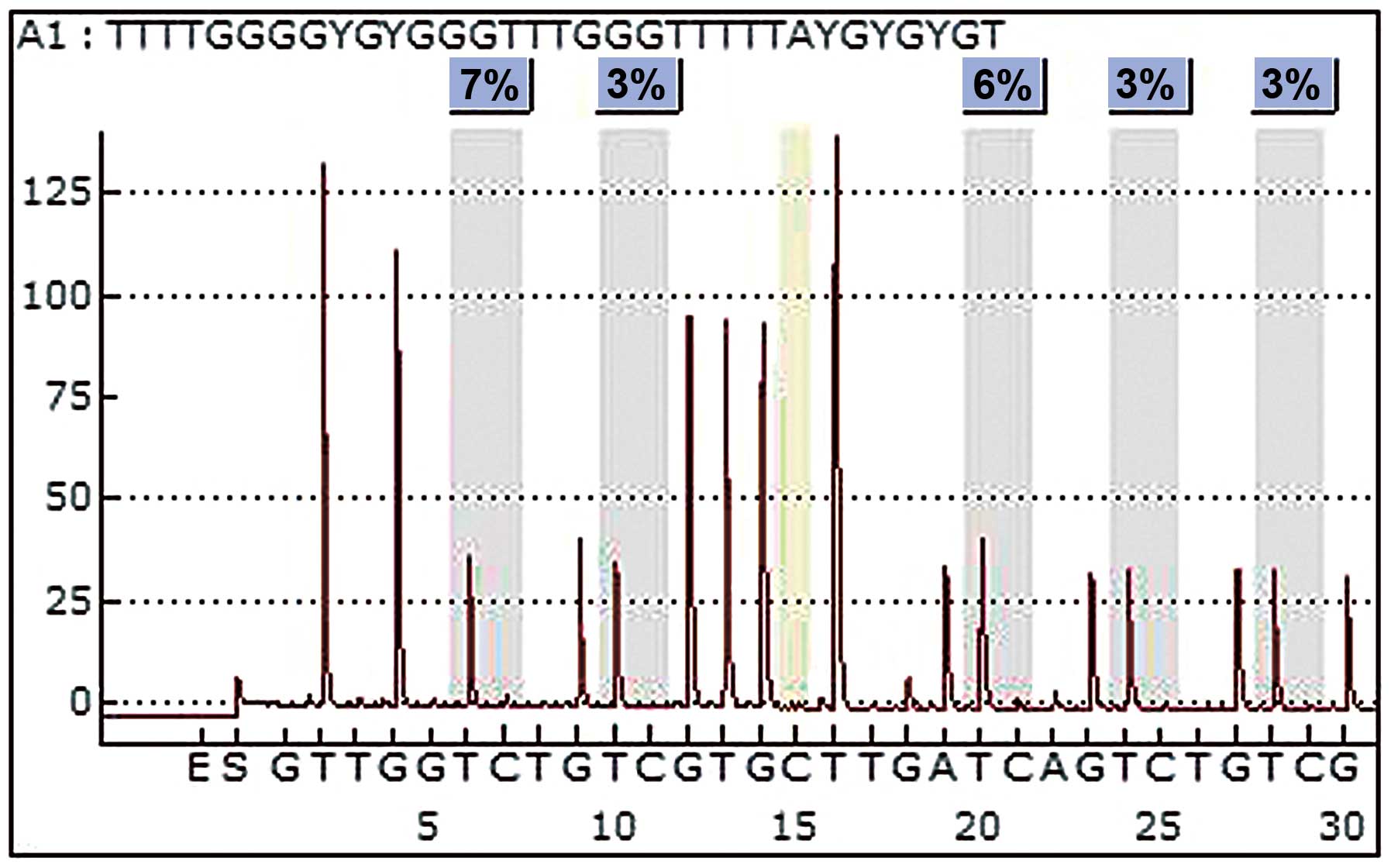
for DNA sequencing was 5′-GTAAGTTTAAGTGGAATATTGT-3′. The
methylation index (MtI) of the FHIT gene in each sample was
calculated as the average value of
mC/(mC + C) for all
examined CpGs in target regions (Fig.
2). All experiments included a negative control without
template.
Statistical analysis
Statistical analyses were performed using SPSS
version 15.0 (SPSS, Inc., Chicago, IL, USA). A one-sample
Kolmogorov-Smirnov test was used to evaluate the fitness to normal
distribution of continuous parameters. Association between the
methylation status of the FHIT gene and its expression was
assessed using the Student’s t-test or non-parametric Mann-Whitney
U test. Associations between the FHIT gene methylation
status and the clinicopathological characteristics were assessed
using the Student’s t-test or the non-parametric Mann-Whitney U
test for categorical variables, and correlation between 2
continuous variables was assessed using correlation analysis. A
comparison of the mean methylation level of the FHIT gene
across the subtypes was performed using the ANOVA or the
Kruskal-Wallis test. The relationship between the Fhit expression
and the clinicopathological characteristics of the patients was
analyzed using the Chi-square test or the Fisher’s exact test for
categorical data and the Student’s t-test or the non-parametric
Mann-Whitney U test for continuous data. Unconditional logistic
regression was used to assess odds ratios (ORs) and 95% confidence
intervals (CIs). All tests were 2-sided and a P-value of <0.05
was considered to indicate a statistically significant
difference.
Results
Clinicopathological characteristics
Clinical and pathological characteristics of
patients are shown in Table I. The
average age of the 60 patients with breast cancer was 51.77±13.22
years (range, 26–90 years). Twenty-nine patients (48.3%) were ER
positive and 30 patients (50.0%) were HER2 positive. Twenty-nine
patients (48.3%) had stage I disease, 21 patients (35.0%) stage II,
6 patients (10.0%) stage III and 4 patients (6.7%) stage IV.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Clinicopathological
variables | Value |
|---|
| Age (years), mean
(range) | 51.77±13.22
(26–90) |
| Menopausal status,
n (%) |
|
Pre-menopausal | 27 (45.8) |
|
Post-menopausal | 32 (54.2) |
| Tumor size (cm),
mean (range) | 1.80±0.93
(0.10–4.50) |
| Histological grade,
n (%) |
| I | 13 (21.7) |
| II | 11 (18.3) |
| III | 36 (60.0) |
| Nodal involvement,
n (%) |
| Negative | 40 (69.0) |
| Positive | 18 (31.0) |
| Distant metastasis,
n (%) |
| Negative | 58 (96.7) |
| Positive | 2 (3.3) |
| Molecular subtype,
n (%) |
| Luminal A | 15 (25.0) |
| Luminal B | 15 (25.0) |
| HER2 | 15 (25.0) |
| Basal-like | 15 (25.0) |
| Lymphovascular
invasion, n (%) |
| Negative | 39 (66.1) |
| Positive | 20 (33.9) |
| ER, n (%) |
| Negative | 31 (51.7) |
| Positive | 29 (48.3) |
| PR, n (%) |
| Negative | 33 (55.0) |
| Positive | 27 (45.0) |
| HER2
overexpression, n (%) |
| Negative | 30 (50.0) |
| Positive | 30 (50.0) |
| Ki-67, n (%) |
| <14 | 25 (41.7) |
| ≥14 | 35 (58.3) |
| FHIT
methylation frequency (%), mean | 3.43±0.97 |
| Fhit expression, n
(%) |
| Negative | 7 (12.8) |
| Positive | 48 (87.3) |
Methylation status of FHIT gene and its
expression in breast cancer
Of the 60 patients studied, 58 patients (96.7%)
showed aberrant methylation of the FHIT gene in
pyrosequencing analysis. The mean methylation level of the
FHIT gene was 3.43±0.97%. The methylation frequency of the
FHIT gene showed no significant differences according to
molecular subtypes of breast cancer (P=0.367).
Expression of Fhit protein was analyzed by
immunohistochemical staining on breast cancer TMA from 60 invasive
breast cancer cases. Some of the tissue specimens that were partly
lost during TMA construction or were unavailable were excluded.
According to the criteria for immunohistochemistry evaluation,
positive Fhit expression was observed in 48/55 (87.3%) primary
breast tumor tissue samples.
To determine whether absence or decrease of Fhit
expression in breast cancer correlates with the hypermethylation of
the FHIT gene, we compared Fhit expression with the level of
the FHIT gene methylation. Mean methylation level of the
FHIT gene was slightly higher in the negative Fhit
expression group (3.49%) than that of the positive expression group
(3.41%); however, there was no significant correlation between
methylation of the FHIT gene and its expression in the
present study (P=0.856) (Table
II).
 | Table IICorrelation between the FHIT
gene methylation and its expression. |
Table II
Correlation between the FHIT
gene methylation and its expression.
| FHIT
methylation levels, mean (%) | P-value |
|---|
| Fhit
expression |
| Positive | 3.41±1.01 | 0.856 |
| Negative | 3.49±0.74 | |
Relationship between methylation levels
of the FHIT gene, the Fhit expression and the clinicopathological
features
No significant correlation was found between the
FHIT gene methylation levels and the clinicopathological
features (Table III). Comparing
the methylation levels of the FHIT gene with other markers,
the FHIT gene methylation was significantly associated with
intratumoral inflammation (P=0.037) (Table IV).
 | Table IIIAssociation of methylation levels of
the FHIT gene and Fhit expression with clinicopathological
characteristics. |
Table III
Association of methylation levels of
the FHIT gene and Fhit expression with clinicopathological
characteristics.
| Clinicopathological
features | FHIT
methylation | Fhit
expression |
|---|
|
|
|---|
| Mean levels
(%) | P-value | Negative
expression, n (%) | P-value |
|---|
| Age (years) |
| <50 | 3.25±0.81 | 0.149 | 4 (14.8) | 0.705 |
| ≥50 | 0.62±1.10 | | 3 (10.7) | |
| Menopausal
state |
|
Pre-menopausal | 3.29±0.99 | 0.316 | 2 (8.3) | 0.443 |
|
Post-menopausal | 3.55±0.96 | | 5 (16.7) | |
| Stage |
| I | 3.47±1.00 | 0.203 | 1 (3.7) | 0.244 |
| II | 3.35±0.88 | | 5 (26.3) | |
| III | 4.04±1.20 | | 0 (0.0) | |
| IV | 2.73±0.61 | | 1 (33.3) | |
| Tumor size
(cm) |
| ≤2 | 3.49±1.13 | 0.644 | 1 (2.9) | 0.006 |
| >2 | 3.36±0.71 | | 6 (31.6) | |
| Nodal
involvement |
| Negative | 3.37±0.92 | 0.476 | 5 (13.9) | 0.651 |
| Positive | 3.57±1.14 | | 1 (5.9) | |
| Distant
metastasis |
| Negative | 3.47±0.97 | 0.187 | 6 (11.3) | 0.240 |
| Positive | 2.54±0.83 | | 1 (50.0) | |
| Histological
grade |
| I | 3.56±0.99 | 0.758 | 0 (0.0) | 0.204 |
| II | 3.26±1.01 | | 1 (10.0) | |
| III | 3.44±0.98 | | 6 (17.1) | |
| Lymphovascular
invasion |
| Negative | 3.41±0.91 | 0.788 | 3 (8.3) | 0.205 |
| Positive | 3.48±1.13 | | 4 (22.2) | |
| ER status |
| Negative | 3.47±0.94 | 0.768 | 6 (20.7) | 0.105 |
| Positive | 3.39±1.03 | | 1 (3.8) | |
| PR status |
| Negative | 3.53±0.83 | 0.428 | 6 (19.4) | 0.122 |
| Positive | 3.32±1.13 | | 1 (4.2) | |
| HER2
overexpression |
| Negative | 3.35±0.87 | 0.498 | 5 (19.2) | 0.236 |
| Positive | 3.52±1.09 | | 2 (6.9) | |
| Molecular
subtype |
| Luminal A | 3.42±1.07 | 0.367 | 0 (0.0) | 0.036 |
| Luminal B | 3.23±1.10 | | 1 (6.7) | |
| HER2 | 3.82±1.02 | | 1 (7.1) | |
| Basal-like | 3.28±0.62 | | 5 (35.7) | |
 | Table IVAssociation of methylation levels of
the FHIT gene and Fhit expression with other markers. |
Table IV
Association of methylation levels of
the FHIT gene and Fhit expression with other markers.
| Variables | FHIT
methylation | Fhit
expression |
|---|
|
|
|---|
| Mean levels
(%) | P-value | Negative
expression, n (%) | P-value |
|---|
| Ki-67 |
| <14% | 3.39±0.93 | 0.759 | 1 (4.5) | 0.223 |
| ≥14% | 3.47±1.02 | | 6 (18.2) | |
| Bcl-2 |
| Negative | 3.46±1.01 | 0.545 | 7 (14.0) | 1.000 |
| Positive | 3.18±0.48 | | 0 (0.0) | |
| p53 |
| Negative | 3.50±0.98 | 0.802 | 1 (10.0) | 1.000 |
| Positive | 3.42±0.98 | | 6 (13.3) | |
| p16 |
| Negative | 3.45±0.98 | 0.765 | 4 (12.1) | 0.677 |
| Positive | 3.36±1.08 | | 3 (17.6) | |
| EGFR |
| Negative | 3.43±0.98 | 0.858 | 1 (3.1) | 0.036 |
| Positive | 3.39±0.98 | | 5 (22.7) | |
| Necrosis |
| Negative | 3.52±1.05 | 0.715 | 2 (6.9) | 0.210 |
| Positive | 3.41±1.00 | | 4 (20.0) | |
| Intratumoral
inflammation |
| Negative | 2.89±0.40 | 0.037 | 0 (0.0) | 0.327 |
| Positive | 3.62±1.06 | | 6 (15.0) | |
| Peritumoral
inflammation |
| Negative | 2.86±0.40 | 0.083 | 0 (0.0) | 1.000 |
| Positive | 3.58±1.05 | | 6 (13.6) | |
We correlated Fhit expression with
clinicopathological features and other markers. The results showed
that loss of Fhit expression was associated with large tumor size,
basal-like subtype and positive expression of EGFR (P=0.003,
P=0.026 and P=0.024, respectively) (Tables III and IV). Loss of Fhit expression in
EGFR-positive breast cancer correlated with tumor size >2 cm
(OR=5.33, 95% CI, 1.92–14.79, P=0.003). This was observed in
ER-negative as well as in PR-negative cases (OR=3.14, 95% CI,
1.71–5.79, P=0.005 and OR=3.00, 95% CI, 1.70–5.28, P=0.005,
respectively).
We stratified all cases by the HER2 status, and
evaluated the relationship between loss of Fhit expression with
clinicopathological features and other markers of breast cancer
based on the HER2 status (Table V).
Associations varied somewhat by HER2 status. For HER2-negative
cases, loss of Fhit expression was significantly associated with
tumor size, ER status and Ki-67 labeling index (P=0.005, P=0.042
and P=0.042, respectively), whereas no significant correlation was
found in HER2-positive cases.
 | Table VAssociation of loss of Fhit
expression with clinicopathological features in HER2-positive and
-negative breast cancer patients. |
Table V
Association of loss of Fhit
expression with clinicopathological features in HER2-positive and
-negative breast cancer patients.
| HER2-positive | HER2-negative |
|---|
|
|
|
|---|
| Loss of Fhit
expression, n (%) | OR (95% CI) | P-value | Loss of Fhit
expression, n (%) | OR (95% CI) | P-value |
|---|
| Stage | | | | | | |
| I | 1 (5.9) | | 1.000 | 0 (0.0) | | 0.081 |
| II | 1 (14.3) | | | 4 (33.3) | | |
| III | 0 (0.0) | | | 0 (0.0) | | |
| IV | 0 (0.0) | | | 1 (100.0) | | |
| Tumor size
(cm) | | | | | | |
| ≤2 | 1 (5.0) | 0.711
(0.174–2.903) | 0.532 | 0 (0.0) | | 0.005 |
| >2 | 1 (11.1) | 1.688
(0.375–7.585) | | 5 (50.0) | 4.000
(1.872–8.545) | |
| Nodal
involvement | | | | | | |
| Negative | 2 (9.5) | 1.368
(1.084–1.728) | 1.000 | 3 (20.0) | 1.313
(0.667–2.581) | 0.626 |
| Positive | 0 (0.0) | | | 1 (10.0) | 0.583
(0.100–3.417) | |
| Distant
metastasis | | | | | | |
| Negative | 2 (7.1) | 1.038
(0.964–1.118) | 1.000 | 4 (16.0) | 0.800
(0.516–1.240) | 0.192 |
| Positive | 0 (0.0) | | | 1 (100.0) | | |
| Histological
grade | | | | | | |
| I | 0 (0.0) | | 1.000 | 0 (0.0) | | 0.061 |
| II | 1 (16.7) | | | 0 (0.0) | | |
| III | 1 (5.0) | | | 5 (33.3) | | |
| Lymphovascular
invasion | | | | | | |
| Negative | 1 (5.0) | 0.711
(0.174–2.903) | 0.532 | 2 (12.5) | 0.571
(0.188–1.736) | 0.312 |
| Positive | 1 (11.1) | 1.688
(0.375–7.585) | | 3 (33.3) | 2.000
(0.751–5.329) | |
| ER status | | | | | | |
| Negative | 1 (6.7) | 0.964
(0.230–4.041) | 1.000 | 5 (35.7) | 2.333
(1.424–3.823) | 0.042 |
| Positive | 1 (7.1) | 1.038
(0.246–4.384) | | 0 (0.0) | | |
| PR status | | | | | | |
| Negative | 1 (6.3) | 0.900
(0.216–3.747) | 1.000 | 5 (33.3) | 2.100
(1.341–3.289) | 0.053 |
| Positive | 1 (7.7) | 1.125
(0.264–4.790) | | 0 (0.0) | | |
| Ki-67 | | | | | | |
| <14% | 1 (10.0) | 1.500
(0.340–6.623) | 1.000 | 0 (0.0) | | 0.042 |
| ≥14% | 1 (5.3) | 0.750
(0.183–3.076) | | 5 (35.7) | 2.333
(1.424–3.823) | |
| Bcl-2 | | | | | | |
| Negative | 2 (7.4) | 1.080
(0.971–1.202) | 1.000 | 5 (21.7) | 1.167
(0.980–1.389) | 1.000 |
| Positive | 0 (0.0) | | | 0 (0.0) | | |
| p53 | | | | | | |
| Negative | 0 (0.0) | | 1.000 | 1 (16.7) | 0.840
(0.124–5.688) | 1.000 |
| Positive | 2 (8.0) | 1.174
(1.003–1.374) | | 4 (20.0) | 1.050
(0.637–1.730) | |
| p16 | | | | | | |
| Negative | 2 (10.5) | 1.588
(1.189–2.121) | 0.532 | 2 (14.3) | 0.533
(0.176–1.619) | 0.280 |
| Positive | 0 (0.0) | | | 3 (42.9) | 2.400
(0.791–7.284) | |
| EGFR | | | | | | |
| Negative | 1 (4.8) | 1.350
(1.080–1.688) | 1.000 | 0 (0.0) | | 0.053 |
| Positive | 0 (0.0) | | | 5 (33.3) | 2.100
(1.341–3.289) | |
Discussion
DNA hypermethylation is one of major epigenetic
modifications and plays an important role in silencing tumor
suppressor genes in all types of cancer, including breast cancer
(2). The FHIT gene is a
candidate tumor suppressor, and it has been postulated that the
FHIT gene is involved in breast carcinogenesis (6,7,9,10).
5′CpG island methylation of the FHIT gene has been
investigated in breast cancer and it was demonstrated that
methylation of the FHIT gene is a frequent event in breast
cancer (11,14). While qualitative analysis,
specifically methylation-specific polymerase chain reaction, has
been used in previous studies, the quantitative analysis of
methylation has rarely been studied. We quantitatively analyzed the
promoter methylation status of the FHIT gene in primary
breast cancer by using pyrosequencing. In the present study, 96.7%
of the breast cancer cases had an aberrant methylation of the
FHIT gene and the result reveals that methylation is one of
the major mechanisms in the regulation of the FHIT gene.
Several studies showed that loss of Fhit expression
was significantly correlated with methylation status of the
FHIT gene (11,20). On the other hand, Yang et
al(22) did not find a
significant correlation between the FHIT gene methylation
and Fhit expression, which is consistent with our results. There
are several mechanisms besides hypermethylation by which reduced
Fhit expression can occur, such as LOH (8,9,22),
homozygous deletions (9,10), abnormal transcripts (12), and reduced mRNA expression (13). In addition, Syeed et
al(21) showed the mutations of
the FHIT gene in breast cancer that lead to the reduced
expression level of Fhit. We did not find any association between
the FHIT gene methylation and Fhit expression. Another
possible complicated mechanism for loss of Fhit expression in
breast cancer has been reported (6), however, it is not included in the
present study.
The HER2 gene encoding a transmembrane glycoprotein
that is a member of the EGFR family, is amplified and overexpressed
in 20–30% of invasive breast carcinomas (26). A recent study showed an association
between HER2 status and DNA methylation profiles of breast cancer,
and suggested that differences in DNA methylation profile reflect
the higher aggressiveness of HER2-positive breast cancer (23). However, in the present study, we did
not find an association between methylation status of the
FHIT gene and HER2 status. Several studies have shown that
downregulation of Fhit protein levels not due to promoter
hypermethylation but to Fhit protein post-translational
modification (24,27), and they demonstrated the association
between Fhit expression and HER2 status in breast cancer. Bianchi
et al analyzed the impact of Fhit downregulation due to EGFR
family activation in human breast tumor development and progression
(29), and showed that Fhit protein
levels can be regulated by Fhit proteasome degradation mediated by
EGF-dependent activation of EGFR family members, including HER2
(27). In the present study, we
showed that Fhit expression does not correlate with HER2
overexpression. However, when stratifying the cases by HER2 status,
loss of Fhit expression was associated with poor prognostic markers
such as large tumor size, negative ER status and high Ki-67
labeling index. Our results suggest cross-regulation between HER2
overexpression and loss of Fhit expression in breast cancer, which
is relevant to the results of a previous study (29).
It has been postulated that aberrant Fhit expression
is associated with pathogenesis and prognostic markers in breast
cancer (12,14,30–32).
Research on the FHIT gene has demonstrated that Fhit
interacts with different proteins through different pathways
(6). Although the exact
clinicopathological significance of loss of Fhit expression in
breast cancer is not known, several studies have indicated that it
is associated with increased tumor size (30), increased histological grade, ER
negativity, increased tumor proliferation index, increased p53
expression, increased expression of Ki-67 and decreased expression
of Bcl-2 (31). In our study, we
correlated the expression of Fhit with clinocopathological
characteristics as well as other prognostic markers. Loss of Fhit
expression was correlated with poor prognostic markers such as
large tumor size, basal-like subtype and positive expression of
EGFR.
Estrogen has been implicated in the etiology of
breast cancer (33) and hormone
receptor (HR) status, defined as ER and/or PR status, have been
used as prognostic markers in breast cancer. Recent advances in
molecular profiling and DNA methylation analysis have suggested
DNA-based surrogate markers for expression status (34). Methylation in breast cancer has been
linked to the hormone regulation. Previous studies showed that gene
expression profiles were different according to the HR status of
breast cancer (35,36), and other studies suggested that DNA
methylation profiles of breast cancer are associated with HR
biology (29,37). However, in the present study, we did
not find an association between methylation status of the
FHIT gene and HR status. When stratifying the cases by HR
status, there was no association between methylation status of the
FHIT gene and clinicopathological features, whereas loss of
Fhit expression was associated with large tumor size in ER-negative
as well as PR-negative cases.
To the best of our knowledge, the present study is
the first report that quantitatively analyzed the promoter
methylation status of the FHIT gene by using pyrosequencing
in primary breast cancer and correlated the quantitative data on
the levels of the FHIT gene methylation with its protein
expression. Pyrosequencing analysis can provide reproducible
measurements of average methylation levels in sequential CpG sites,
thus, this method is rapid and accurate (38). On the other hand, limitations of our
study include relatively small number of sample size and absence of
control group, including normal or benign breast tissue. In
addition, we did not perform survival analysis due to short
follow-up period. Further studies in larger cohorts with longer
follow-up are required to clarify the predictive and prognostic
value of the FHIT gene methylation and Fhit expression in
breast cancer.
In conclusion, our study revealed that loss of Fhit
expression in breast cancer is associated with poor prognostic
features, although there is no significant association between the
FHIT gene methylation and Fhit expression. We found that in
HER2-negative breast cancer, loss of Fhit expression was associated
with poor prognostic features. These results support the
possibility of potential complementation between HER2 and the Fhit
pathway (29). The clinical
significance of our findings requires further evaluation in larger
cohorts with longer follow-up.
Acknowledgements
The present study was supported by research grants
from the Catholic University of Daegu in 2011.
References
|
1
|
Dworkin AM, Huang TH and Toland AE:
Epigenetic alterations in the breast: implications for breast
cancer detection, prognosis and treatment. Semin Cancer Biol.
19:165–171. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hinshelwood RA and Clark SJ: Breast cancer
epigenetics: normal human mammary epithelial cells as a model
system. J Mol Med. 86:1315–1328. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Widschwendter M and Jones PA: DNA
methylation and breast carcinogenesis. Oncogene. 21:5462–5482.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pogribny IP and Beland FA: DNA
hypomethylation in the origin and pathogenesis of human diseases.
Cell Mil Life Sci. 66:2249–2261. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Balch C, Montgomery JS, Paik HI, et al:
New anti-cancer strategies: epigenetic therapies and biomarkers.
Front Biosci. 10:1897–1931. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wali A: FHIT: Doubts are clear now. Sci
World J. 10:1142–1151. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ingvarsson S: FHIT alterations in breast
cancer. Semin Cancer Biol. 11:361–366. 2001. View Article : Google Scholar
|
|
8
|
Ingvarsson S, Sigbjornsdottir BI, Huiping
C, Jonasson JG and Agnarsson BA: Alterations of the FHIT gene in
breast cancer: association with tumour progression and patient
survival. Cancer Detect Prev. 25:292–298. 2001.PubMed/NCBI
|
|
9
|
Ahmadian M, Wistuba II, Fong KM, et al:
Analysis of the FHIT gene and FRA3B region in sporadic breast
cancer, preneoplastic lesions, and familial breast cancer probands.
Cancer Res. 57:3664–3668. 1997.PubMed/NCBI
|
|
10
|
Negrini M, Monaco G, Vorchovsky I, et al:
The FHIT gene at 3p14.2 is abnormal in breast carcinomas. Cancer
Res. 56:3173–3179. 1996.PubMed/NCBI
|
|
11
|
Zöchbauer-Müller S, Fong KM, Maitra A, et
al: 5′ CpG island methylation of the FHIT gene is correlated
with loss of gene expression in lung and breast cancer. Cancer Res.
61:3581–3585. 2001.
|
|
12
|
Hayashi S, Tanimoto K, Hajiro-Nakanishi K,
et al: Abnormal FHIT transcripts in human breast carcinomas: a
clinicopathological and epidemiological analysis of 61 Japanese
cases. Cancer Res. 57:1981–1985. 1997.PubMed/NCBI
|
|
13
|
Ingvarsson S, Agnarsson BA,
Sigbjornsdottir BI, et al: Reduced Fhit expression in sporadic and
BRCA2-linked breast carcinomas. Cancer Res. 59:2682–2689.
1999.PubMed/NCBI
|
|
14
|
Gatalica Z, Lele SM, Rampy BA and Norris
BA: The expression of Fhit protein is related inversely to disease
progression in patients with breast carcinoma. Cancer.
88:1378–1383. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Naqvi RA, Hussain A, Raish M, et al:
Specific 5′ CpG island methylation signatures of FHIT and
p16 genes and their potential diagnostic relevance in Indian
breast cancer patients. DNA Cell Biol. 27:517–525. 2008.
|
|
16
|
Maruyama R, Toyooka S, Toyooka KO, et al:
Aberrant promoter methylation profile of bladder cancer and its
relationship to clinicopathological features. Cancer Res.
61:8659–8663. 2001.PubMed/NCBI
|
|
17
|
Maruyama R, Toyooka S, Toyooka KO, et al:
Aberrant promoter methylation profile of prostate cancers and its
relationship to clinicopathological features. Clin Cancer Res.
8:514–519. 2002.PubMed/NCBI
|
|
18
|
Virmani AK, Muller C, Rathi A,
Zoechbauer-Mueller S, Mathis M and Gazdar AF: Aberrant methylation
during cervical carcinogenesis. Clin Cancer Res. 7:584–589.
2001.PubMed/NCBI
|
|
19
|
Kim H, Kwon YM, Kim JS, et al:
Tumor-specific methylation in bronchial lavage for early detection
on non-small cell lung cancer. J Clin Oncol. 22:2363–2370. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Raish M, Dhillon VS, Ahmad A, et al:
Promoter hypermethylation in tumor suppressing genes p16 and FHIT
and their relationship with estrogen receptor and progesterone
receptor status in breast cancer patients from Northern India.
Trans Oncol. 2:264–270. 2009. View Article : Google Scholar
|
|
21
|
Syeed N, Husain SA, Sameer AS, Chowdhri NA
and Siddiqi MA: Mutational and promoter hypermethylation status of
FHIT gene in breast cancer patients of Kashmir. Mutation
Res. 707:1–8. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang Q, Nakamura M, Nakamura Y, et al:
Two-hit inactivation of FHIT by loss of heterozygosity and
hypermethylation in breast cancer. Clin Cancer Res. 8:2890–2893.
2002.
|
|
23
|
Fiegl H, Millinger S, Goebel G, et al:
Breast cancer DNA methylation profiles in cancer cells and tumor
stroma: association with HER-2/neu status in primary breast cancer.
Cancer Res. 66:29–33. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bianchi F, Magnifico A, Olgiati C, et al:
FHIT-proteasome degradation caused by mitogenic stimulation of the
EGF receptor family in cancer cells. Proc Natl Acad Sci USA.
103:18981–18986. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Goldhirsch A, Wood WC, Coates AS, et al:
Strategies for subtypes - dealing with the diversity of breast
cancer: highlights of the St. Gallen International Expert Consensus
on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol.
22:1736–1747. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Menard S, Tagliabue E, Campiglio M and
Pupa SM: Role of HER2 gene overexpression in breast carcinoma. J
Cell Physiol. 182:150–162. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pekarsky Y, Garrison PN, Palamarchuk A, et
al: Fhit is a physiological target of the protein kinase Src. Proc
Natl Acad Sci USA. 101:3775–3779. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Campan M, Weisenberger DJ and Laird PW:
DNA methylation profiles of female steroid hormone-driven human
malignancies. Curr Top Microbiol Immunol. 310:141–178.
2006.PubMed/NCBI
|
|
29
|
Bianchi F, Tagliabue E, Ménard S and
Campiglio M: Fhit expression protects against HER2-driven breast
tumor development. Cell Cycle. 6:643–646. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Campiglio M, Pekarsky Y, Menard S,
Tagliabue E, Pilotti S and Croce CM: FHIT loss of function in human
primary breast cancer correlates with advanced stage of the
disease. Cancer Res. 59:3866–3869. 1999.PubMed/NCBI
|
|
31
|
Yang Q, Yoshimura G, Suzuma T, et al:
Clinicopathological significance of fragile histidine triad
transcription protein expression in breast carcinoma. Clin Cancer
Res. 7:3869–3873. 2001.PubMed/NCBI
|
|
32
|
Arun B, Kilic G, Yen C, et al: Loss of
FHIT expression in breast cancer is correlated with poor prognostic
markers. Cancer Epidemiol Biomarkers Prev. 14:1681–1685. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Key TJ and Pike MC: The role of oestrogens
and progestagens in the epidemiology and prevention of breast
cancer. Eur J Cancer Clin Oncol. 24:29–43. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
35
|
Creighton CJ, Kent Osborne C, van de
Vijver MJ, et al: Molecular profiles of progesterone receptor loss
in human breast tumors. Breast Cancer Res Treat. 114:287–299. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li L, Lee KM, Ha W, et al: Estrogen and
progesterone receptor status affect genome-wide DNA methylation
profile in breast cancer. Hum Mol Genet. 19:4273–4277. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Widschwendter M, Siegmund KD, Muller HM,
et al: Association of breast cancer DNA methylation profiles with
hormone receptor status and response to tamoxifen. Cancer Res.
64:3807–3813. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tost J and Gut IG: Analysis of
gene-specific DNA methylation patterns by pyrosequencing
technology. Methods Mol Biol. 373:89–102. 2007.PubMed/NCBI
|