Introduction
Hepatocellular carcinoma (HCC) is one of the most
common cancers and the third leading cause of cancer-related
mortality worldwide. Its incidence and mortality rate in Asia are
high and have also rapidly increased in the United States (1–3). Due
to its characteristic fast growth and early metastasis, the overall
3-year survival rate of HCC patients after resection is 35–62% and
the 5-year survival rate is 17–50%. Without treatment, the survival
rate is <10%; with recurrence, it is 70% (4–7). HCC
is one of the most aggressive and intractable malignant tumors and,
unfortunately, the mechanisms contributing to its carcinogenesis
and progression are still poorly understood.
Chromosomal instability (CIN) leading to aneuploidy
is a characteristic feature of various types of cancer (8–11).
Kinetochore, a structure composed of proteins such as centromere
protein (CENP)-A, CENP-B, CENP-C, CENP-E, CENP-F, CENP-H and
CENP-I/MIS6 (12–14), is assembled on centromeres on which
spindle microtubules attach during mitosis to pull apart the sister
chromatids. Normal expression of kinetochore is vital to mitosis
(15); however, disruption of core
centromere proteins may induce an increase in CIN and tumorigenesis
(16). CENP-H, with an apparent
molecular mass of 33 kDa, was first isolated from mouse and human
CENP-H protein was shown to localize in the inner plate with CENP-A
and CENP-C (17,18). It is a fundamental component of the
active centromere complex. Accumulating evidence indicates that
CENP-H is frequently upregulated in cancers, and its overexpression
and mislocation are associated with the development of aneuploidy,
a hallmark of malignancy. Studies have shown that CENP-H is
significantly associated with human nasopharyngeal carcinoma,
esophageal carcinoma, tongue cancer, breast cancer, non-small cell
lung cancer and gastric cancer (19–22).
Increased expression of CENP-H increases the proliferative activity
of human oral squamous cells (23),
and depletion of CENP-H through RNA interference produces severe
mitotic phenotypes such as misaligned chromosomes and multipolar
spindles (24). Research has also
revealed that CENP-H is closely associated with colorectal cancer
and that its overexpression induces aneuploidy (25).
In the present study, we first confirmed the
differential expression of CENP-H in 10 pairs of HCC samples and
corresponding adjacent non-cancerous samples using reverse
transcription-polymerase chain reaction (RT-PCR), real-time
quantitative PCR (qPCR) and western blotting. We used
immunohistochemistry to compare the expression of CENP-H with
clinicopathological features and overall survival of patients with
HCC. Finally, an immunoflurorescence assay was used to detect and
localize the expression of CENP-H in HCC cells.
Materials and methods
Patient specimens
From January 2009 to December 2010, after obtaining
approval from the Human Subjects Committee of the First Affiliated
Hospital of Xi’an Jiaotong University and informed consent of the
patients, HCC tissues (including adequately sized tumor tissue
samples and the corresponding adjacent non-cancerous tissue samples
obtained 5–10 cm from the tumor) were obtained from patients
undergoing liver resection for hepatic cancer at the Department of
Hepatobiliary Surgery, First Affiliated Hospital of Xi’an Jiaotong
University. All tissues were obtained within 30 min following
excision from the patient’s body. A part of the sample was cut into
small pieces, snap-frozen in liquid nitrogen immediately and stored
at −80°C to prepare for analysis of CENP-H gene expression, the
other part was fixed with 4% formalin and paraffin-embedded for
histological studies. Of the 60 patients, there were 49 men and 11
women. Median age at the time of surgery was 52 years (range, 25–74
years). The histological type in all 60 patients was hepatocellular
carcinoma, diagnosed according to the World Health Organization
histological classification of tumors of the liver and intrahepatic
bile ducts (2000)(26). The disease
stages of all the patients were classified or reclassified
according to the classification of the International Union Against
Cancer (27) and the Chinese
Anti-Cancer Association, as outlined in Table I. Clinical information of the
samples is described in detail in Table II. Patients with preoperative
anticancer treatment or with evidence of other malignancies were
excluded from the study. All patients were followed up, and the
median duration of follow-up was 20 months (range, 2–39
months).
 | Table IThe Chinese 2001 staging system. |
Table I
The Chinese 2001 staging system.
| Stage | Description |
|---|
| Ia | Signal tumor, the
maximum diameter ≤3 cm; no tumor thrombus; no intraperitoneal lymph
node or distant metastasis; Child A. |
| Ib | Signal or two tumors
located in hemihepatis, the sum of maximum diameters ≤5 cm; no
thrombus; no intraperitoneal lymph node or distant metastasis;
Child A. |
| IIa | Signal or two tumors
located in hemihepatis, the sum of maximum diameters ≤10 cm, or the
sum of maximum diameters of two tumors ≤5 cm located in the left
and right half of liver respectively; no thrombus; no
intraperitoneal lymph node or distant metastasis; Child A. |
| IIb | Signal or two
tumors located in hemihepatis, the sum of maximum diameters >10
cm, or the sum of maximum diameters of two tumors >5 cm located
in the left and right half of liver respectively, or more than two
tumors; no thrombus; no intraperitoneal lymph node or distant
metastasis; Child A.
Regardless of the tumors; there is thrombus of branch of portal
vein, hepatic vein, or bile duct; no intraperitoneal lymph node or
distant metastasis; Child A.
Regardless of the tumors; no thrombus; no intraperitoneal lymph
node or distant metastasis; Child B. |
| IIIa | Regardless of the
tumors; there is one of the next two cases: thrombus of main portal
vein or inferior vena cava, metastasis of intraperitoneal lymph
node or distant metastasis; Child A/B. |
| IIIb | Whatever the
others; Child C. |
 | Table IIAssociations between CENP-H protein
expression and the clinicopathological features of HCC cases. |
Table II
Associations between CENP-H protein
expression and the clinicopathological features of HCC cases.
| | CENP-H-positive
cases, n (%) | |
|---|
| |
| |
|---|
|
Characteristics | Cases N=60 | Low (33) | High (27) | P-value |
|---|
| Age (years) | | | | 1.000 |
| >60 | 9 | 5 (55.6) | 4 (44.4) | |
| ≤60 | 51 | 28 (54.9) | 23 (45.1) | |
| Gender | | | | 0.519 |
| Male | 49 | 26 (53.1) | 23 (46.9) | |
| Female | 11 | 7 (63.6) | 4 (36.4) | |
| Histopathological
grade | | | | 0.001 |
| I–II | 37 | 27 (73.0) | 10 (27.0) | |
| III–IV | 23 | 6 (26.1) | 17 (73.9) | |
| TNM clinical
stage | | | | 0.002 |
| I–II | 34 | 25 (73.5) | 9 (26.5) | |
| III–IV | 26 | 8 (30.8) | 18 (69.2) | |
| Lymph node
metastasis | | | | 0.739 |
| Negative | 49 | 26 (53.1) | 23 (46.9) | |
| Positive | 11 | 7 (63.6) | 4 (36.4) | |
| Distant
metastasis | | | | 0.085 |
| Negative | 57 | 33 (57.9) | 24 (42.1) | |
| Positive | 3 | 0 (0) | 3 (100) | |
| Tumor size
(cm) | | | | 0.032 |
| ≤5 | 23 | 17 (73.9) | 6 (26.1) | |
| >5 | 37 | 16 (43.2) | 21 (56.8) | |
| Venous
invasion | | | | 0.129 |
| Negative | 29 | 19 (65.5) | 10 (34.5) | |
| Positive | 31 | 14 (45.2) | 17 (54.8) | |
| Chinese clinical
stage | | | | 0.008 |
| I | 12 | 9 (75) | 3 (25) | |
| II | 29 | 19 (65.5) | 10 (34.5) | |
| III | 19 | 5 (26.3) | 14 (73.7) | |
RNA isolation, RT-PCR and real-time
qPCR
Total RNA was extracted from HCC tumors and the
corresponding non-tumor tissues with TRIzol reagent (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer’s instructions.
First-strand cDNA was synthesized from total RNA with
PrimeScript® RT Master Mix [Takara Biotechnology
(Dalian) Co., Ltd., Dalian, China].
The PCR primers sets were as follows: i) CENP-H,
forward 5′-CAGTCTAGTGTGCTCATGGAT-3′ and reverse 5′-TCCA
TCTGTAGGTTTTGTCG-3′; ii) glyceraldehyde-3-phosphate dehydrogenase
(GAPDH), forward 5′-CAAGCTCATTTCC TGGTATGAC-3′ and reverse
5′-CAGTGAGGGTCTCTCTC TTCCT-3′. The RT-PCR conditions included an
initial denaturation step for 5 min at 94°C followed by 30 cycles
of amplification: 94°C for 30 sec, 55°C for 30 sec and 72°C for 30
sec. After the last cycle, a final extension was performed at 72°C
for 10 min, and the RT-PCR products were separated by
electrophoresis on 1.5% agarose gels.
Real-time quantitative qPCR of CENP-H cDNA was
carried out (Bio-Rad Laboratories, Hercules, CA, USA) with
SYBR® Priemix Ex Taq™ II (Takara Biotechnology). The
cycling conditions were as follows: initial denaturation at 95°C
for 30 sec, and then 40 cycles of denaturation at 95°C for 5 sec,
annealing and elongation at 60°C for 30 sec. Bio-Rad CFX Manager
2.1 software was used for analysis of qPCR. The housekeeping gene
GAPDH was used as an internal control for both RT-PCR and real-time
qPCR. The optimization and synthesis of primers was carried out by
Sangon Biotech Co., Inc. (Shanghai, China). Serial dilutions of the
template cDNA were made for reactions to optimize the PCR products
within the linear range.
Protein extraction and western
blotting
Frozen tissue samples were first solubilized 1 h in
lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China)
in ice using homogenizers, then centrifugated (14,000 × g;
Eppendorf, Germany) 30 min at 4°C. After denaturation, equal
amounts of supernatant proteins were separated electrophoretically
on 10% SDS/polyacrylamide gels (SDS-PAGE) and transferred onto
polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA)
in a tank transfer apparatus (Bio-Rad). The membranes were blocked
with 5% skim milk in Tris-buffered saline with Tween (TBS-T) for 2
h, then incubated with mouse anti-CENP-H antibody diluted 1:200
(Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight at 4°C.
The next day, horseradish peroxidase-conjugated anti-mouse
immunoglobulin G (HRP; Santa Cruz Biotechnology) diluted 1:5,000 in
phosphate-buffered saline (PBS pH 7.4) was used as the secondary
antibody. Antigens on the membrane were detected with enhanced
chemiluminescence horseradish peroxidase (HRP) substrate
(Millipore) according to the manufacturer’s instructions. The
intensity of each band was measured using Image Lab 4.0 (Bio-Rad).
To confirm equal loading, mouse anti-β-actin antibody (Santa Cruz
Biotechnology) diluted 1:1,000 was used as the primary
antibodies.
Immunohistochemistry
CENP-H expression was detected by
immunohistochemistry through a standardized streptavidin-peroxidase
(SP) method. For immunohistochemistry, 4-μm paraffin sections were
adhered to glass slides, which then were deparaffinized in fresh
xylene and rehydrated by passage through an ethanol series.
Following antigen retrieval in citrate buffer (0.01 M, pH 6.0) in a
pressure cooker for 1–2 min after air jetting, slides were first
cooled to room temperate and washed by distilled water and PBS
twice successively, then incubated for 10 min with 0.3% (v/v)
hydrogen peroxide to block activity of endogenous peroxidase. After
blockage with 10% normal goat serum for 10 min, the slides were
incubated with mouse monoclonal anti-CENP-H (1:40 dilution; Santa
Cruz Biotechnology) overnight at 4°C. The next day, the slides were
washed in PBS three times and incubated with biotinylated goat
anti-mouse IgG (1:100 dilution; Beijing Zhongshan Golden Bridge
Biotechnology Co., Ltd., Beijing, China) as the secondary
antibodies for 20 min. After being washed with PBS and incubated
with peroxidase-conjugated streptavidin (1:100 dilution; Beijing
Zhongshan Golden Bridge Biotechnology) for 10 min, each slide was
colored with 100 μl 0.02% 3,3′-diaminobenzidine (DAB) (Sigma, St.
Louis, MO, USA). Finally, all the paraffin sections were rinsed by
running water to terminate coloration, counterstained with
hematoxylin, differentiated through 0.1% hydrochloric acid (HCl),
washed with PBS, dehydrated in graded ethanol and coverslipped with
neutral gum. All the incubations were carried out in a humidified
chamber, and non-immune mouse serum replaced the primary antibody
as the negative control. The slides were read and scored by two
independent experiments under a microscope (Olympus Optical Co.,
Ltd., Tokyo, Japan). According to Guo et al(21), the extent of staining and the
proportion of stained cells were used as criteria of evaluation.
First, according to the intensity of staining, the cells were
scored on a scale of 1, no staining; 2, weak staining (light
yellow); 3, moderate staining (yellowish brown); 4, strong staining
(brown). Second, according to the percentage of positively stained
tumor cells, the proportion of CENP-H-positive cells varied from 0
to 100%: <5% of the cells, 1; 6–35% of the cells, 2; 36–70% of
the cells, 3; >71% of the cells, 4. If the final score,
calculated by multiplying the above two scores, was ≥4, the tumor
was considered to have high expression; otherwise, the tumor was
considered to have low expression.
Cell culture
The liver cancer cell line Hep3B obtained from the
Transform Medical Center of Xi’an Jiaotong University was cultured
in Dulbecco’s modified Eagle’s medium (DMEM)(Invitrogen)
supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA)
and 1% penicillin/streptomycin, and incubated at 37°C in a
humidified 5% CO2 atmosphere.
Immunocytofluorescence staining
To observe the expression and localization of CENP-H
in Hep3B cells, 1.6×105 cells were fixed by acetone, and
incubated with mouse monoclonal anti-CENP-H primary antibody (1:40
dilution; Santa Cruz Biotechnology) overnight after antigen
retrieval and blockage, followed by fluorescein isothiocyanate
(FITC), conjugated rabbit anti-mouse IgG (1:100 dilution; Beijing
Zhongshan Golden Bridge Biotechnology).
4,6-Diamidino-2-phenylindole (DAPI) (Sigma) was used to stain the
nucleus. Fluorescent imaging was observed with a fluorescence
microscope (Leica QFISH; Leica Microsystems, Tokyo, Japan).
Statistical analysis
All statistical analyses were carried out using SPSS
19.0 statistical software (SPSS, Inc., Chicago, IL, USA). CENP-H
protein and mRNA levels were determined by the t-test. Chi-square
and Fisher’s exact tests were used to analyze the relationship
between CENP-H protein expression and the clinicopathological
characteristics. Survival rate was calculated by the Kaplan-Meier
method, and differences were examined by the log-rank test. Factors
found to be associated with survival rate were then selected for a
stepwise Cox’s multivariate proportional hazard model to determine
their prognostic values. P<0.05 was considered to indicate a
statistically significant result.
Results
Upregulation of CENP-H mRNA and protein
levels in HCC tissues
To investigate whether CENP-H is upregulated in the
HCC tissue, western blot analysis was performed in 10 matched pairs
of HCC tissues and corresponding non-cancerous tissues from the
same patient. The result revealed that CENP-H was highly
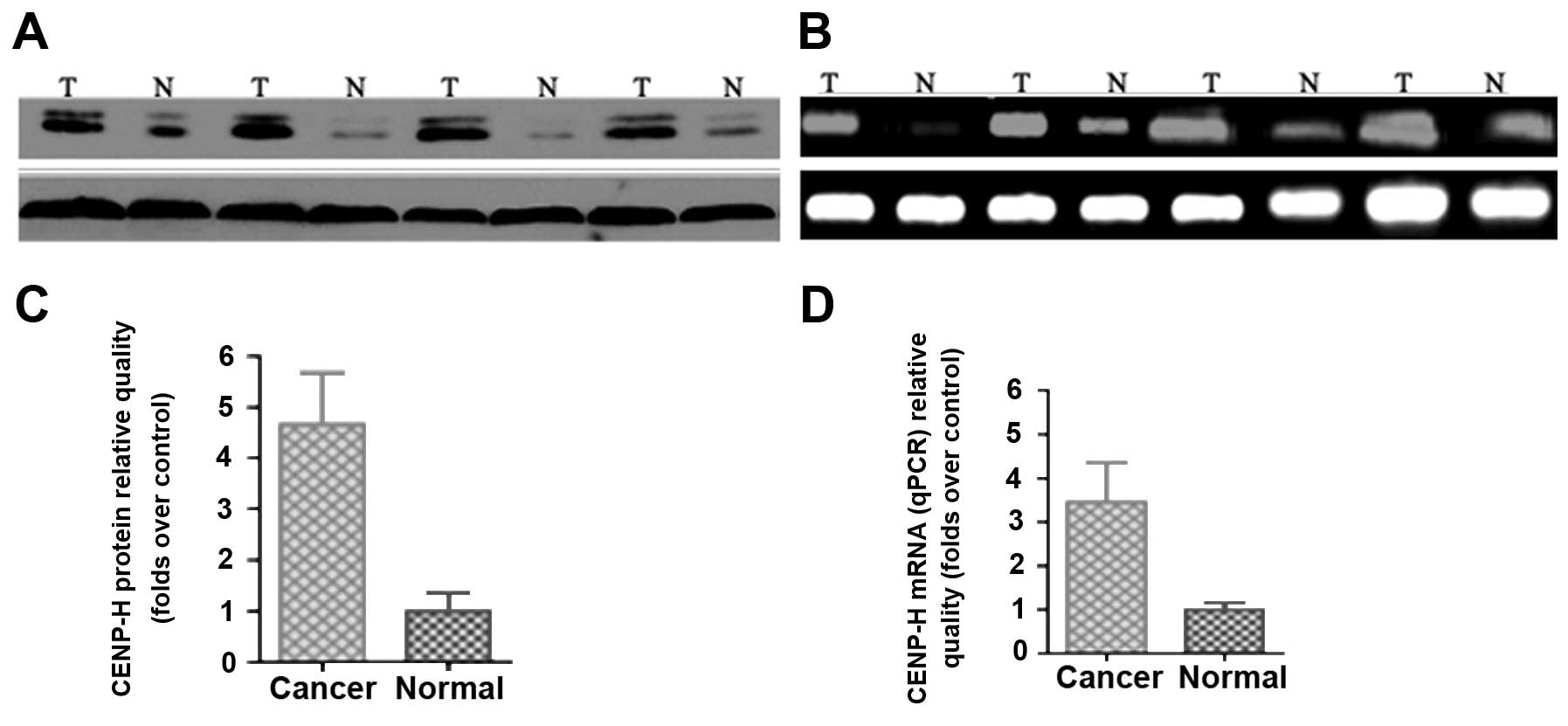
upregulated in all 10 pairs of HCCs (Fig. 1A). The relative expression of CENP-H
protein in tumor samples compared with adjacent non-cancerous
samples varied from 1.75 to 13.13.
To determine whether upregulation of CENP-H is the
result of increased transcription, CENP-H mRNA levels in HCC
tissues and corresponding non-cancerous tissues were examined using
RT-PCR and real-time qPCR. All HCC tissues showed higher expression
of CENP-H mRNA when compared with that in non-cancerous tissues
(Fig. 1C). Therefore, relative mRNA
levels in tumor samples correlated well with the relative protein
levels (Fig. 1B and C). These
results revealed that CENP-H was upregulated at both the mRNA and
protein levels in clinical HCC tissues and that the overexpression
of CENP-H may occur at the transcription level.
Immunohistochemistry of CENP-H in HCC
samples and non-cancerous samples
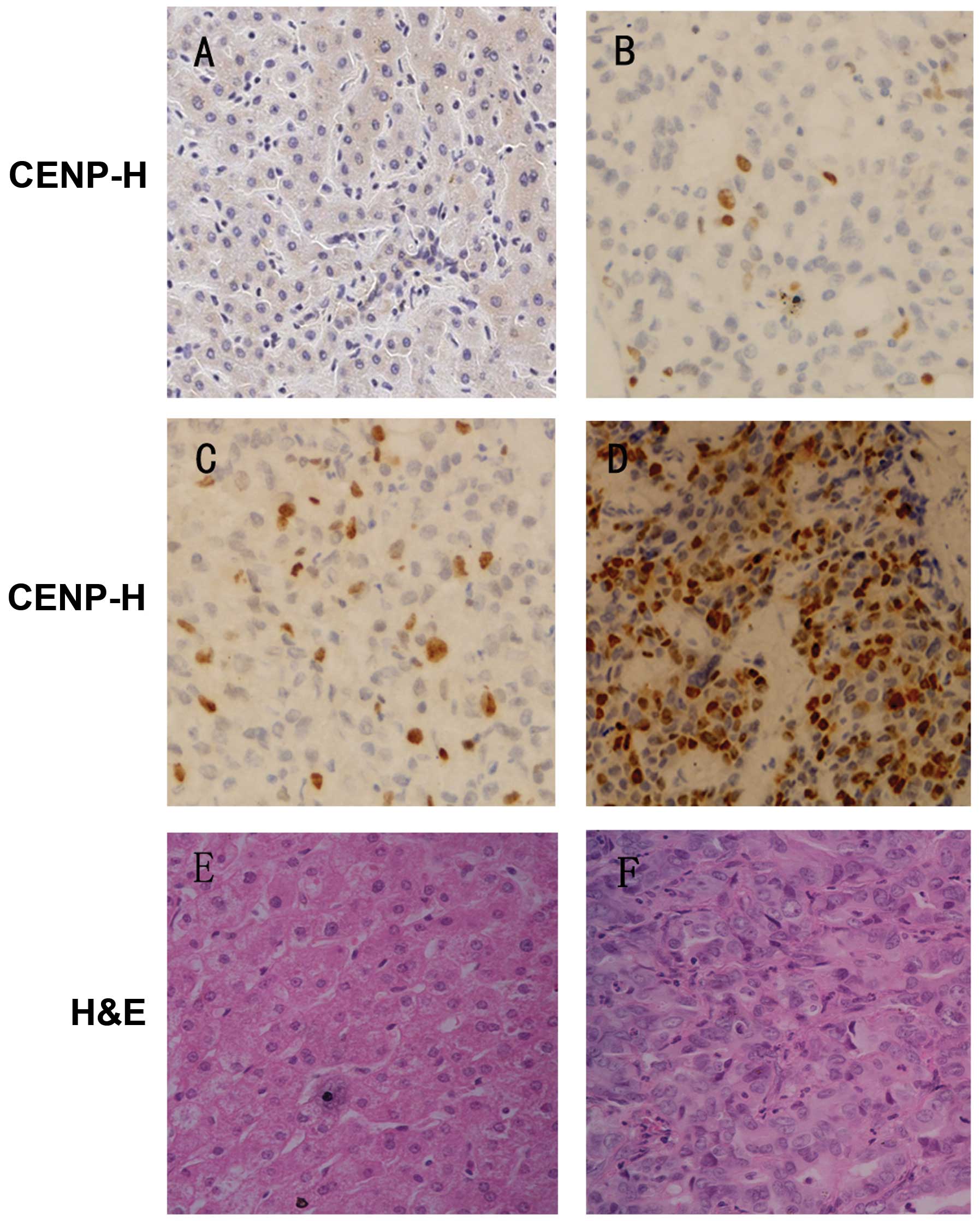
To further study the expression and subcellular
location of CENP-H protein, immunohistochemical analysis was
performed in 60 pairs of paraffin-embedded HCC samples and
corresponding adjacent non-cancerous samples. We observed that
CENP-H expression was significantly higher in HCC tissues (38/60,
63.3%; Fig. 2B–D) than that in the
corresponding adjacent non-cancerous tissues (21/60, 35%; Fig. 2A) (P=0.003). Moreover, the
expression of CENP-H appeared to increase with histological grade
of HCC; well-differentiated HCC samples had the lowest expression
of CENP-H (Fig. 2B), and moderately
differentiated samples had higher expression (Fig. 2C), while the expression of poorly
differentiated samples was the highest (Fig. 2D). CENP-H was predominantly
localized in the nucleus.
Relationship of CENP-H expression with
clinicopathological features
After we performed immunohistochemistry, we
investigated the correlation between the expression of CENP-H
protein and the clinicopathological features in 60 HCC cases. As
shown in Table II, statistical
analysis revealed no statistical correlation between the CENP-H
protein level and age, gender, lymph node metastasis, distant
metastasis and venous invasion. However, the expression of CENP-H
protein was closely related to histological grade, with higher
histological grade associated with a higher frequency of CENP-H
overexpression in HCC patients (P=0.001). In addition, statistics
showed a significant difference in CENP-H expression in patients
categorized according to tumor size (P=0.032). Therefore,
expression of CENP-H protein was statistically correlated to both
TNM stage (P=0.002) and Chinese (P=0.008) clinical stage.
Expression and localization of CENP-H
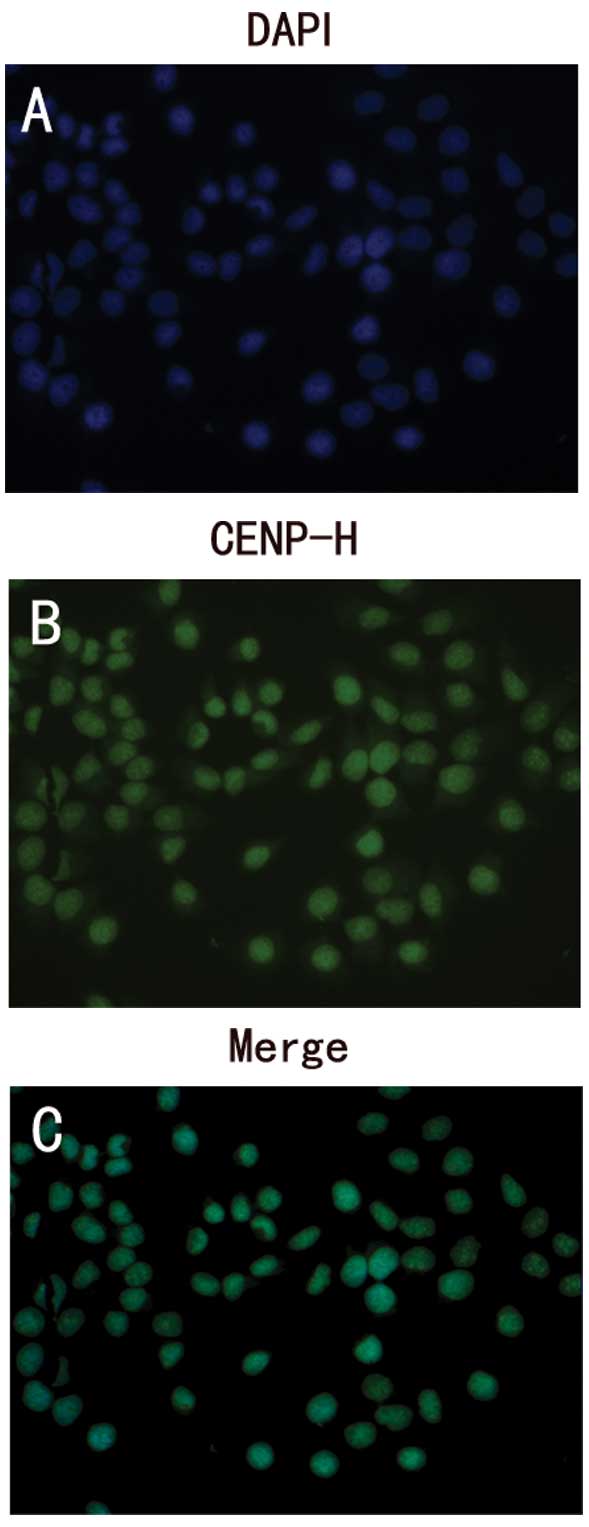
protein in Hep3B cells
To study the expression and localization of CENP-H
protein in HCC cells, Hep3B cells were stained with anti-human
CENP-H monoclonal antibody using an immunofluorescence assay.
Consistent with the results of the immunohistochemical analysis
CENP-H was present in the nucleus of Hep3B cells (Fig. 3).
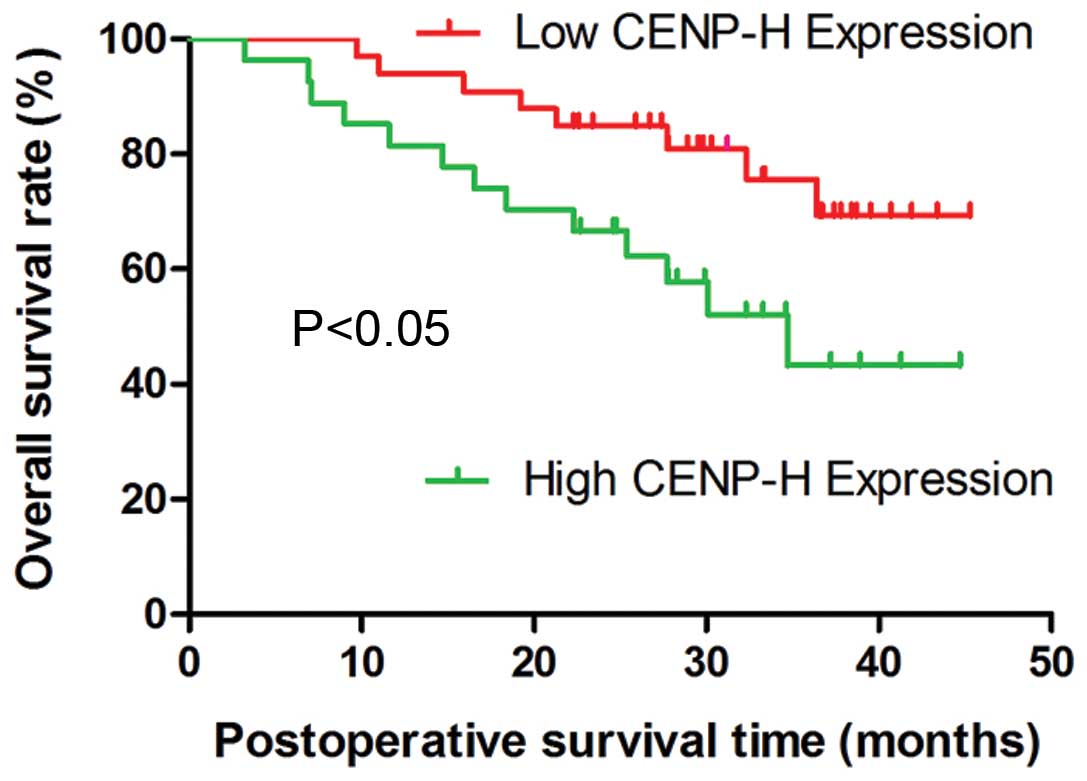
Survival analysis
To determine the effect of classic
clinicopathological characteristics and CENP-H expression on
survival in HCC, we analyzed the cumulative survival of patients
through Kaplan-Meier analysis (Fig.
4). A log-rank test showed that the low CENP-H expression group
had longer survival time, whereas the high CENP-H expression group
was associated with shorter survival (P<0.05).
Multivariate analysis in our study revealed that
venous invasion (P=0.037), advanced TNM stage (P=0.026) and Chinese
stage (P=0.020) were independent prognostic factors for overall
survival time in patients with HCC. Overexpression of CENP-H
protein was also an independent prognostic factor for overall
survival (P=0.046). Tumor size and other clinical parameters were
not independent prognostic factors.
Discussion
HCC is characterized by rapid growth, early
metastasis, a high recurrence rate and poor patient prognosis
(4,5,7). The
present study was designed to detect the expression of CENP-H in
HCC samples to determine its clinical significance for HCC patients
and to better define its potential role as a prognostic factor.
CENP-H, located on the inner centromere plate, has been found to
play a fundamental role in the organization and function of the
active human centromere complex (17,18).
In the present study, we first revealed the differential expression
of CENP-H at both the mRNA and protein levels in HCC tissues and
adjacent non-cancerous tissues. In addition, upregulation of the
CENP-H protein level was consistent with the change in the
corresponding mRNA level, indicating that the overexpression occurs
at the transcriptional level. The results of the
immunohistochemical analysis also confirmed the overexpression of
CENP-H protein in the HCC tissues, consistent with the results of
the western blotting. The present study suggests that high CENP-H
expression is closely correlated with worse histological grade,
advanced TNM and Chinese clinical stage, and larger tumor size. In
contrast, the Chi-square and Fisher’s exact test did not indicate a
positive correlation between CENP-H expression and age, gender,
lymph node metastasis, distant metastasis or venous invasion.
Regarding the localization of CENP-H, immunohistochemistry of
paraffin-embedded tissues and immunofluorescence of Hep3B cells
showed nuclear staining, which is consistent with other reports
(19,25).
The equal distribution of genetic material to the
two daughter cells is the ultimate goal of mitotic cell division,
which is ensured by the correct assembly of kinetochore at each
centromere locus (28,29). Deregulation of centromere proteins
is believed to be related to aneuploidy and carcinogenesis
(10,30). CENP-A has been shown to be
overexpressed and mistargeted in human colon cancer cells (31), which can lead to kinetochore
malfunction (14). Li et
al(32) demonstrated that
CENP-A was upregulated in HCC, the overexpression of which prompted
HCC cell proliferation, while the depletion of CENP-A inhibited HCC
cell growth and induced apoptosis. In addition, CENP-A has been
associated with cancers such as breast cancer and lung
adenocarcinoma (33,34). At the same time, CENP-E, CENP-F, and
inner centromere protein have been demonstrated to contribute to
carcinogenesis (35–38). All observations suggest that
deregulation of centromere proteins is common in the development
and progression of various tumors.
CENP-H has been shown to be overexpressed in several
types of tumors. Tomonaga et al(25) found that CENP-H was upregulated in
both primary colorectal cancer tissues and CIN cell lines and that
overexpression induces aneuploidy and interphase micronuclei, a
characteristic of chromosome missegregation. CENP-H is also
confirmed to be a prognostic marker for nasopharyngeal carcinoma,
tongue cancer, esophageal carcinoma, and human non-small cell lung
cancer (19–22). Fukagawa et al(17) discovered that, in the absence of
CENP-H, chicken DT40 cells were arrested in metaphase and died of
chromosome missegregation. Similarly, Orthaus et al(24) observed that knockdown of CENP-H in
human HEp-2 cells induced aberrant mitotic phenotypes and decreased
the number of living cells. Similarly, the present study found
CENP-H to be upregulated in HCC and its overexpression was
correlated with the overall survival rate. All these findings
illustrate that CENP-H plays an important role in tumorigenesis and
progression.
Centrosome aberration can induce genetic instability
in HCC (39). CENP-A, another inner
centromere protein, has been observed to be amplified in HCC
(32). Similarly, we found that
CENP-H was overexpressed in HCC tissues. According to Tomonaga
et al(25), the deregulation
of CENP-H prevented it from localizing to the centromere by
depleting the factors that recruit it to the centromere. As a
consequence, functional CENP-H protein decreased, and the normal
kinotechore assembly was disrupted. Meanwhile, depletion of
functional CENP-H reduces hBubR1 activation through mislocalizing
kinetochore-associated microtubule motor protein CENP-E (24), which stimulates the kinase activity
of hBubR1 and is required for the establishment and maintenance of
the checkpoint in mitosis. Defects in checkpoints are closely
associated with tumorigenesis (15,29).
All of these findings provide the theoretical basis that
overexpression of CENP-H results in defects in checkpoints and CIN
and eventually plays a key role in the development and progression
of HCC. However, further studies are needed to investigate the
specific mechanisms of overexpression of CENP-H in HCC and how it
contributes to tumorigenesis and development.
In summary, CENP-H is upregulated in HCC and is
closely related to larger tumor size, worse histological grade and
advanced TNM and Chinese stages. Although the mechanisms require
further elucidation, the overexpression of CENP-H may be used as a
novel biomarker for HCC prognosis.
Acknowledgements
We thank the staff of the Transform Medical Center,
Xi’an Jiaotong University, for their technical assistance.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar
|
|
2
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar
|
|
3
|
Altekruse SF, McGlynn KA and Reichman ME:
Hepatocellular carcinoma incidence, mortality, and survival trends
in the United States from 1975 to 2005. J Clin Oncol. 27:1485–1491.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Khorsandi SE and Heaton N: Contemporary
strategies in the management of hepatocellular carcinoma. HPB Surg.
2012:1540562012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Llovet JM, Fuster J and Bruix J:
Intention-to-treat analysis of surgical treatment for early
hepatocellular carcinoma: resection versus transplantation.
Hepatology. 30:1434–1440. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Franco D, Capussotti L, Smadja C, et al:
Resection of hepatocellular carcinomas. Results in 72 European
patients with cirrhosis. Gastroenterology. 98:733–738.
1990.PubMed/NCBI
|
|
7
|
Schwartz M, Roayaie S and Konstadoulakis
M: Strategies for the management of hepatocellular carcinoma. Nat
Clin Pract Oncol. 4:424–432. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rajagopalan H and Lengauer C: Aneuploidy
and cancer. Nature. 432:338–341. 2004. View Article : Google Scholar
|
|
9
|
Ricke RM and van Deursen JM: Aneuploidy in
health, disease, and aging. J Cell Biol. 201:11–21. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cimini D and Degrassi F: Aneuploidy: a
matter of bad connections. Trends Cell Biol. 15:442–451. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bakhoum SF and Compton DA: Chromosomal
instability and cancer: a complex relationship with therapeutic
potential. J Clin Invest. 122:1138–1143. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liang QJ, Lu XF, Cheng XL, et al: The
active expression of CenpB, a constitutive protein in the
centromeres of chromosomes, in breast cancer tissues. Yi Chuan Xue
Bao. 31:236–240. 2004.(In Chinese).
|
|
13
|
Sugata N, Munekata E and Todokoro K:
Characterization of a novel kinetochore protein, CENP-H. J Biol
Chem. 274:27343–27346. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yuen KW, Montpetit B and Hieter P: The
kinetochore and cancer: what’s the connection? Curr Opin Cell Biol.
17:576–582. 2005.
|
|
15
|
Bakhoum SF and Compton DA: Kinetochores
and disease: keeping microtubule dynamics in check! Curr Opin Cell
Biol. 24:64–70. 2012.PubMed/NCBI
|
|
16
|
Kramer A, Neben K and Ho A: Centrosome
replication, genomic instability and cancer. Leukemia. 16:767–775.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fukagawa T, Mikami Y, Nishihashi A, et al:
CENP-H, a constitutive centromere component, is required for
centromere targeting of CENP-C in vertebrate cells. EMBO J.
20:4603–4617. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sugata N, Li S, Earnshaw WC, et al: Human
CENP-H multimers colocalize with CENP-A and CENP-C at active
centromere - kinetochore complexes. Hum Mol Genet. 9:2919–2926.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liao WT, Wang X, Xu LH, et al: Centromere
protein H is a novel prognostic marker for human nonsmall cell lung
cancer progression and overall patient survival. Cancer.
115:1507–1517. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liao WT, Song LB, Zhang HZ, et al:
Centromere protein H is a novel prognostic marker for
nasopharyngeal carcinoma progression and overall patient survival.
Clin Cancer Res. 13:508–514. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo XZ, Zhang G, Wang JY, et al:
Prognostic relevance of Centromere protein H expression in
esophageal carcinoma. BMC Cancer. 8:2332008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liao WT, Yu CP, Wu DH, et al: Upregulation
of CENP-H in tongue cancer correlates with poor prognosis and
progression. J Exp Clin Cancer Res. 28:742009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shigeishi H, Higashikawa K, Ono S, et al:
Increased expression of CENP-H gene in human oral squamous cell
carcinomas harboring high-proliferative activity. Oncol Rep.
16:1071–1075. 2006.PubMed/NCBI
|
|
24
|
Orthaus S, Ohndorf S and Diekmann S: RNAi
knockdown of human kinetochore protein CENP-H. Biochem Biophys Res
Commun. 348:36–46. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tomonaga T, Matsushita K and Ishibashi M:
Centromere protein H is up-regulated in primary human colorectal
cancer and its overexpression induces aneuploidy. Cancer Res.
65:4683–4689. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hamilton SR and Aaltonen LA: World Health
Organization Classification of Tumors: Pathology and Genetics of
Tumours of the Digestive System. LARC Press; Lyon: 2000
|
|
27
|
Sobin LH and Fleming ID: TNM
Classification of Malignant Tumors, fifth edition (1997). Union
Internationale Contrele Cancer and the American Joint Committee on
Cancer. Cancer. 80:1803–1804. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cleveland DW, Mao Y and Sullivan KF:
Centromeres and kinetochores: from epigenetics to mitotic
checkpoint signaling. Cell. 112:407–421. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schmidt JC, Arthanari H and Boeszoermenyi
A: The kinetochore-bound Ska1 complex tracks depolymerizing
microtubules and binds to curved protofilaments. Dev Cell.
23:968–980. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kops GJ, Weaver BA and Cleveland DW: On
the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev
Cancer. 5:773–785. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tomonaga T, Matsushita K, Yamaguchi S, et
al: Overexpression and mistargeting of centromere protein-A in
human primary colorectal cancer. Cancer Res. 63:3511–3516.
2003.PubMed/NCBI
|
|
32
|
Li Y, Zhu Z, Zhang S, et al:
ShRNA-targeted centromere protein A inhibits hepatocellular
carcinoma growth. PLoS One. 6:e177942011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
McGovern SL, Qi Y, Pusztai L, et al:
Centromere protein-A, an essential centromere protein, is a
prognostic marker for relapse in estrogen receptor-positive breast
cancer. Breast Cancer Res. 14:R722012. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu Q, Qian YM, Zhao XL, et al: Expression
and prognostic significance of centromere protein A in human lung
adenocarcinoma. Lung Cancer. 77:407–414. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wood KW, Lad L, Luo L, et al: Antitumor
activity of an allosteric inhibitor of centromere-associated
protein-E. Proc Natl Acad Sci USA. 107:5839–5844. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
O’Brien SL, Fagan A, Fox EJ, et al: CENP-F
expression is associated with poor prognosis and chromosomal
instability in patients with primary breast cancer. Int J Cancer.
120:1434–1443. 2007.PubMed/NCBI
|
|
37
|
Shigeishi H, Mizuta K, Higashikawa K, et
al: Correlation of CENP-F gene expression with tumor-proliferating
activity in human salivary gland tumors. Oral Oncol. 41:716–722.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Barbanis S, Ioannou M, Kouvaras E, et al:
INCENP (inner centromere protein) is overexpressed in high grade
non-Hodgkin B-cell lymphomas. Pathol Oncol Res. 15:11–17. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nakajima T, Moriguchi M, Mitsumoto Y, et
al: Centrosome aberration accompanied with p53 mutation can induce
genetic instability in hepatocellular carcinoma. Mod Pathol.
17:722–727. 2004. View Article : Google Scholar : PubMed/NCBI
|