Introduction
Lung cancer is the leading cause of cancer-related
deaths worldwide, and especially in China. The majority of these
deaths are due to non-small cell lung cancer (NSCLC), which is the
most common histologic type of lung cancer (1). It is current practice to treat
advanced NSCLC with platinum based chemotherapy, although treatment
outcomes are particularly poor (2,3).
Therefore, target therapy for patients with advanced NSCLC are
currently being evaluated.
The epidermal growth factor receptor (EGFR)
is a receptor tyrosine kinase (TK) that is frequently overexpressed
and plays a central role in the development of NSCLC (4,5).
Abnormal activation of EGFR can promote tumor cell
proliferation, differentiation and migration. EGFR tyrosine kinase
inhibitors (EGFR-TKIs), such as gefitinib and erlotinib, which
target EGFR, have demonstrated promising outcomes in the
treatment of NSCLC patients (6–8). The
efficacy of EGFR-TKIs is associated with Asian race, shows gender
specificity to women, non-smokers and adenocarcinoma histology
(9). Furthermore, an association
between mutations in the EGFR TK domain and sensitivity to
EGFR-TKIs has been previously reported (6,10).
EGFR mutations are located in EGFR exons 18
to 21 (9) and most mutations are
observed as in-frame deletions in exon 19 and a point mutation
L858R in exon 21 (11). Thus,
testing for EGFR mutations may be prognostically important
to identify potential responders who would benefit from treatment
with EGFR-TKIs. This is particularly true for Chinese NSCLC
patients with high EGFR mutation rates (12). The samples used for EGFR
mutations are usually from resected tumor tissues, which could be
stably and easily detected. It is difficult to obtain sufficient
tumor tissues with advanced NSCLC, thus alternative specimens need
to be established for testing EGFR mutations.
Malignant pleural effusion is a common complication
of lung cancer. It is present in ~15% of lung cancer patients and
in ~10–50% of patients at the time of diagnosis (13). In about half of NSCLC patients with
a pleural effusion, most effusions are determined to be malignant
consistent with the progress of the disease. As sampling of pleural
effusion fluid is usually a standard and uncomplicated procedure,
which is also non-invasive and repeatable, we hypothesized that
genetic alterations in the pleural effusion fluid of NSCLC patients
could provide useful guidelines with regard the response to
EGFR-TKIs therapy.
In the present study, we used two approaches to
detect major EGFR mutations in malignant pleural effusions
from 24 patients presenting with advanced NSCLC and compared the
acquired results. The relationship between EGFR mutations
with the efficacy of gefitinib was also evaluated.
Patients and methods
Patients
Cytologically or pathologically confirmed pleural
effusions were obtained from 24 Chinese patients presenting with
advanced NSCLC. Jinan General Hospital of PLA approved this study,
and written informed consent was obtained from all participants.
Eligibility criteria included patients with stage IIIB–IV, ECOG
performance status (PS) of 0–3, and a life expectancy of at least 3
months. The records of all patients consisted of age, gender,
smoking habit, histological type of NSCLC and treatment. The
response of the patients to treatment with gefitinib was evaluated
in accordance with the ‘Response Evaluation Criteria in Solid
Tumors (RECIST)’ guidelines (14).
No research results were entered into the records of any of the
patients whatsoever or released to the patient or the physician of
the patient. Each specimen was only labeled by a serial number
without any identification.
Collection of pleural effusion fluid and
DNA extraction
Pleural effusion fluid was collected from patients
in heparinized tubes between 20th February and 22nd June 2012. No
particular collection method was used. A 30 ml volume sample of the
fluid was centrifuged at 250 × g for 10 min at room temperature,
and the cell pellets were stored at −80°C until used. Genomic DNA
in the cell pellets was extracted by DNeasy tissue kits (Qiagen,
Germany), and according to the manufacturer's protocol. The
concentration and purity of extracted DNA were assessed by
spectrophotometry (Nanodrop, ADx, China).
Polymerase chain reaction amplification
and direct sequencing
Exons 19, 20 and 21 of the EGFR gene were
amplified by polymerase chain reaction (PCR). The primers specific
for EGFR were designed using Primer Designer Software
(primer premier 5.0). The sequences of primers for EGFR exon
19 to 21 are described in Table I.
Each 50 μl reaction specimen contained 2 μl of template DNA, 0.25
μl of Ampli Taq Gold DNA polymerace (Roche, USA), 5 μl of
10X PCR buffer, and 10 μM of forward and reverse primer. The same
PCR program was used for all amplicons: 95°C for 3 min; 32 cycles
of 95°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec; 72°C for 10
min. After PCR assay had completed, the resultant amplicons were
further purified by QIAquick PCR purification kit (Qiagen), and
subjected to sequencing analysis in both sense and antisense
directions.
 | Table IPrimers used for EGFR mutation
screening by direct sequencing. |
Table I
Primers used for EGFR mutation
screening by direct sequencing.
| Sense primer | Antisense primer |
|---|
| Exon 19 |
CCAGCAATATCAGCCTTAGGTG |
GGGGAGGGAGTTATACCCACTA |
| Exon 20 |
GTCACTTCACAGCCCTGCGTA |
GTCACTTCACAGCCCTGCGTA |
| Exon 21 |
CTTGGAGGACCGTCGCTTG |
GAGAGACTGAAACCTAACATTTGCTA |
ADx-ARMS for the detection of EGFR
mutations
We used an EGFR Gene 4 Mutations Diagnostic
kit (ADx, Xiamen, China), which combines the two technologies of
ARMS and Bi-loop Probe, to detect mutations in real-time PCR
reactions. All reactions proceeded in 25 μl volumes according to
the manufacturer's protocol. Real-time PCR was performed using the
Mx3000P™ real-time PCR system (Agilent, Germany) under the
following conditions: initial denaturation at 95°C for 5 min, 15
cycles of 95°C for 25 sec, 64°C for 20 sec, 72°C for 20 sec, and 31
cycles of 95°C for 25 sec, 60°C for 35 sec (with fluorescence
collection, set to FAM and HEX), and finally 72°C for 20 sec. Data
were analyzed using Stratagene Mxpro software. The threshold cycle
(Ct) was defined as the cycle at the highest peak of the curve,
which represents the point of maximum curvature of the growth
curve. Positive results were defined as Ct <26. Analysis of each
sample was carried out in duplicate, and the whole test process
required only 90 min. The EGFR mutation kit is intended for
detection of the major somatic mutations in EGFR.
Statistical analysis
SPSS statistical software (version 13.0) was used
for statistical analysis. The Chi-square test was used to compare
the sensitivity between direct sequencing and ADx-ARMS. Two-sided
P-values of <0.05 were considered statistically significant.
Results
Patients
Sixteen male and 8 female patients were enrolled for
the study. The median age was 58 years. Fourteen patients had no
history of cigarette smoking; the ten current smokers were all male
(Table).
Results of direct sequencing
analysis
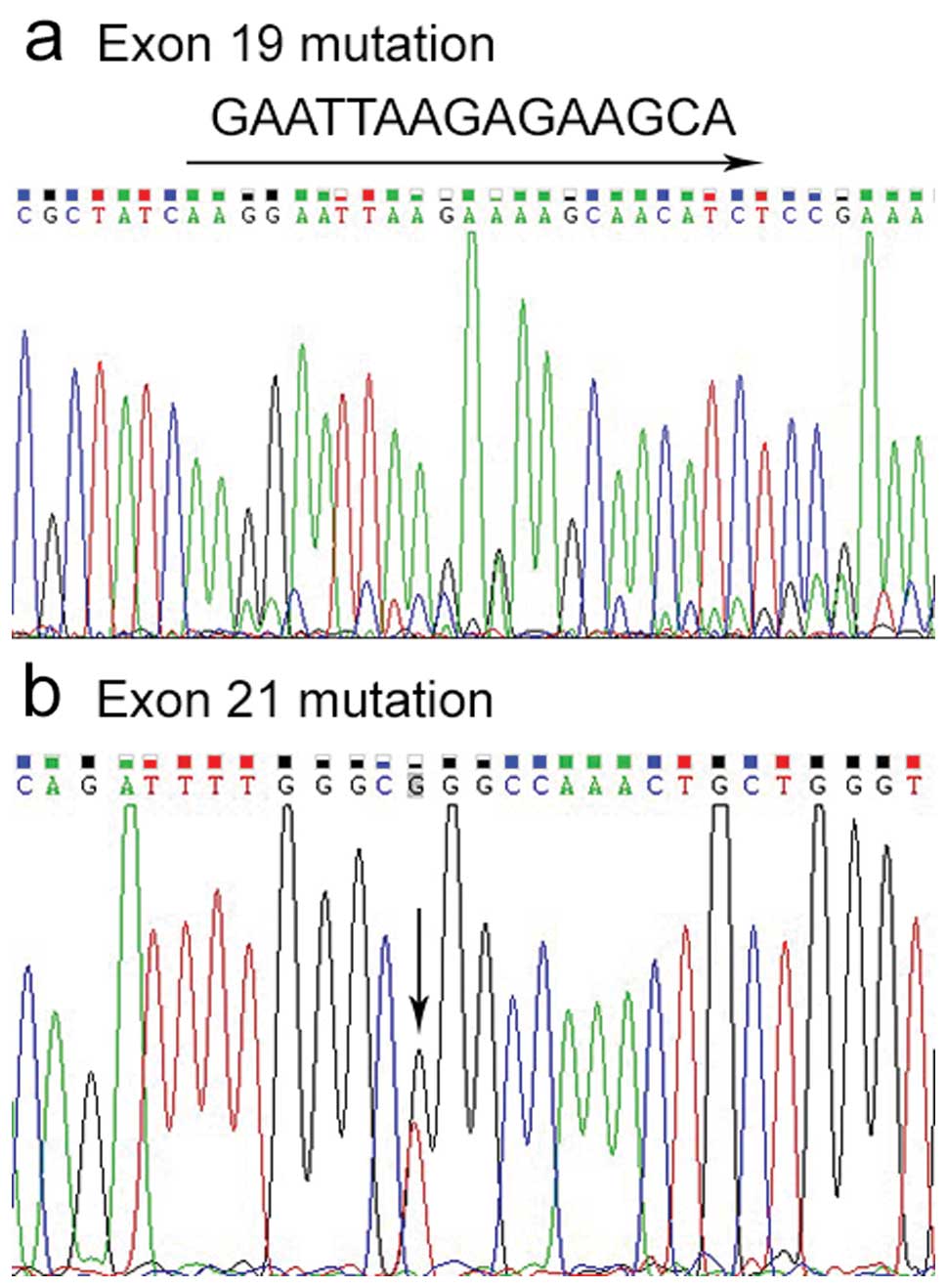
EGFR mutations were observed in 10 samples by
direct sequencing of DNA, 4 deletions in exon 19, and 6 L858R
mutations in exon 21 (Fig. 1). We
did not detect any mutations in exon 20 (data not shown).
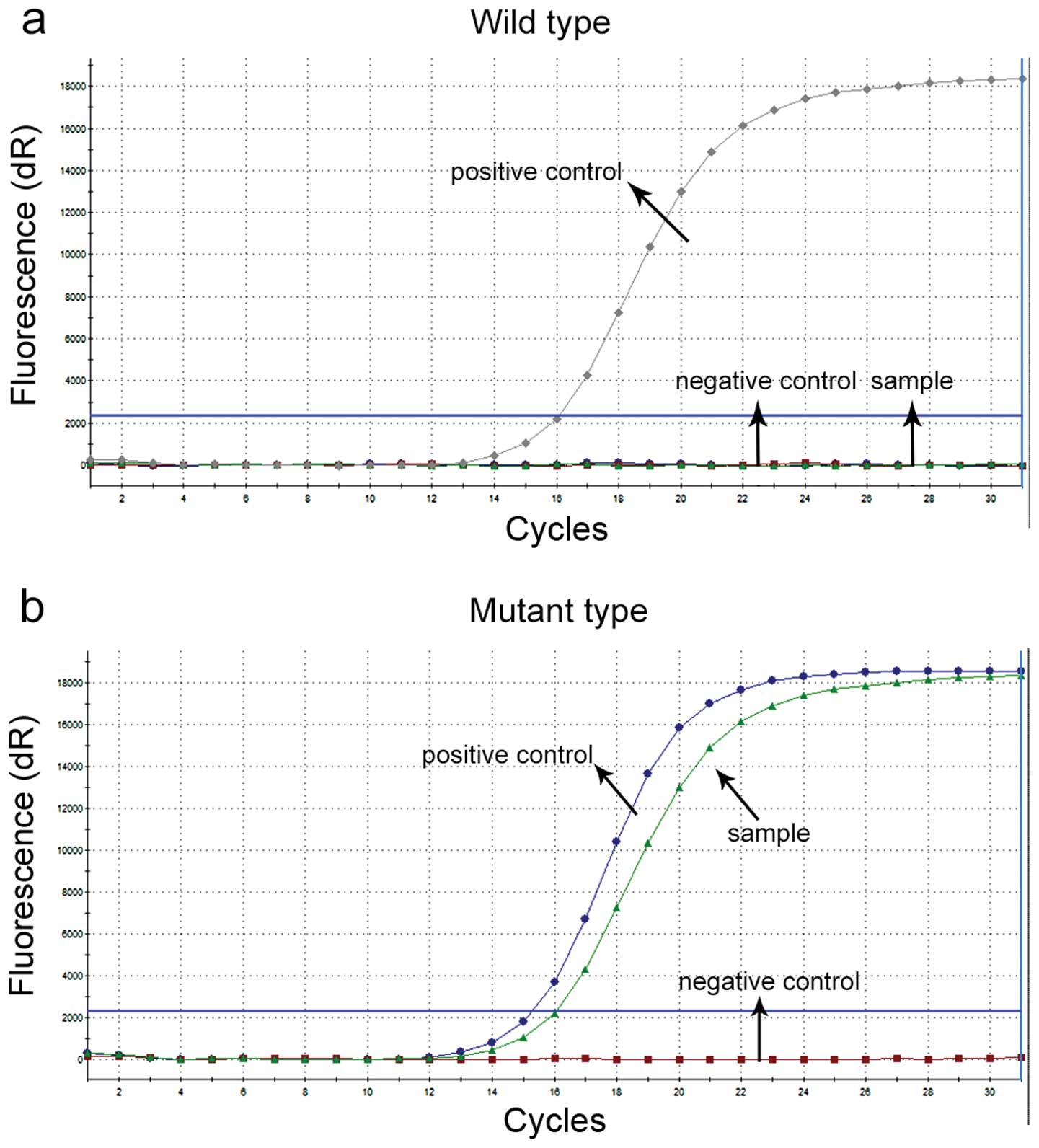
ADx-ARMS analysis
ADx-ARMS analysis of EGFR mutations are shown
in Fig. 2. The wild-type showed one
increased curve, which was the positive control, and the mutant
type showed two increased curves, which were the mutant and
positive control curves, respectively. Using the EGFR
Mutations Diagnostic kit, 6 deletion mutations in exon 19, and 8
L858R mutations in exon 21 of EGFR were detected. We confirmed that
there was no mutation in exon 20 (not shown).
Comparison between direct sequencing and
ADx-ARMS
We found gene mutations in EGFR in only 10
patients by the direct sequencing assay. Thus, direct gene
sequencing was less sensitive than ADx-ARMS analysis. In 24
patients, EGFR mutations were detected in 14 samples (58.3%) by
ADx-ARMS, while 10 mutations (41.7%) were detected by direct
sequencing. However, no significant difference was seen between
these approaches (χ2=1.333, P=0.248). Among the test
results of 24 patients, there was an 83.3% concordance between
direct sequencing and ADx-ARMS. Four EGFR mutation-negative
samples found by direct sequencing were mutation-positive by
ADx-ARMS.
Correlation between EGFR mutation and
clinical response
For patients treated with gefitinib, EGFR
mutations were detected in cells from malignant pleural effusions
in ten of the 18 patients (Table
III). Among those 10 EGFR mutant samples, 8 patients
achieved partial response, and 2 presented with stable disease
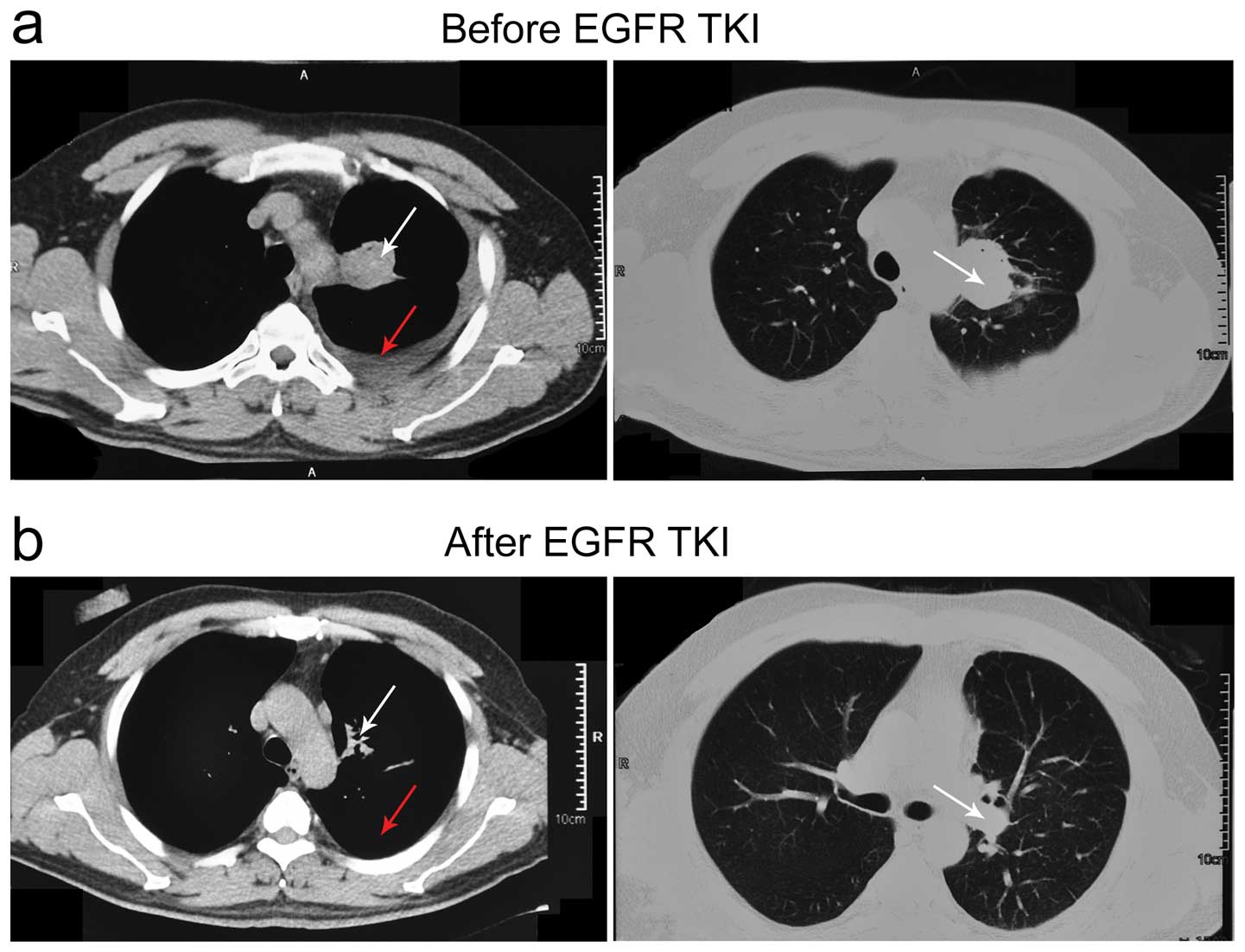
after 28 days of gefitinib therapy. In the 8 patients who partially
responded, 6 of them showed decreased levels of pleural effusion,
and reduced size of the tumor (Fig.
3). Six of the eight patients who had no demonstrable
EGFR mutations progressed to develop the disease. While
defining a patient with partial response as a responder, the
frequency of EGFR mutations was significantly higher in
gefitinib responders (8/9) than was found in non-responders (2/9,
P=0.02).
 | Table IIIEGFR mutations and the response
treated with gefitinib in 18 patients. |
Table III
EGFR mutations and the response
treated with gefitinib in 18 patients.
| No. of patients | Response | Gender | Age (yrs.) | EGFR
mutation |
|---|
| 1 | PR | F | 61 | Exon 21 |
| 22 | PR | F | 49 | Exon 21 |
| 5 | SD | M | 77 | Exon 19 |
| 16 | PR | M | 84 | Exon 19 |
| 7 | PR | M | 51 | Exon 21 |
| 8 | PR | F | 59 | Exon 19 |
| 11 | PR | F | 65 | Exon 21 |
| 2 | PR | M | 81 | Exon 19 |
| 9 | PR | M | 73 | Exon 21 |
| 3 | SD | M | 51 | Exon 21 |
| 14 | PD | M | 63 | WT |
| 17 | PD | M | 49 | WT |
| 19 | SD | F | 52 | WT |
| 24 | PD | F | 73 | WT |
| 20 | PD | F | 61 | WT |
| 23 | PD | M | 73 | WT |
| 12 | PR | M | 61 | WT |
| 4 | PD | M | 55 | WT |
Discussion
In this study, we demonstrated the feasibility of
using DNA from malignant pleural effusion as an alternative to
tumor samples for the detection of EGFR mutations from
advanced NSCLC patients.
We used the pleural effusion samples to detect
EGFR mutation status and compared two methods: i) gene
sequencing and, ii) ADx-ARMS. We also showed that patients with
mutant EGFR had a better response to treatment with EGFR-TKIs. In
our study, the response rate was 80% (8 of 10 patients achieved
partial response) in EGFR mutation patients, while
EGFR wild-type patients had only a 12.5% response rate (1 of
8 patients achieved partial response). Patients with mutant
EGFR, had a response rate which was significantly higher
than patients with wild-type EGFR (P<0.05). The data are
in agreement to other previously reported studies (15–17).
Direct gene sequencing has been regarded as a
gold-standard method for gene mutation analysis in the last
decades. Direct sequencing usually requires sufficient tumor tissue
as the testing sample with a sensitivity of ~30% (18,19).
However, it is challenging to obtain sufficient tissue for gene
sequencing in advanced NSCLC. In addition, gene sequencing is both
time-consuming and technically demanding (17). Many studies have shown that gene
sequencing is unable to provide satisfactory data for the detection
of pleural effusion fluid samples that contain mixtures of DNA from
normal cells (20,21), thus it cannot be widely used in
clinical practice. Therefore, alternative clinical samples with
more sensitive methodological approaches are urgently needed for
individualized therapy of EGFR-TKIs.
Pleural effusion fluid, which has DNA from tumor
cell pellets or the free DNA from the tumor provide a good
alternative (17,20,22).
The advantage of collecting free DNA or cell pellets is that it is
a relatively simple approach, it is non-invasive and a repeatable
technique. Thus, it could dynamically guide clinical approaches.
Due to different methods and the selectivity of lung cancer
patients with pleural effusion fluid, the frequency of mutant
EGFR is in the range of 12.5–73% (17,20,21,23–25).
In our study, the frequency of EGFR mutations
(deletion mutations and L858R mutations) detected by sequencing and
by ADx-ARMS was found to be 41.7% and 58.3%, respectively. ADx-ARMS
appeared to be the more sensitive approach as compared with direct
sequencing in this study. The mutations detected by ADx-ARMS
consisted of an in-frame deletion in exon 19 (E746_A750 del:
2235_2249del15 and 2236_2250del15), an insertion mutation in exon
20 (T790M), and a point mutation in exon 21 (L858R). Other deletion
patterns in exon 19 and other mutations in the tyrosine kinase
domain of EGFR could not be detected by this assay.
Among the 24 patients, there was 83.3% concordance
between direct sequencing and ADx-ARMS. Our findings of a
correlation between EGFR mutations and tumor response to
therapy with TKIs was consistent with previous studies (15,16).
Due to the small number of our samples, the EGFR mutation
rate showed no significant difference between these two methods
(χ2=1.333, P=0.248). At this point, it is worthwhile
mentioning two limitations of our study. One is that we did not
compare EGFR mutations between effusion cells and primary
tumors, the main reason being that some tumor samples were not
available. In addition, our results need further study based on the
relationship between EGFR mutations and progressive-free
survival and overall survival.
In summary, the clinical responses of NSCLC to
EGFR-targeted therapy are closely associated with EGFR
sensitive mutations. Screening of EGFR mutations by the
ADx-ARMS approach using malignant pleural effusion as the source
specimen is more sensitive and faster as compared with traditional
gene sequencing approaches. These observations support the utility
of this technology in routine clinical practice, an approach that
can benefit patients presenting with advanced NSCLC.
References
|
1
|
No authors listed. Chemotherapy in
non-small cell lung cancer: a meta-analysis using updated data on
individual patients from 52 randomised clinical trials. Non-small
Cell Lung Cancer Collaborative Group. BMJ. 311:899–909. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Haura EB: Treatment of advanced
non-small-cell lung cancer: a review of current randomized clinical
trials and an examination of emerging therapies. Cancer Control.
8:326–336. 2001.PubMed/NCBI
|
|
3
|
Schiller JH: Current standards of care in
small-cell and non-small-cell lung cancer. Oncology. 61(Suppl 1):
3–13. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ohsaki Y, Tanno S, Fujita Y, et al:
Epidermal growth factor receptor expression correlates with poor
prognosis in non-small cell lung cancer patients with p53
overexpression. Oncol Rep. 7:603–607. 2000.PubMed/NCBI
|
|
5
|
Nicholson RI, Gee JM and Harper ME: EGFR
and cancer prognosis. Eur J Cancer. 37(Suppl 4): 9–15. 2001.
View Article : Google Scholar
|
|
6
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Paez JG, Janne PA, Lee JC, et al: EGFR
mutations in lung cancer: correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Han SW, Kim TY, Hwang PG, et al:
Predictive and prognostic impact of epidermal growth factor
receptor mutation in non-small-cell lung cancer patients treated
with gefitinib. J Clin Oncol. 23:2493–2501. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shigematsu H, Lin L, Takahashi T, et al:
Clinical and biological features associated with epidermal growth
factor receptor gene mutations in lung cancers. J Natl Cancer Inst.
97:339–346. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Carey KD, Garton AJ, Romero MS, et al:
Kinetic analysis of epidermal growth factor receptor somatic mutant
proteins shows increased sensitivity to the epidermal growth factor
receptor tyrosine kinase inhibitor, erlotinib. Cancer Res.
66:8163–8171. 2006. View Article : Google Scholar
|
|
11
|
Borras E, Jurado I, Hernan I, Gamundi MJ,
Dias M, Marti I, Mane B, Arcusa A, Agundez JA, Blanca M and
Carballo M: Clinical pharmacogenomic testing of KRAS, BRAF and EGFR
mutations by high resolution melting analysis and ultra-deep
pyrosequencing. BMC Cancer. 11:4062011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou C, Wu YL, Chen G, et al: Erlotinib
versus chemotherapy as first-line treatment for patients with
advanced EGFR mutation-positive non-small-cell lung cancer
(OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase
3 study. Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fenton KN and Richardson JD: Diagnosis and
management of malignant pleural effusions. Am J Surg. 170:69–74.
1995. View Article : Google Scholar
|
|
14
|
Therasse P, Arbuck SG, Eisenhauer EA, et
al: New guidelines to evaluate the response to treatment in solid
tumors. European Organization for Research and Treatment of Cancer,
National Cancer Institute of the United States, National Cancer
Institute of Canada. J Natl Cancer Inst. 92:205–216. 2000.
View Article : Google Scholar
|
|
15
|
Giaccone G, Herbst RS, Manegold C, et al:
Gefitinib in combination with gemcitabine and cisplatin in advanced
non-small-cell lung cancer: a phase III trial - INTACT 1. J Clin
Oncol. 22:777–784. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Herbst RS, Giaccone G, Schiller JH, et al:
Gefitinib in combination with paclitaxel and carboplatin in
advanced non-small-cell lung cancer: a phase III trial - INTACT 2.
J Clin Oncol. 22:785–794. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jian G, Songwen Z, Ling Z, et al:
Prediction of epidermal growth factor receptor mutations in the
plasma/pleural effusion to efficacy of gefitinib treatment in
advanced non-small cell lung cancer. J Cancer Res Clin Oncol.
136:1341–1347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bosari S, Marchetti A, Buttitta F, et al:
Detection of p53 mutations by single-strand conformation
polymorphisms (SSCP) gel electrophoresis. A comparative study of
radioactive and nonradioactive silver-stained SSCP analysis. Diagn
Mol Pathol. 4:249–255. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fan X, Furnari FB, Cavenee WK, et al:
Non-isotopic silver-stained SSCP is more sensitive than automated
direct sequencing for the detection of PTEN mutations in a mixture
of DNA extracted from normal and tumor cells. Int J Oncol.
18:1023–1026. 2001.PubMed/NCBI
|
|
20
|
Kimura H, Fujiwara Y, Sone T, et al: EGFR
mutation status in tumour-derived DNA from pleural effusion fluid
is a practical basis for predicting the response to gefitinib. Br J
Cancer. 95:1390–1395. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kimura H, Kasahara K, Kawaishi M, et al:
Detection of epidermal growth factor receptor mutations in serum as
a predictor of the response to gefitinib in patients with
non-small-cell lung cancer. Clin Cancer Res. 12:3915–3921. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kubo A, Koh Y, Kawaguchi T, et al:
Malignant pleural effusion from lung adenocarcinoma treated by
gefitinib. Intern Med. 50:745–748. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang X, Zhao Y, Wang M, et al: Detection
and comparison of epidermal growth factor receptor mutations in
cells and fluid of malignant pleural effusion in non-small cell
lung cancer. Lung Cancer. 60:175–182. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Han HS, Lim SN, An JY, et al: Detection of
EGFR mutation status in lung adenocarcinoma specimens with
different proportions of tumor cells using two methods of
differential sensitivity. J Thorac Oncol. 7:355–364. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsai TH, Wu SG, Chang YL, et al: Effusion
immunocytochemistry as an alternative approach for the selection of
first-line targeted therapy in advanced lung adenocarcinoma. J
Thorac Oncol. 7:993–1000. 2012. View Article : Google Scholar : PubMed/NCBI
|