Introduction
Hematological malignancies are a form of cancer that
includes leukemia and lymphoma (1).
Leukemia is one of the cancers that causes extensive mortality in
the human population. It is known that leukemia starts in
hematopoietic elements primarily in the bone marrow, the soft
tissue inside most bones, then progresses to the blood, lymph nodes
or other organs leading to serious health problems. In Taiwan, the
2009 Report of the Department of Health, R.O.C. (Taiwan) indicated
that ~4 out of 100,000 individuals die each year of leukemia, and
it is the 11th most common malignancy (2). At present, the treatment of patients
with leukemia includes bone marrow transplant, radiotherapy and
chemotherapy (3,4). However, these treatments are still
unsatisfactory, and the exact mechanisms involved in leukemia are
unclear.
Propofol (2,6-diisopropylphenol), is commonly used
as an intravenous sedative-hypnotic (anesthetic). It has analgesic
effects in inflammation-induced pain and facilitated pain (5) and has been clinically used for
patients in rapid emergence after cessation of infusion (6). Propofol contains cardiac protection
during ischemia-reperfusion (7–9), and
it has been reported that propofol upregulates AQP1 expression
(10) and inhibits nitric oxide
synthase (11) in
endotoxemia-mediated lung injury. However, animal studies suggest
that propofol protects against endotoxemia-induced lung and kidney
injuries (12,13).
A previous study found that propofol induces
apoptosis, which is dependent on the mechanism that activates both
the cell surface death receptor pathway and the mitochondrial
pathway. It was also found that propofol may trigger
neurodegeneration in neurons during development and its derivative
may affect neuronal injury, including apoptotic cell death
(14). It was also demonstrated
that propofol inhibited the decarboxylase activity in
three-dimensional primary cell cultures of fetal rat telencephalon
(15). Recently, we found that
propofol induced apoptosis in murine leukemia RAW264.7 cells in
vitro through altered levels of apoptosis-associated proteins
resulting in induction of apoptotic gene expression and inhibition
of cell growth (13).
It was reported that the neuro-degeneration in
age-related diseases, cerebral ischemia and brain trauma is
associated with DNA damage (16),
and if agents can induce DNA damage in cells, this may lead to cell
mutations and to the development of malignancy. Although numerous
studies indicate that propofol induces cell death, no information
has addressed the effects of propofol-induced DNA damage in murine
leukemia cells. Therefore, in the present study, we investigated
the effects of propofol on DNA damage and expression (mRNA) of DNA
repair-associated genes in murine leukemia RAW264.7 cells. The
results revealed that propofol induced DNA damage and inhibited the
expression of DNA damage and repair-associated genes in RAW264.7
cells in vitro.
Materials and methods
Chemicals and reagents
Propofol was obtained from B. Braun Melsungen AG
(Schwarzenberger Weg, Melsungen, Germany). A stock solution of
propofol was prepared in phosphate-buffered saline (PBS), and an
equal volume of PBS (0.1%) was added to the controls. RPMI-1640
medium, fetal bovine serum (FBS), L-glutamine and
penicillin-streptomycin were obtained from
Gibco®/Invitrogen Life Technologies (Grand Island, NY,
USA).
Cell culture and treatment
The RAW264.7 murine leukemia cell line was obtained
from the Food Industry Research and Development Institute (Hsinchu,
Taiwan). The RAW264.7 cells were maintained in RPMI-1640 medium
with 2 mM L-glutamine, supplemented with 10% heat-inactivated FBS
and 100 U/ml penicillin and 100 μg/ml streptomycin, and incubated
at 5% CO2 and 37°C and 90% relative humidity. Propofol
was diluted in sterile PBS before addition to the culture. The
cells which were treated with PBS served as the control and were
assigned to have 100% survival.
Cell viability analysis
Approximately 2×105 cells/well of
RAW264.7 cells in 12-well plates were maintained for 24 h in an
incubator. Cells in each well were incubated with propofol at the
final concentrations of 0, 25, 50, 100 and 200 μg/ml, vehicle (1 μl
PBS) or 0.1% hydrogen peroxide (H2O2,
positive control) for 48 h. Cells were also treated with 100 μg/ml
propofol for 24, 48 and 72 h. Cells from each treatment group were
stained with 4 μg/ml propidium iodide (PI) (Sigma-Aldrich Corp.,
St. Louis, MO, USA) and analyzed by FACSCalibur flow cytometry
(Becton-Dickinson, San Jose, CA, USA). Cell viability was
calculated as previously described (13,17).
Measurement of DNA damage by comet
assay
RAW264.7 cells (2×105/well) in 12-well
plates were incubated with 0, 25, 50, 100 and 200 μg/ml of
propofol, vehicle (1 μl PBS) or 0.1% H2O2 for
48 h, or cells were exposed to 100 μg/ml propofol for 24, 48 and 72
h in RPMI-1640 medium and grown at 37°C in 5% CO2 and
95% air. Cells from each treatment group were harvested by
centrifugation for examination of DNA damage using the comet assay
as previously described (18,19).
The TriTek CometScore™ software image analysis system (TriTek
Corp., Sumerduck, VA, USA) was used to calculate and quantify the
comet tail length as previously described (18,19).
Evaluation of DNA fragmentation by DNA
gel electrophoresis
RAW264.7 cells at a density of 2×106
cells/well in 6-well plates were incubated with propofol (25, 50,
100 and 200 μg/ml) or vehicle (1 μl PBS) for 48 h at 37°C in 5%
CO2 and 95% air. Cells from each treatment group were
individually isolated, and then DNA was extract using a DNA
isolation kit (Genemark Technology Co., Ltd., Tainan, Taiwan)
(20,21). DNA electrophoresis was carried out
on 1.5% agarose gel in Tris/boric acid buffer at 15 V for 2 h, and
DNA was stained with ethidium bromide (EtBr; Sigma-Aldrich Corp.)
and then examined and photographed under UV light box as previously
described (18–20).
Analysis of gene expression by real-time
PCR assay
RAW264.7 cells at a density of 2×106/well
in 6-well plates were incubated with 100 μg/ml propofol or without
for 48 h. The total RNA from each sample was extracted using the
Qiagen RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA) as
previously described (18,20). Briefly, the High Capacity cDNA
reverse transcription kit was used for RNA samples which were
reverse-transcribed for 30 min at 42°C according to the standard
protocol of the supplier (Applied Biosystems, Carlsbad, CA, USA).
The quantitative PCR from each sample was performed under the
following conditions: 2 min at 50°C, 10 min at 95°C, and 40 cycles
of 15 sec at 95°C, 1 min at 60°C using 1 μl of the cDNA
reverse-transcribed as described above, 2X SYBR-Green PCR Master
Mix (Applied Biosystems) and 200 nM of forward and reverse primers
as shown in Table I. Finally, the
DNA sequence was evaluated using the Primer Express software and
each assay was run on an Applied Biosystems 7300 real-time PCR
system in triplicates to ensure reproducibility. Expression
fold-changes were derived using the comparative CT
method (22).
 | Table IPrimer sequences used in the
real-time PCR. |
Table I
Primer sequences used in the
real-time PCR.
| Primer name | Primer
sequence |
|---|
|
DNA-PK-F |
TTCAAGACTTCAACCGCTTT |
|
DNA-PK-R |
TGCGGCTGGGTCAAGTGT |
| BRCA1-F |
TTGAAGTCAAAGGAGACGTTG |
| BRCA1-R |
TCTTTGGGCATGTTGGTGAA |
| MGMT-F |
TCCCTTGCCTGCTCTCCAT |
| MGMT-R |
AACCATTTCTCCGAATTTCACAA |
| p53-F |
GGGTTAGTTTACAATCAGCCACATT |
| p53-R |
GGGCCTTGAAGTTAGAGAAAATTCA |
| GAPDH-F |
GGTGGACCTCATGGCCTACA |
| GAPDH-R |
CAGCAACTGAGGGCCTCTCT |
Analysis of protein expression by Western
blot assay
RAW264.7 cells (2×106/well) were placed
in a 6-well plate and then propofol was added to the cells at final
concentrations of 2.5, 5 and 10 μM, while DMSO (solvent) alone was
added to the wells as a vehicle control. Cells were incubated with
propofol in medium with 10% FBS at 37°C for 0, 6, 12 and 24 h.
Cells were harvested and resuspended in lysis buffer [ice-cold 50
mM potassium phosphate buffer (pH 7.4) containing 2 mM EDTA and
0.1% Triton X-100]. The collected cells were sonicated and
centrifugated at 13,000 × g for 20 min at 4°C to remove cell
debris, and the supernatant was collected for determination of
total protein concentration using a Bio-Rad protein assay kit
(Bio-Rad Laboratories, Hercules, CA, USA) with bovine serum albumin
(BSA) as the standard. SDS gel electrophoresis and western blotting
were performed as previously described (22,23)
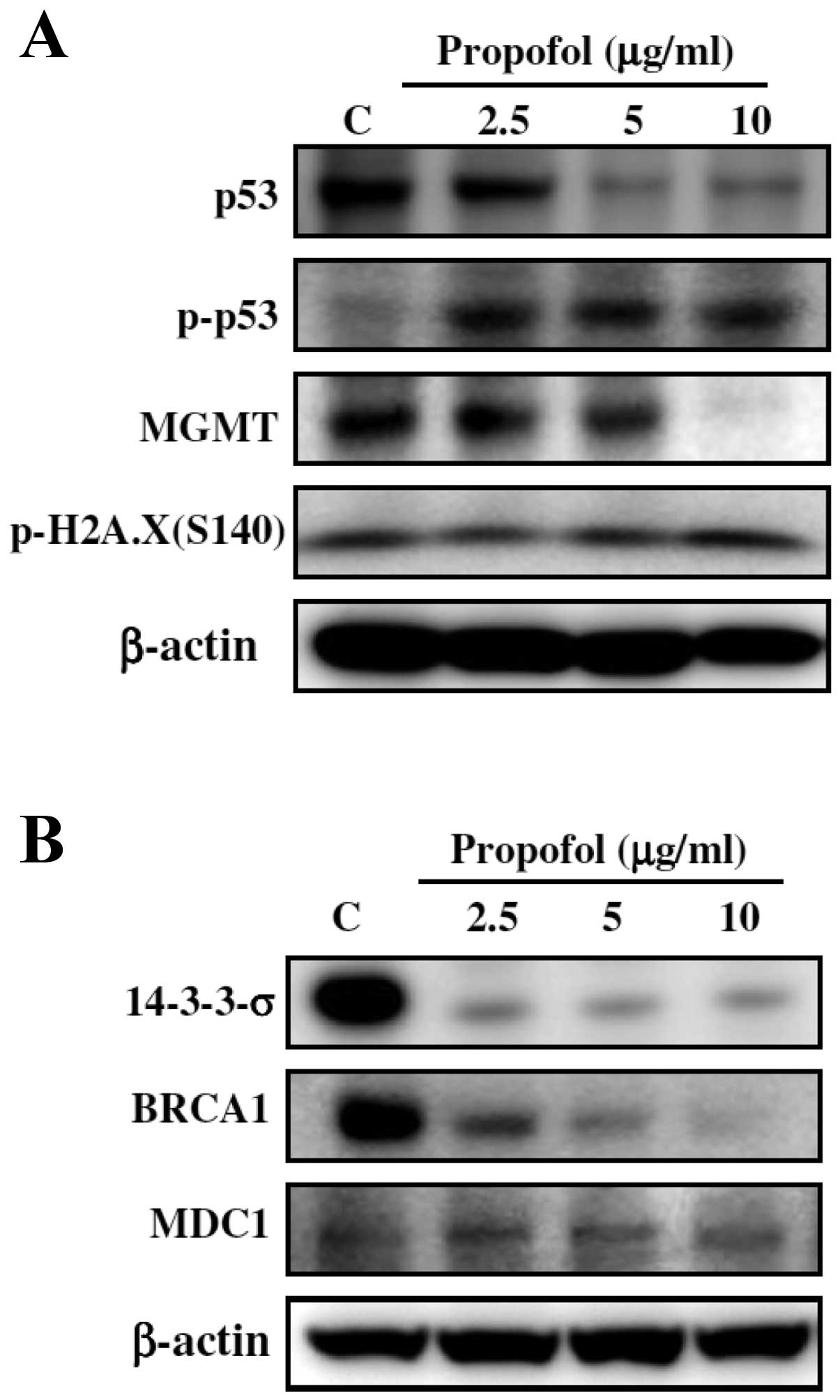
for determining the effects of propofol on the protein levels of
p53, p-p53, MGMT, p-H2A.X(S140), 14-3-3-σ, BRCA1 and MDC1.
Statistical analysis
All data are presented as the means ± SD, and the
Student’s t-test was used to analyze differences between the
propofol-treated and untreated (control) groups. All statistical
analyses were performed, and P<0.05 was considered to indicate a
statistically significant result.
Results
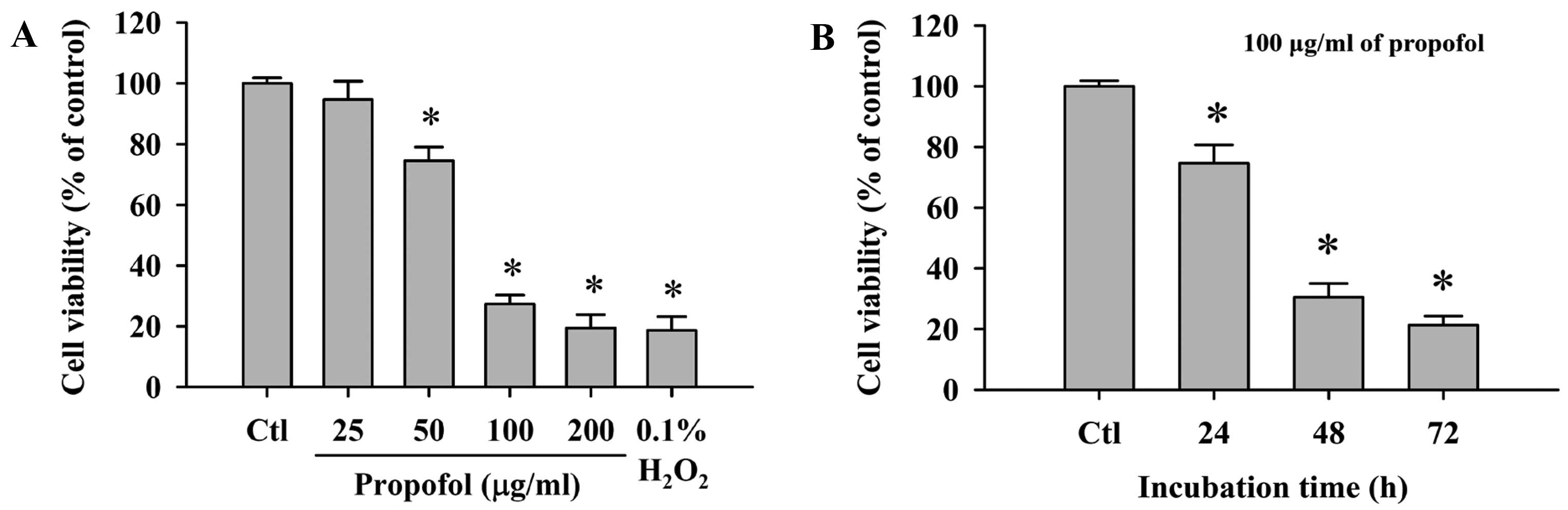
Effect of propofol on the viability of
RAW264.7 cells
Cells were treated with 0, 25, 50, 100 and 200 μg/ml
of propofol for 48 h or treated with 100 μg/ml propofol for 0, 24,
48 and 72 h. All cells from each treatment group were collected and
then flow cytometric assay was used to evaluate the percentage of
viable RAW264.7 cells. Propofol decreased the percentage of viable
cells when compared to the control in a dose-dependent (Fig. 1A) and time-dependent manner
(Fig. 1B).
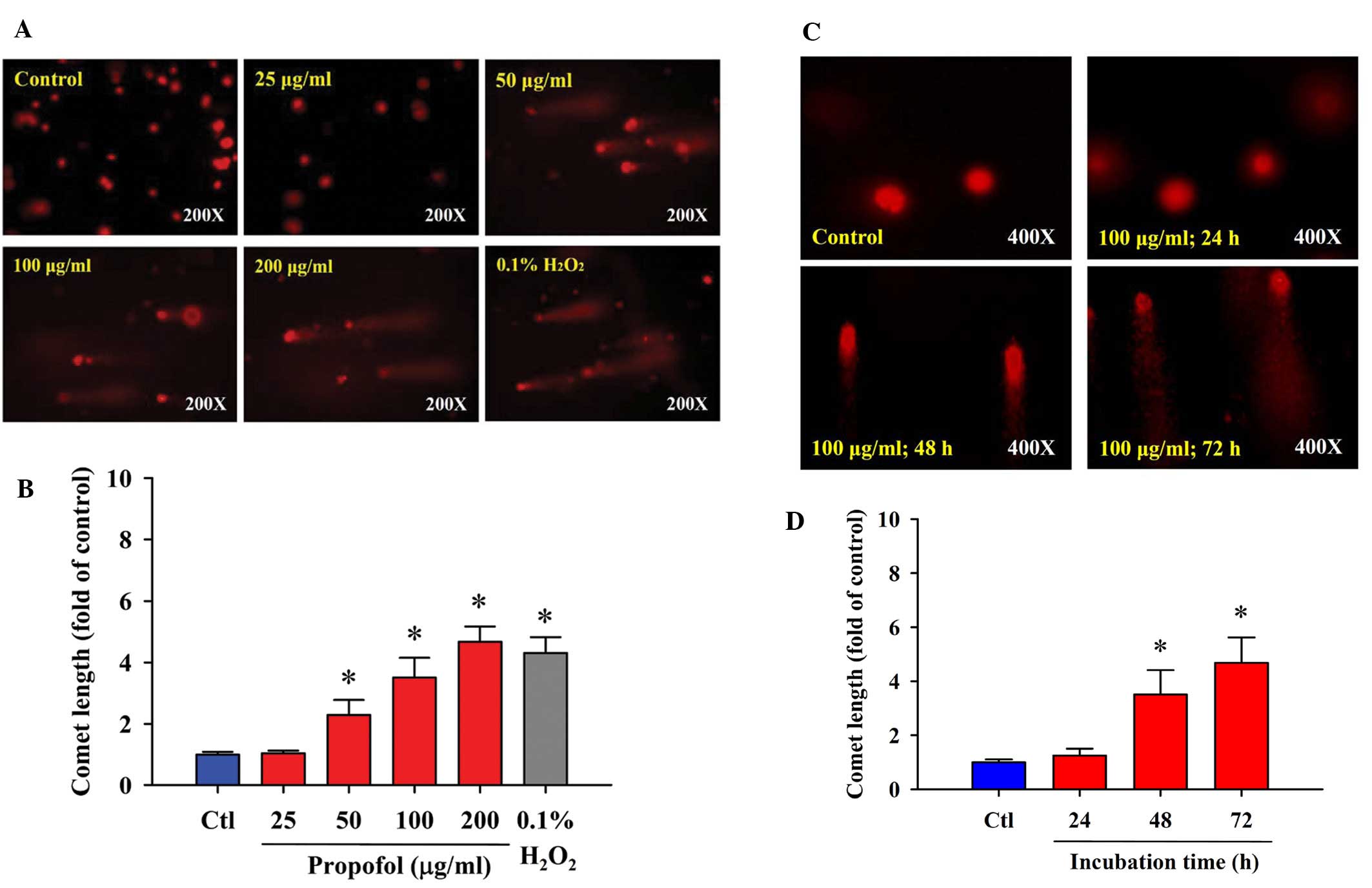
Propofol induces DNA damage in RAW264.7
cells
We investigated whether propofol induces DNA damage
in RAW264.7 cells in vitro. The comet assay was selected for
determining DNA damage. The results indicated that propofol induced
DNA damage in RAW264.7 cells. Higher concentrations of propofol led
to a longer DNA migration smear (comet tail), and these effects
occurred in a dose- (Fig. 2A and B)
and time-dependent manner (Fig. 2C and
D). As shown in Fig. 2A, 0.1%
H2O2 induced the occurrence of a comet tail,
and H2O2 is well documented as a highly
reactive oxygen species and it has been used as a positive control
for numerous studies involving agent-induced DNA damage.
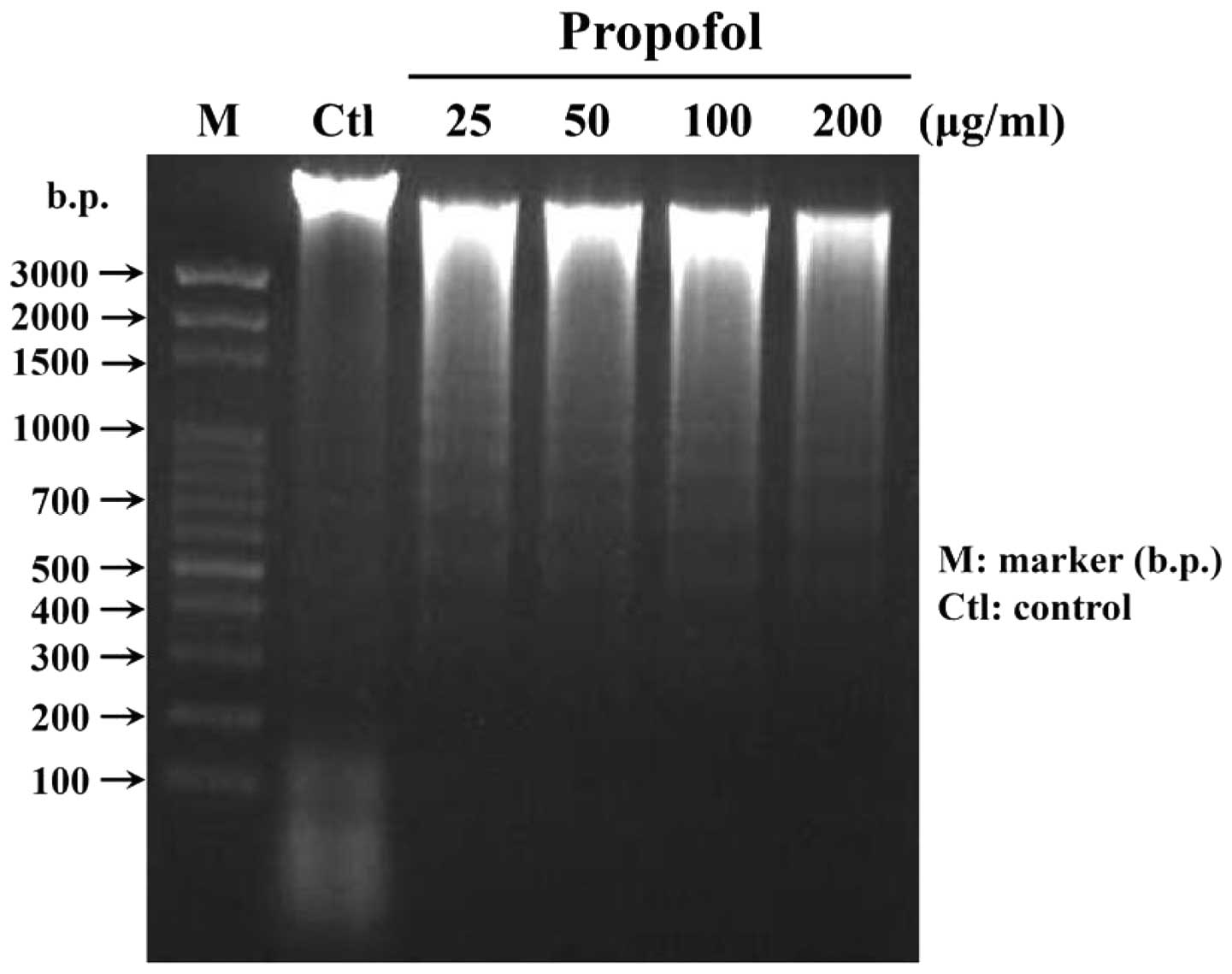
Propofol induces DNA fragmentation in
RAW264.7 cells
As shown in Fig. 2,
propofol induced DNA damage in RAW264.7 cells, and therefore, DNA
gel electrophoresis was used to investigate whether propofol causes
DNA fragmentation in RAW264.7 cells. Cells were treated with
various concentrations of propofol for 48 h, and then DNA was
isolated from each treatment group. DNA fragmentation was assessed
by DNA gel electrophoresis. As shown in Fig. 3, propofol induced DNA fragmentation
in RAW264.7 cells.
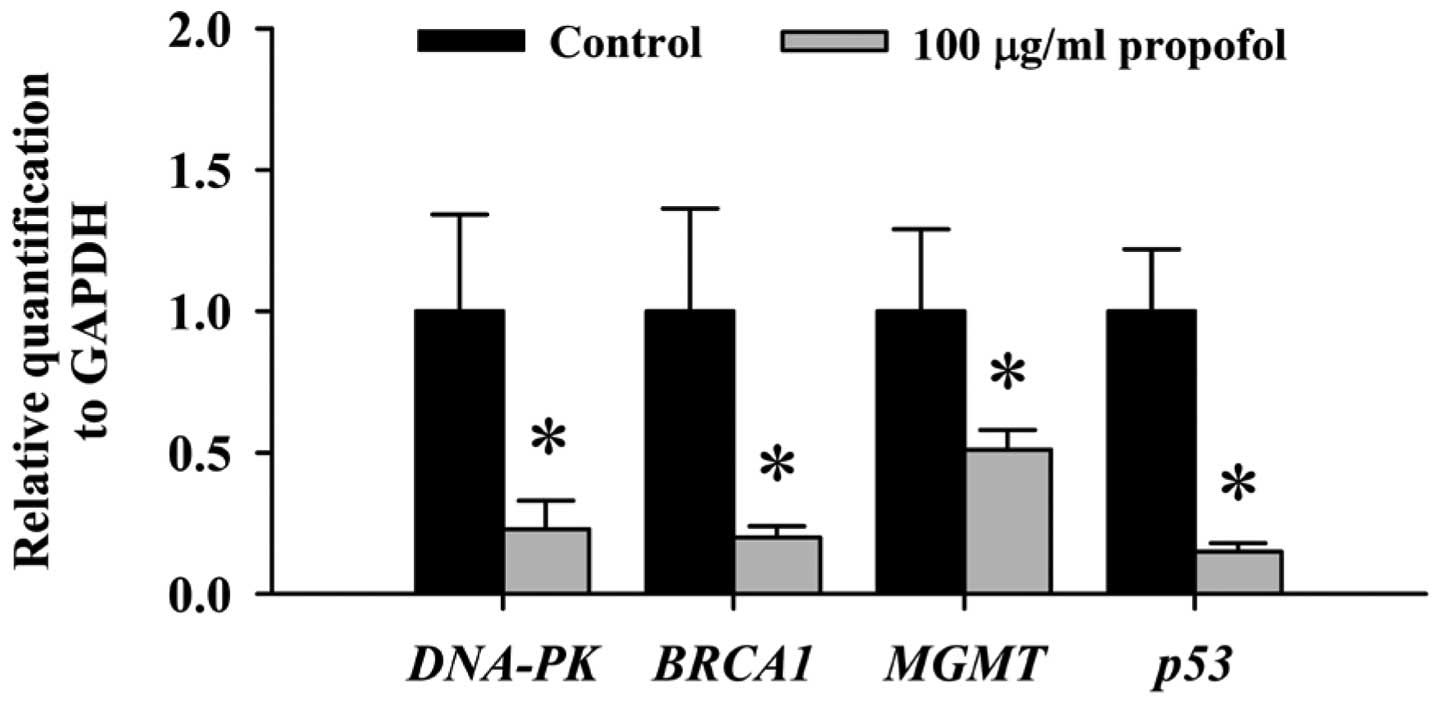
Effects of propofol on expression of DNA
damage and repair-associated genes in RAW264.7 cells
Based on the above results from the comet assay and
DNA gel electrophoresis, we found that propofol induced DNA damage
and fragmentation in RAW264.7 cells. Thus, we further investigated
the effects of propofol on the expression of DNA damage and
repair-associated genes in RAW264.7 cells. mRNA expression of all
examined genes associated with DNA damage and repair including
DNA-PK, BRCA1, MGMT and p53 was decreased following 48 h of
exposure to propofol (Fig. 4).
Effects of propofol on the expression of
DNA damage and repair-associated proteins in RAW264.7 cells
To further confirm whether propofol inhibits DNA
repair gene expression and whether it also affects the associated
protein expression, RAW264.7 cells were treated with propofol and
then harvested for western blotting. As shown in Fig. 5, levels of p53, MGMT, 14-3-3-σ,
BRCA1 and MDC1 proteins were decreased while p-p53 and
p-H2A.X(S140) were increased in the RAW264.7 cells following
exposure to propofol.
Discussion
Cytotoxic drugs and radiation are well-known
DNA-damaging agents commonly used for cancer therapy and associated
with the development of therapy-related myeloid neoplasms (23). Based on reports from other
investigators, propofol was found to induce cytotoxic effects on
human leukemia HL-60 cells via induction of apoptosis (24). Our previous study found that
propofol induced apoptosis in murine leukemia RAW264.7 cells
(13). However, there is no
available information that has addressed propofol-induced DNA
damage in murine leukemia cells. In the present study, we confirmed
that propofol has cytotoxic effects on RAW264.7 cells, and we also
demonstrated that propofol induced DNA damage (Fig. 2A and C) and DNA fragmentation
(Fig. 3) and these effects were
dose-dependent, and associated with loss of cell viability
(Fig. 1). Furthermore, western
blotting assay was used to examine the expression of DNA
repair-associated protein. The results indicated that levels of
p53, MGMT, 14-3-3-σ, BRCA1 and MDC1 proteins were decreased while
p-p53 and p-H2A.X(S140) were increased in RAW264.7 cells following
exposure to propofol. In the present study, the dose of propofol
for inducing DNA damage was 25 μg/ml which is lower than that of
other reports showing the blood concentration of propofol to be
<25 μg/ml during and after anesthesia (24). Possibly the in vitro studies
used culture plates, and the propofol directly affected the
cells.
In the present study, the comet assay revealed that
propofol induced nuclear DNA damage. It has been reported that
various types of DNA damage are associated with cancer development,
and therefore, mutations resulting from DNA damage are considered
to be a hallmark of cancer (25).
Thus, whether or not propofol is a carcinogen requires further
investigation. However, numerous studies have used the comet assay
to examine whether agents induce DNA damage based on its high
sensitive technique for DNA damage assessment (26–28).
Other investigators also showed that the comet assay can measure
the strand-break formation during the process of excision repair of
DNA (29,30).
In our previous study, we found that
propofol-induced cell death was mediated through induction of
apoptosis in RAW264.7 cells, as determined by DAPI staining
(13). In the present study, we
also confirmed that propofol caused DNA fragmentation as determined
by DNA gel electrophoresis (Fig.
3). Our previous study showed that propofol induced apoptosis
through the activation of caspase-3 in RAW264.7 cells, not via
reactive oxygen species (ROS) (13). This is in agreement with other
reports which found that propofol was capable of scavenging
hydrogen peroxide (H2O2) (31). Thus, we suggest that
propofol-induced DNA damage occurs independently of ROS production.
Further studies are needed to establish the role of the interaction
of propofol with DNA in cancer cells.
Agent-induced DNA damage can result in the loss of
DNA repair capacity and accumulation of DNA damage. It was reported
that agent-induced DNA damage can be reduced by DNA repair system
through eliminating DNA lesions (32–34).
In the present study, propofol-induced DNA damage in RAW264.7 cells
is not clear. Therefore, to our knowledge this is the first report
on propofol-induced DNA damage in murine leukemia cells. The
mechanism involved in the DNA damage observed in the present study
remains to be determined.
The results from real-time PCR revealed that
propofol inhibited expression of DNA repair-associated genes
including DNA-PK, BRCA-1, MGMT and p53 (Fig. 4) dose-dependently in the RAW264.7
cells. Importantly, it has been reported that if agents cause DNA
damage in checkpoints of the cell cycle, then there are signal
transduction pathways involved in the cell cycle and cellular
responses to DNA damage for maintaining genomic integrity (35–37).
In human breast and ovarian cancer, it was reported that BRCA1
(tumor suppressor) plays critical roles in DNA repair, cell cycle
checkpoint control and maintenance of genomic stability (38). DNA-PK plays an important role in DNA
damage repair (39). Furthermore,
MGMT reduces cytotoxicity of therapeutic or environmental
alkylating agents (40,41). Another report demonstrated that
anesthesia with propofol did not directly influence the expression
of the DNA repair genes hOGG1 and XRCC1 in blood cells (42). Herein, we demonstrated that propofol
inhibited expression of several of the DNA repair-associated genes
including DNA-PK, BRCA-1 and MGMT in RAW264.7 cells. Furthermore,
western blotting showed that propofol inhibited levels of DNA
repair-associated proteins such as MGMT, 14-3-3-σ, BRCA1 and MDC1,
and DNA damage-associated p53 proteins. Notably, propofol promoted
p53 phosphorylation.
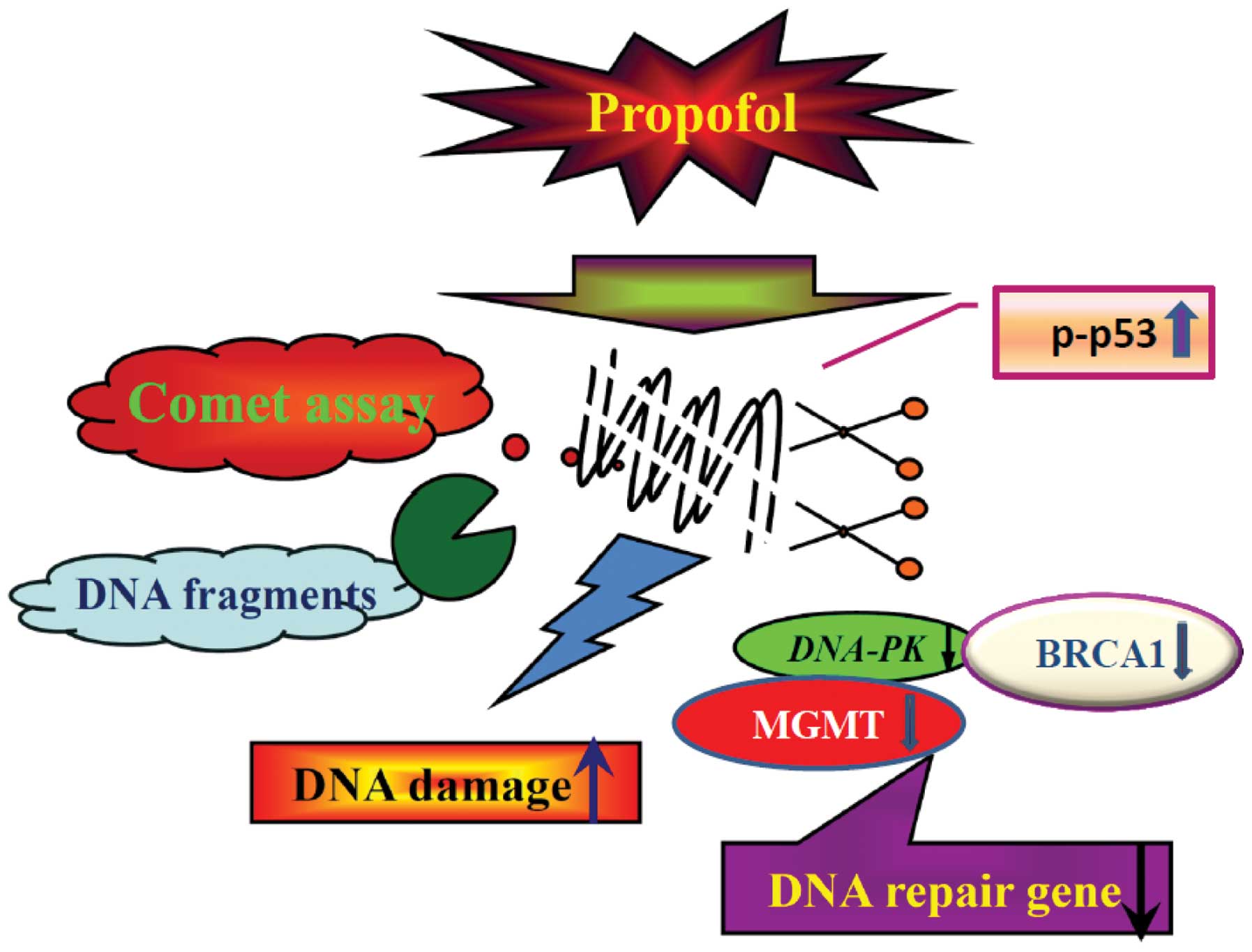
Our proposed flow chart for propofol-induced DNA
damage in murine leukemia RAW264.7 cells is illustrated in Fig. 6, which shows that propofol induces
DNA damage followed by the inhibition of expression (mRNA) of DNA
repair-associated genes including DNA-PK, BRCA1, MGMT and p53 which
then leads to DNA damage as shown by the comet assay and
quantitative real-time PCR analysis.
Acknowledgements
The present study was supported by the Taiwan
Department of Health, China Medical University Hospital, Cancer
Research Center of Excellence (grant no. DOH101-TD-C-111-005).
References
|
1
|
Lichtman MA: Battling the hematological
malignancies: the 200 years’ war. Oncologist. 13:126–138.
2008.PubMed/NCBI
|
|
2
|
Lin JP, Yang JS, Lin JJ, et al: Rutin
inhibits human leukemia tumor growth in a murine xenograft model in
vivo. Environ Toxicol. 27:480–484. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fotoohi AK, Assaraf YG, Moshfegh A, et al:
Gene expression profiling of leukemia T-cells resistant to
methotrexate and 7-hydroxymethotrexate reveals alterations that
preserve intracellular levels of folate and nucleotide
biosynthesis. Biochem Pharmacol. 77:1410–1417. 2009. View Article : Google Scholar
|
|
4
|
Nau KC and Lewis WD: Multiple myeloma:
diagnosis and treatment. Am Fam Physician. 78:853–859.
2008.PubMed/NCBI
|
|
5
|
Nishiyama T, Matsukawa T and Hanaoka K:
Intrathecal propofol has analgesic effects on inflammation-induced
pain in rats. Can J Anaesth. 51:899–904. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
de Ruijter W, Stienen GJ, van Klarenbosch
J and de Lange JJ: Negative and positive inotropic effects of
propofol via L-type calcium channels and the sodium-calcium
exchanger in rat cardiac trabeculae. Anesthesiology. 97:1146–1155.
2002.PubMed/NCBI
|
|
7
|
Kato R and Foex P: Myocardial protection
by anesthetic agents against ischemia-reperfusion injury: an update
for anesthesiologists. Can J Anaesth. 49:777–791. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ko SH, Yu CW, Lee SK, et al: Propofol
attenuates ischemia-reperfusion injury in the isolated rat heart.
Anesth Analg. 85:719–724. 1997.PubMed/NCBI
|
|
9
|
Kokita N, Hara A, Abiko Y, Arakawa J,
Hashizume H and Namiki A: Propofol improves functional and
metabolic recovery in ischemic reperfused isolated rat hearts.
Anesth Analg. 86:252–258. 1998.PubMed/NCBI
|
|
10
|
Gao J, Zhao W, Xiang D and Shi Y: Effects
of propofol on the expression of aquaporin-1 in
lipopolysaccharide-activated rat lung microvessel endothelial
cells. Chin J Critl Care Med. 26:670–672. 2006.
|
|
11
|
Chen HI, Hsieh NK, Kao SJ and Su CF:
Protective effects of propofol on acute lung injury induced by
oleic acid in conscious rats. Crit Care Med. 36:1214–1221. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cui WY, Tian AY and Bai T: Protective
effects of propofol on endotoxemia-induced acute kidney injury in
rats. Clin Exp Pharmacol Physiol. 38:747–754. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu RS, Liu KC, Tang NY, et al: cDNA
microarray analysis of the gene expression of murine leukemia RAW
264.7 cells after exposure to propofol. Environ Toxicol. 8:471–478.
2011.PubMed/NCBI
|
|
14
|
Sagara Y, Hendler S, Khoh-Reiter S, et al:
Propofol hemisuccinate protects neuronal cells from oxidative
injury. J Neurochem. 73:2524–2530. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Honegger P, Pardo B and Monnet-Tschudi F:
Muscimol-induced death of GABAergic neurons in rat brain
aggregating cell cultures. Brain Res Dev Brain Res. 105:219–225.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dallas ML, Boyle JP, Milligan CJ, et al:
Carbon monoxide protects against oxidant-induced apoptosis via
inhibition of Kv2.1. FASEB J. 25:1519–1530. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu CC, Yang JS, Huang AC, et al:
Chrysophanol induces necrosis through the production of ROS and
alteration of ATP levels in J5 human liver cancer cells. Mol Nutr
Food Res. 54:967–976. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chiang JH, Yang JS, Ma CY, et al:
Danthron, an anthraquinone derivative, induces DNA damage and
caspase cascade-mediated apoptosis in SNU-1 human gastric cancer
cells through mitochondrial permeability transition pores and
Bax-triggered pathways. Chem Res Toxicol. 24:20–29. 2011.
View Article : Google Scholar
|
|
19
|
Liu KC, Ho HC, Huang AC, et al: Gallic
acid provokes DNA damage and suppresses DNA repair gene expression
in human prostate cancer PC-3 cells. Environ Toxicol. Sep
2–2011.(Epub ahead of print). View Article : Google Scholar
|
|
20
|
Kuo CL, Wu SY, Ip SW, et al: Apoptotic
death in curcumin-treated NPC-TW 076 human nasopharyngeal carcinoma
cells is mediated through the ROS, mitochondrial depolarization and
caspase-3-dependent signaling responses. Int J Oncol. 39:319–328.
2011.
|
|
21
|
Lu HF, Lai TY, Hsia TC, et al: Danthron
induces DNA damage and inhibits DNA repair gene expressions in GBM
8401 human brain glioblastoma multiform cells. Neurochem Res.
35:1105–1110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu FS, Yang JS, Yu CS, et al: Safrole
induces apoptosis in human oral cancer HSC-3 cells. J Dent Res.
90:168–174. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Voso MT, D’Alo F, Greco M, et al:
Epigenetic changes in therapy-related MDS/AML. Chem Biol Interact.
184:46–49. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsuchiya M, Asada A, Arita K, et al:
Induction and mechanism of apoptotic cell death by propofol in
HL-60 cells. Acta Anaesthesiol Scand. 46:1068–1074. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Loeb KR and Loeb LA: Significance of
multiple mutations in cancer. Carcinogenesis. 21:379–385. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ashby J, Tinwell H, Lefevre PA and Browne
MA: The single cell gel electrophoresis assay for induced DNA
damage (comet assay): measurement of tail length and moment.
Mutagenesis. 10:85–90. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Donatus IA, Sardjoko and Vermeulen NP:
Cytotoxic and cytoprotective activities of curcumin. Effects on
paracetamol-induced cytotoxicity, lipid peroxidation and
glutathione depletion in rat hepatocytes. Biochem Pharmacol.
39:1869–1875. 1990.
|
|
28
|
Pool-Zobel BL, Lotzmann N, Knoll M, et al:
Detection of genotoxic effects in human gastric and nasal mucosa
cells isolated from biopsy samples. Environ Mol Mutagen. 24:23–45.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Olive PL, Banath JP and Durand RE:
Detection of etoposide resistance by measuring DNA damage in
individual Chinese hamster cells. J Natl Cancer Inst. 82:779–783.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tice RR, Andrews PW and Singh NP: The
single cell gel assay: a sensitive technique for evaluating
intercellular differences in DNA damage and repair. Basic Life Sci.
53:291–301. 1990.PubMed/NCBI
|
|
31
|
Gulcin I, Alici HA and Cesur M:
Determination of in vitro antioxidant and radical scavenging
activities of propofol. Chem Pharm Bull. 53:281–285. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Epe B: Role of endogenous oxidative DNA
damage in carcinogenesis: what can we learn from repair-deficient
mice? Biol Chem. 383:467–475. 2002.PubMed/NCBI
|
|
33
|
Jiang MR, Li YC, Yang Y and Wu JR: c-Myc
degradation induced by DNA damage results in apoptosis of CHO
cells. Oncogene. 22:3252–3259. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Marnett LJ: Oxyradicals and DNA damage.
Carcinogenesis. 21:361–370. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cimprich KA and Cortez D: ATR: an
essential regulator of genome integrity. Nat Rev Mol Cell Biol.
9:616–627. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu Y and Kulesz-Martin M: p53 protein at
the hub of cellular DNA damage response pathways through
sequence-specific and non-sequence-specific DNA binding.
Carcinogenesis. 22:851–860. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lou Z, Minter-Dykhouse K, Wu X and Chen J:
MDC1 is coupled to activated CHK2 in mammalian DNA damage response
pathways. Nature. 421:957–961. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Choi JH, Sancar A and Lindsey-Boltz LA:
The human ATR-mediated DNA damage checkpoint in a reconstituted
system. Methods. 48:3–7. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Venkitaraman AR: Cancer susceptibility and
the functions of BRCA1 and BRCA2. Cell. 108:171–182. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jesien-Lewandowicz E, Jesionek-Kupnicka D,
Zawlik I, et al: High incidence of MGMT promoter methylation
in primary glioblastomas without correlation with TP53 gene
mutations. Cancer Genet Cytogenet. 188:77–82. 2009.
|
|
41
|
Mi J, Dziegielewski J, Bolesta E,
Brautigan DL and Larner JM: Activation of DNA-PK by ionizing
radiation is mediated by protein phosphatase 6. PLoS One.
4:e43952009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Braz MG, Braz LG, Mazoti MA, et al: Lower
levels of oxidative DNA damage and apoptosis in lymphocytes from
patients undergoing surgery with propofol anesthesia. Environ Mol
Mutagen. 53:70–77. 2012. View Article : Google Scholar : PubMed/NCBI
|