Introduction
Head and neck squamous cell carcinomas (HNSCCs) are
a group of common malignant cancers and account for more than
550,000 cases annually worldwide, and involve the oral cavity,
larynx, hypopharynx and oropharynx (1). Tobacco use and alcohol consumption are
two well-established risk factors, while epidemiological evidence
suggests that a subgroup of HNSCCs results from the infection of
high-risk types of human papillomavirus (HPV), particularly HPV16
(2,3).
One of the most frequent alteration in HNSCCs is
perturbation of the p53 pathway which regulates the cell cycle to
conserve genomic stability and prevent mutations (4). Most of the TP53 mutations occur
in exon 5–8. Traditional risk factors such as tobacco use lead to
abrogation of p53 by mutation. In contrast, HPV obviates the need
for mutation by encoding the E6 oncoprotein, which promotes
degradation of wild-type p53 in the presence of the E6-AP complex
(5), causing perturbation of cell
cycle regulation. It has been found that HNSCCs with HPV infection
show a lower frequency of mutations in TP53 when compared to
HNSCCs without HPV infection (6,7).
However, whether this phenomenon exists in Chinese patients is not
definitive.
The TP53 gene has more than 200
single-nucleotide polymorphisms (SNPs), most of which have no
biological effects. However, SNP of codon 72 (SNP72) within exon 4,
which encodes either proline (Pro, by codon CCC) or arginine (Arg,
by codon CGC), appears to be able to influence the function of p53.
Compared to TP53 encoding proline, TP53 encoding
arginine is more effective at inducing apoptosis and preventing
cells from neoplastic development (8,9).
Although the association of the SNP72 with various types of cancers
including lung and cervical cancer has been reported (10,11),
the effect of the polymorphisms on HNSCCs with HPV infection
remains uncertain.
In the present study, we analyzed mutations of
TP53, the phenotype of SNP72 and the expression profiles of
p53 in 93 Chinese patients with HNSCCs. Collectively with the HPV
infection status and pathological and clinical data as previously
described (12), possible
correlations among the mutations in TP53, the phenotype of
SNP72, the expression profiles of p53, HPV infection, and the
pathological and clinical features were comprehensively analyzed in
the group of laryngeal carcinomas and other HNSCCs (from other
sites).
Materials and methods
Patients and specimens
A total of 93 patients with malignant squamous
tumors were enrolled in the present study. The specimens consisted
of 64 laryngocarcinoma and 29 other HNSCCs (5 oropharyngeal, 15
hypopharynx and 9 lip carcinoma). Details of the cases are provided
in Table I. The professions of the
patients varied and included laborers, teachers, medical staff and
clients. The permanent residences of the patients were widely
distributed throughout China. All samples were surgically removed
and were conventionally fixed in 10% formalin and
paraffin-embedded. Pathological assays verified that all cancers
were squamous cell cancers. The pathological and clinical grades
were assessed by pathologists and surgeons at the Peking University
Cancer Hospital and Institute, respectively.
 | Table IClinicopathological features and p53
status in the HNSCC patients. |
Table I
Clinicopathological features and p53
status in the HNSCC patients.
| Patient
characteristics | N | TP53
mutation n/N (%) | p53 IHC positive
n/N (%) |
|---|
| All patients | 93 | 35/93 (37.6) | 34/93 (36.6) |
| Gender | | | |
| Male | 84 | 33/84 (39.3) | 32/84 (38.1) |
| Female | 9 | 2/9 (22.2) | 2/9 (22.2) |
| Age (years) | | | |
| <40 | 5 | 2/5 (40.0) | 3/5 (60.0) |
| 40–65 | 69 | 26/69 (37.7) | 24/69 (34.8) |
| >65 | 19 | 7/19 (36.8) | 7/19 (36.8) |
| Anatomical
diagnosis | | | |
| Larynx | 64 | 22/64 (34.4) | 20/64 (31.3) |
| Other
carcinomas | 29 | 13/29 (44.8) | 14/29 (48.3) |
| Pathological
grades | | | |
| 1 (carcinoma in
situ) | 2 | 1/2 (50.0) | 0/2 (0) |
| 2 (poorly
diff.) | 15 | 5/15 (33.3) | 7/15 (46.7) |
| 3 (moderately
diff.) | 50 | 19/50 (38.0) | 19/50 (38.0) |
| 4 (highly
diff.) | 26 | 9/26 (34.6) | 8/26 (30.8) |
| Clinical stage | | | |
| I | 28 | 12/28 (42.9) | 13/28 (46.4) |
| II | 35 | 15/35 (42.9) | 9/35 (25.7) |
| III | 25 | 7/25 (28.0) | 11/25 (44.0) |
| IV | 5 | 1/5 (20.0) | 1/5 (20.0) |
DNA extraction
Total DNAs from the tumor tissues were extracted
from the paraffin-embedded tissue blocks with a commercial genomic
DNA extraction FFPE kit (Qiagen). Briefly, 4 to 5 formalin-fixed,
paraffin-embedded (FFPE) sections (10 μm) were soaked in xylene and
vortexed vigorously for at least 1 h. Then the pellet was acquired
and purified in accordance with the kit protocol. The DNA
extraction was evaluated by electrophoresis on 2% agarose gel.
Quality of the extracted DNAs was assessed with a settled PCR
protocol with a pair of β-actin-specific primers.
PCR protocol for SNP72 in exon 4 and
exons 5–8 of P53
SNP72 in exon 4 and exons 5–8 of the TP53
gene was amplified using polymerase chain reaction (PCR) in a
Bio-Rad S1000 Thermal Cycler. The primer sequences and amplicons
are listed in Table II. The
reaction mixtures consisted of a total of 50 μl containing 1 μl of
gDNA, 20 pmol of sense and antisense primers, 21 μl RNase-free
water and 25 μl 2X Taq MasterMix (CW0682; CWBIO, China). Touchdown
method was adopted to increase the specificity of the PCR products.
Details of the PCR condition were as follows: denaturing at 95°C
for 30 sec, annealing at 65°C for 45 sec with a decrease of 1°C
every cycle in the first 10 cycles and 55°C for the other 35
cycles, and extension at 72°C for 45 sec. All PCR assays were
carefully carried out in the PCR laboratory with 4 separated rooms
to avoid DNA contamination.
 | Table IIPrimer sequences and amplicons of
SNP72 and exon 5–8 in the TP53 gene. |
Table II
Primer sequences and amplicons of
SNP72 and exon 5–8 in the TP53 gene.
| Primer pairs
(5′-3′) | Amplicons (bp) |
|---|
| SNP72 | F:
TTGCCGTCCCAAGCAATGGATGA | 199 |
| R:
TCTGGGAAGGGACAGAAGATGAC | |
| Exon 5 | F:
TGTTCACTTGTGCCCTGACT | 268 |
| R:
CAGCCCTGTCGTCTCTCCAG | |
| Exon 6 | F:
GCCTCTGATTCCTCACTGAT | 181 |
| R:
TTAACCCCTCCTCCCAGAGA | |
| Exon 7 | F:
CTTGCCACAGGTCTCCCCAA | 237 |
| R:
AGGGGTCAGAGGCAAGCAGA | |
| Exon 8 | F:
TTCCTTACTGCCTCTTGCTT | 231 |
| R:
AGGCATAACTGCACCCTTGG | |
Direct sequencing
The PCR products were analyzed on 2% agarose gel and
recovered from the gel using the QIAquick Gel extraction kit (cat.
no. 28706; Qiagen, Germany) according to the manufacturer’s
instructions. Direct sequencing was performed using the same PCR
primers by the ABI PRISM™ 3730xl DNA analyzer.
Immunohistochemical (IHC) assay
Paraffin sections (5 μm) were routinely
deparaffinized in xylene for 5 min twice and gradually rehydrated.
Sections were quenched for endogenous peroxidases in 3%
H2O2 in methanol for 15 min, pretreated with
enzyme digestion antigen retrieval for 1 min. After blocking in 1%
normal goat serum to avoid nonspecific binding, the sections were
incubated overnight at 4°C with a 1:500-diluted mAb for p53
(Novus). The sections were then incubated for 60 min with
1:1,000-diluted HRP-conjugated goat anti-mouse secondary antibody
(Vector Laboratories, USA), and visualized by incubation with
3,3-diaminobenzidine tetrahydrochloride (DAB). The slices were
dehydrated and mounted in Permount. For the negative controls, the
primary antibody for p53 was replaced with mouse non-immuno IgG.
Images were captured with a DP70 digital camera mounted on a BX5
microscope (Olympus Optical, Japan) (12). The slides were analyzed separately
by 2 independent observers blinded to the clinical data. The
immunoreactivity in the malignant cells in each section was graded
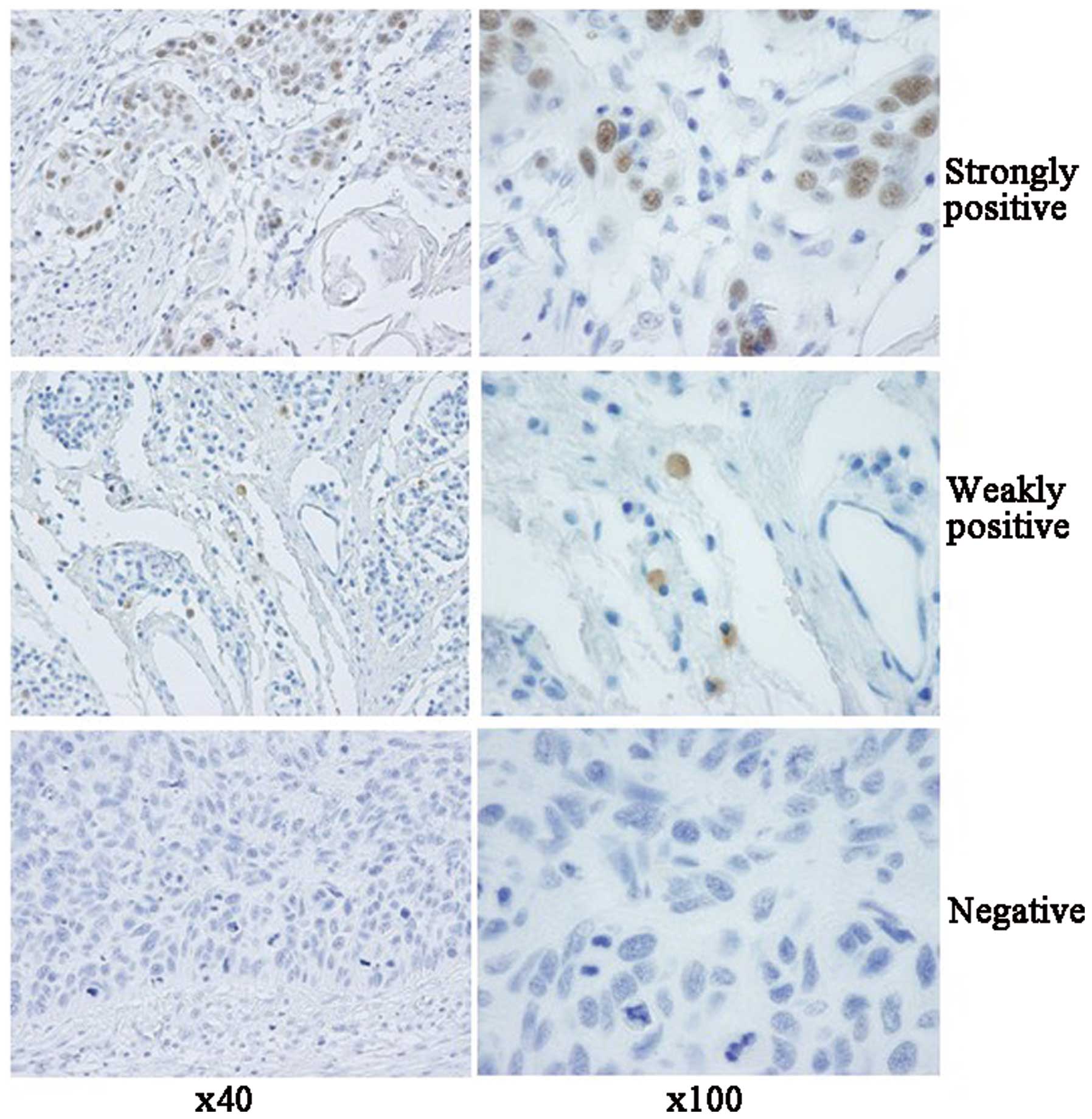
according to the number of positively stained nuclei: <1% as
negative, ≥1 and ≤10% as weakly positive, >10% as strongly
positive.
Prediction of p53 protein function for
the different mutations in each Transcriptional activity (TA)
class
Functional classification was based on the overall
TA of 8 different promoters (WAF1, MDM2, BAX, H1433s, AIP1, GADD45,
NOXA and P53R2) as measured by Kato et al(13). For each mutant, the median of the 8
promoter-specific activities was calculated, and missense mutations
were classified as ‘non-functional’ (median ≤20), ‘partially
functional’ (median >20 and ≤75), ‘functional’ (median >75
and ≤140) and ‘supertrans’ (median >140).
Sorting intolerant from tolerant (SIFT)
class
SIFT (14) predicts
whether an amino acid substitution affects protein function based
on sequence homology and the physical properties of amino acids.
Briefly, SIFT searches for similar sequences to the query sequence,
chooses closely related sequences that may share similar function
to the query sequence, obtains the alignment of these chosen
sequences, and calculates normalized probabilities for all possible
substitutions from the alignment. Positions with normalized
probabilities <0.05 are predicted to be deleterious; those ≥0.05
are predicted to be tolerated.
Statistical analysis
Statistical analysis was performed using Chi-square
and Fisher’s exact tests for correlations between groups in regards
to HPV infection, SNP72 phenotype, P53 mutations and IHC.
Mann-Whitney U test was used for the relationship between SNP72 and
age. Probability values of <0.05 were considered to indicate
statistically significant results. All statistical analyses were
performed by SPSS 20 (IBM, US).
Statement of ethics
Written consent for further investigation and
publication was obtained from the patients or the patients’
relatives, respectively. Usage of the stored human samples in this
study was approved by the Ethics Committees of Peking University
Cancer Hospital and Institute and the National Institute for Viral
Disease Prevention and Control, China CDC.
Results
Mutations in the TP53 gene in laryngeal
carcinoma and other HNSCCs
Altogether, 45 mutations within the TP53 gene
were found in the tissues of 35 cases (37.6%) out of the 93
recruited patients by direct sequencing, including 30 mutations in
34.4% (22/64) of patients in the group of laryngeal carcinoma and
15 mutations in 44.8% (13/29) of patients in the group of other
HNSCCs. The detailed information of the individual mutations is
documented in Tables I and III. Most of the mutations (93.3%, 42/45)
were missense and only 3 were nonsense. Six mutations generated
stop codons, 2 deletions and 1 insertion led to frameshift
mutations. The mutations at codon 245 within exon 7 and codon 271
within exon 8 were most frequently observed; both were detected in
4 cases. A mutation at codon 266 was observed in 3 cases and
mutations at codons 248, 267 and 298 were detected in 2 cases,
respectively. Meanwhile, most of the patients (28/35, 80%) had only
one mutation in the TP53 gene. Only one patient had 4
mutations, 1 case had 3 mutations and 5 cases had 2 mutations. Two
cases contained 2 mutations in the same exon region. Additionally,
the locations of the mutations were frequently distributed at the
hypervariable region of TP53.
 | Table IIIIndividual TP53 mutations and
the predicted functional changes of p53 proteins. |
Table III
Individual TP53 mutations and
the predicted functional changes of p53 proteins.
| Group | Sample no. | Exon | Mutated codon | Mutation by
sequencing | Amino acid
change | Change in
properties | Motif
structure | Protein
function |
|---|
|
|---|
| TA class | SIFT class |
|---|
| Larynx | 20 | 8 | 281 | GAC>AAC | Asp>Asn | Acid>polar | LSH | NF | D |
| 23 | 7 | 249 | AGG>ACG | Arg>Thr |
Alkaline>polar | L3 | NF | D |
| 37 | 5 | 163 | TAC>TGC | Tyr>Cys |
Aromatic>sulfurated | S4 | NF | D |
| 38 | 5 | 158 | CGC>GGC | Arg>Gly |
Alkaline>hydrophobic | S4 | NF | D |
| 7 | 248 | CGG>CAG | Arg>Gln |
Alkaline>polar | L3 | NF | D |
| 41 | 5 | 145 | CTG>ATG | Leu>Met |
Hydrophobic>polar | S3 | F | N |
| 46 | 7 | 245 | GGC>TGC | Gly>Cys |
Hydrophobic>polar | L3 | NF | D |
| 50 | 6 | 194 | CTT>CAT | Leu>His |
Hydrophobic>alkaline | L2 | NF | D |
| 7 | 245 | GGC>AGC | Gly>Ser |
Hydrophobic>polar | L3 | NF | D |
| 63 | 5 | 151 | CCC>TCC | Pro>Ser |
Hydrophobic>polar | L | NF | D |
| 64 | 7 | 242 | TGC>TTC | Cys>Phe |
Polar>hydrophobic | L3 | NF | D |
| 65 | 8 | 266 | GGA>GAA | Gly>Glu |
Hydrophobic>acid | S10 | NF | D |
| 66 | 8 | 298 | GAG>TAG | Glu>stop | NA | C-term | NF | D |
| 69 | 5 | 175 | CGC>CAC | Arg>His | No change | L2 | NF | D |
| 70 | 5 | 179 | CAT>CGT | His>Arg | No change | L2 | NF | D |
| 8 | 298 | GAG>GGG | Glu>Gly |
Acid>hydrophobic | C-term | F | N |
| 74 | 6 | 207 | GAT>AAT | Asp>Asn | Acid>polar | S6 | F | D |
| 79 | 7 | 259 | GAC>AAC | Asp>Asn | Acid>polar | L | PF | N |
| 81 | 7 | 245 | GGC>GAC | Gly>Asp |
Hydrophobic>acid | L3 | NF | D |
| 83 | 7 | Intron | AGGTC>AGATC | | NA | NA | NA | NA |
| 8 | 266 | GGA>TGA | Gly>stop | NA | S10 | ND | ND |
| 8 | 267 | CGG>CGT | Arg>Arg | No | change | S10 | NA |
| 84 | 4 | 47 | CCG>TCG | Pro>Ser |
Hydrophobic>polar | N-term | S | N |
| 85 | 4 | 51 | GAA>TAA | Glu>stop | NA | N-term | ND | ND |
| 7 | Intron | ACCCT>ACTCT | | NA | NA | NA | NA |
| 8 | 266 | GGA>TGA | Gly>stop | NA | S10 | ND | ND |
| 8 | 267 | CGG>CGT | Arg>Arg | No | change | S10 | NA |
| 86 | 5 | 166 | 7 base
deletion | Frameshift
mutation | NA | L2 | NA | NA |
| 88 | 7 | 251 | ATC>ATT | Lle>Lle | No change | S9 | NA | NA |
| 92 | 8 | 303 | AGC>AAC | Ser>Asn | No change | C-term | F | N |
| Other
carcinomas | 1 | 8 | 271 | GAG>TTG | Glu-Val |
Acid>hydrophobic | S10 | NF | D |
| 2 | 8 | 271 | GAG>TTG | Glu-Val |
Acid>hydrophobic | S10 | NF | D |
| 5 | 7 | 241 | 1 base insert | Frameshift
mutation | / | L3 | NA | NA |
| 8 | 5 | 168 | CAC>CCC | His>Pro |
Alkaline>hydrophobic | L2 | NF | D |
| 9 | 7 | 245 | GGC>GAC | Gly>Asp |
Hydrophobic>acid | L3 | NF | D |
| 11 | 8 | 274 | GTT>TTT | Val>Phe | No change | LSH | NF | D |
| 14 | 6 | 219 | CCC>TCC | Pro>Ser |
Hydrophobic>polar | S7 | NF | D |
| 15 | 5 | 157 | 10 base
deletion | Frameshift
mutation | NA | S4 | NA | NA |
| 8 | 271 | GAG>TTG | Glu-Val |
Acid>hydrophobic | S10 | NF | D |
| 16 | 8 | 271 | GAG>TTG | Glu-Val |
Acid>hydrophobic | S10 | NF | D |
| 17 | 5 | 146 | TGG>TGA | Trp-stop | NA | S3 | ND | ND |
| 22 | 7 | 248 | CGG>CAG | Arg>Gln |
Alkaline>polar | L3 | NF | D |
| Other | 27 | 6 | 220 | TAT>TGT | Tyr>Cys | No change | S | NF | D |
| 7 | Intron | GGTCA>GGCCA | NA | NA | NA | NA | NA |
| 30 | 8 | 286 | GAA>TAA | Glu>stop | NA | LSH | ND | ND |
Based on the structure domains of p53 described in
the IARC p53 database, 6.7% (3/45) of the mutations affected the
LSH (loop-sheet-helix) motif (codons 119–135 and 272–287), 11.1%
(5/45) affected the L2 domain (between codons 164 and 194), which
was needed for the correct folding and stabilization of the central
part of the protein, and 20% (9/45) affected the L3 domain (between
codons 237 and 250), directly involved in the interaction between
the protein and DNA.
With the help of the protocol described by Kato
et al(13) and an online
software, the possible influences of the identified TP53
mutations on p53 protein function were analyzed and are summarized
in Table III. Notably, the two
methodologies revealed similar results. Among the 36
point-mutations, 25 were non-functional mutations that will abolish
the normal function of p53. Four point-mutations were neutral ones
that will not affect the p53 function. Other 5 point-mutations were
stop codon mutations, which were not available by the two methods.
Only 2 point-mutations (at codons 47 and 207) showed different
consequences on p53 function by the two techniques. Additionally,
there were 3 different frameshift insertions whose effects on p53
function were also unable to be predicted by the two
techniques.
The numbers of patients with mutations in the
different exons of the TP53 gene varied largely. Two
patients (2/92, 2.2%) contained mutations in exon 4, 10 (10/85,
11.8%) in exon 5, 4 (4/92, 4.3%) in exon 6, 14 (14/89, 15.7%) in
exon 7 and 13 (13/92, 14.1%) in exon 8. Although various mutation
rates were detected in exons, there was no significant difference
in TP53 mutations between the laryngeal carcinoma group and
the other HNSCC group.
Correlation of the TP53 mutation status
with IHC for p53 protein
The expression profiles of p53 protein in sections
of the tumor samples were analyzed by p53-specific IHC. Out of 93
tested primary HNSCCs, 34 showed positive staining for p53 protein.
The brown color staining was distributed prominently in the
malignant cells, and was localized in the cell nuclei (Fig. 1). Based on the semi-quantitative
protocol for p53 immunostaining, the expression profiles of p53
were classified into strongly positive, weakly positive and
negative. Twenty-two of the 34 (64.7%, 22/34) p53-positive cases
were characterized as strongly positive, while 12 (35.3%, 12/34)
were weakly positive. In the group of laryngeal carcinomas, 12 of
64 (18.8%) cases were strongly positive for p53, 8 cases were
weakly positive (12.5%) and 44 were negative (68.8%). In the other
HNSCC group, 10 of 29 (34.5%) cases were strongly positive for p53,
4 cases were weakly positive (13.8%) and 15 were negative (51.7%).
Despite a higher frequency of cases with strongly positivity for
p53 in the other HNSCC group, no statistical difference in p53
expression profile was observed between the two groups.
In the cases with strong positivity for p53, 63.6%
(14/22) of the cases contained mutation(s) in the TP53 gene,
while 33.3% (4/12) of the cases with weak positivity for p53
presented with TP53 mutations. Further analyses revealed
that mutations in exon 5 (P<0.05) and exon 7 (P<0.01) had a
significant correlation with p53 positivity in the IHC assays in
the laryngeal carcinoma group, as well as in the context of all
tested patients (P<0.05), but not in that of the other HNSCC
group (Table IV). No significant
difference between the mutations in the other exons of TP53
and positivity for p53 in IHC was noted in all the tested groups
(Table IV).
 | Table IVRelationship between mutations in
TP53 exon 4–8 and p53 IHC. |
Table IV
Relationship between mutations in
TP53 exon 4–8 and p53 IHC.
| | p53 IHC |
|---|
| |
|
|---|
| | All samples | Larynx | Other HNSCCs |
|---|
| |
|
|
|
|---|
| | Pos (%) | Neg (%) | Pos (%) | Neg (%) | Pos (%) | Neg (%) |
|---|
| Exon 4 | Pos (%) | 2/92 (2.2) | 0/92 (0.0) | 2/63 (3.2) | 0/63 (0.0) | 0/29 (0.0) | 0/29 (0.0) |
| Neg (%) | 32/92 (34.8) | 58/92 (63.0) | 18/63 (28.6) | 43/63 (68.2) | 14/29 (48.3) | 15/29 (51.7) |
| Exon 5 | Pos (%) | 7/85 (8.2)b | 3/85 (3.5) | 5/57 (8.8)b | 2/57 (3.5) | 2/28 (7.1) | 1/28 (3.6) |
| Neg (%) | 26/85 (30.6) | 49/85 (57.7) | 14/57 (24.5) | 36/57 (63.2) | 12/28 (42.9) | 13/28 (46.4) |
| Exon 6 | Pos (%) | 2/92 (2.2) | 2/92 (2.2) | 1/63 (1.6) | 1/63 (1.6) | 1/29 (3.4) | 1/29 (3.4) |
| Neg (%) | 32/92 (34.8) | 56/92 (60.8) | 19/63 (30.1) | 42/63 (66.7) | 13/29 (44.9) | 14/29 (48.3) |
| Exon 7 | Pos (%) | 9/89 (10.1)b | 5/89 (5.6) | 7/60 (11.7)a | 3/60 (5.0) | 2/29 (6.9) | 2/29 (6.9) |
| Neg (%) | 23/89 (25.9) | 52/89 (58.4) | 11/60 (18.3) | 39/60 (65.0) | 12/29 (41.4) | 13/29 (44.8) |
| Exon 8 | Pos (%) | 5/92 (5.4) | 8/92 (8.7) | 2/63 (3.2) | 5/63 (7.9) | 3/29 (10.3) | 3/29 (10.3) |
| Neg (%) | 29/92 (31.5) | 50/92 (54.4) | 18/63 (28.6) | 38/63 (60.3) | 11/29 (38.0) | 12/29 (41.4) |
Correlation of p53 positivity and HPV E6
positivity in the IHC assays
Our previous study of the 93 patients with head and
neck carcinomas demonstrated that 29 samples were HPV16/18
E6-positive in the IHC assays: 18 (18/64, 28.1%) in the laryngeal
carcinoma group and 11 (11/29, 37.9%) in the other HNSCC group
(12). Further analyses of the
possible correlation between HPV E6 positivity and p53 positivity
in the tumor tissues showed that in the laryngeal carcinoma group,
an equal amount of HPV E6-positive cases were p53 positive, while a
higher percentage of HPV E6-negative cases were p53 negative
(Table V). In the group of other
HNSCCs, markedly fewer numbers of patients with HPV E6 positivity
were p53 positive, while a relatively higher percentage of HPV
E6-negative cases were p53 positive (Table V). Moreover, the correlation between
HPV DNA and p53 protein expression was analyzed, and no difference
was found in the laryngeal carcinoma group, the other HNSCCs and
all the patients tested.
 | Table VRelationship between HPV IHC and p53
IHC. |
Table V
Relationship between HPV IHC and p53
IHC.
| | p53 IHC |
|---|
| |
|
|---|
| | All samples | Larynxa | Other
HNSCCsb |
|---|
| |
|
|
|
|---|
| | Pos (%) | Neg (%) | Pos (%) | Neg (%) | Pos (%) | Neg (%) |
|---|
| HPV IHC | Pos (%) | 12/93 (12.9) | 17/93 (18.3) | 9/64 (14.1) | 9/64 (14.1) | 3/29 (10.4) | 8/29 (27.6) |
| Neg (%) | 22/93 (23.7) | 42/93 (45.1) | 11/64 (17.1) | 35/64 (54.7) | 11/29 (37.9) | 7/29 (24.1) |
Correlation of TP53 codon 72 polymorphism
with HNSCCs
The codon 72 of the TP53 gene shows
polymorphisms among human beings. Sequencing assays of the tumor
samples of the 92 HNSCCs found that the numbers of Arg/Arg
homozygotes, Pro/Pro homozygotes and Arg/Pro heterozygotes were
29/92 (31.5%), 17/92 (18.5%) and 46/92 (50.0%), respectively. The
potential correlations of the codon 72 polymorphism with a series
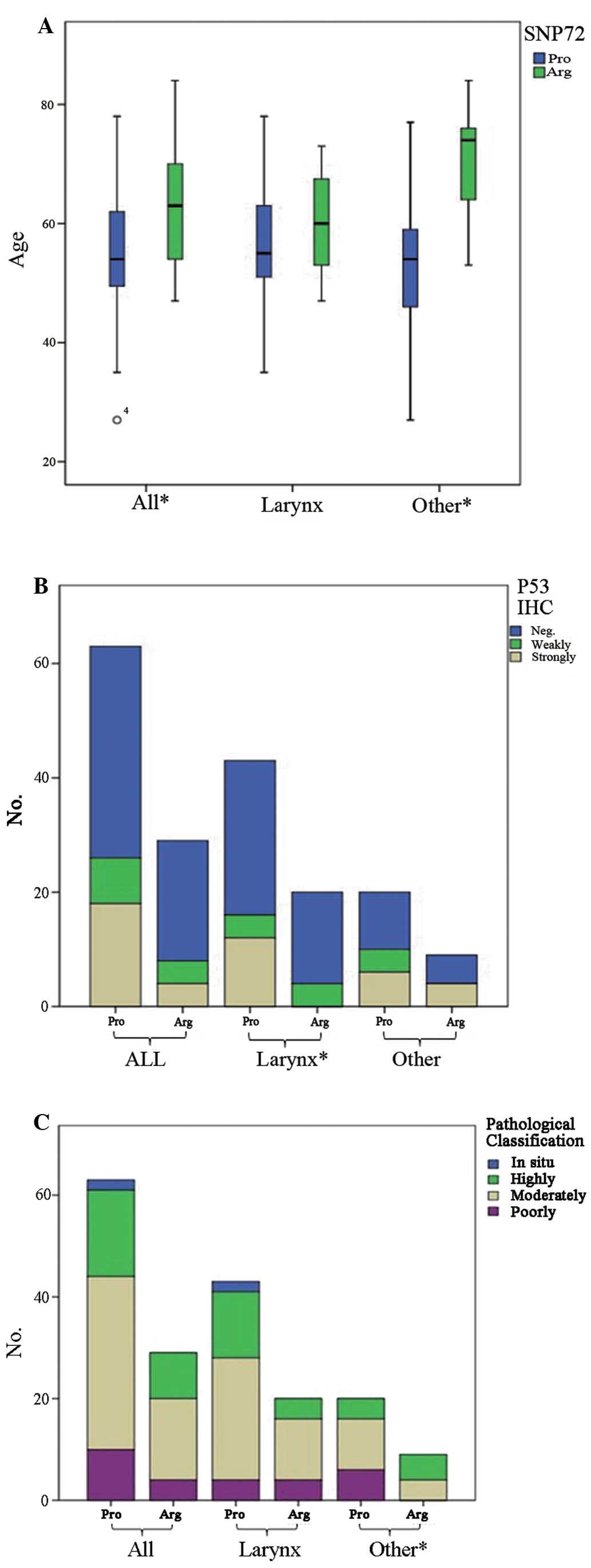
of characteristics of HNSCCs were evaluated. It demonstrated that
the average median age of disease onset (74 years) for patients
with the Arg phenotype was significantly older than the age of
disease onset (54 years) for the patient with the Pro phenotype in
the group of other HNSCCs (P<0.05), whereas the age was slightly
older but without statistical difference in the group of laryngeal
carcinomas with the Arg phenotype (60 years) compared with patients
with the Pro phenotype (55 years) (Fig.
2A). Assessment of the relationship with the pathological
features of HNSCCs revealed that all poorly differentiated SCCs in
the other HNSCC group had the Pro phenotype, while these phenotypes
in poorly differentiated SCCs in the laryngeal carcinoma group were
almost equal (Fig. 2B). Evaluation
of the linkage of codon 72 polymorphism and the p53 expression
profiles found that predominantly more HNSCC cases with the Pro
phenotype had strong staining for p53 in the tumor tissues,
particularly the laryngeal carcinomas in which all p53 strongly
stained cases had the Pro phenotype (Fig. 2B). In contrast, the codon 72
polymorphism showed a weak correlation with the p53 expression
profile in the other HNSCC group, as a similar frequency of cases
with the Pro phenotype was observed in the cases with strong and
weak positivity for p53. The pathological features of the
carcinomas were classified into 4 degrees including in situ,
highly differentiated, moderately differentiated and weakly
differentiated cases. In the group of other HNSCCs, all the tumors
with weak differentiation had the Pro phenotype, but there was no
obvious difference in pathological classification in the laryngeal
carcinoma group (Fig. 2C).
Additionally, no significant association between SNP72 polymorphism
and other factors, including HPV infection, clinical stage, tobacco
use and alcohol consumption was found (data not shown).
Discussion
It has been established that TP53, a
tumor-suppressor gene, plays a key role in organizing cellular
responses to various types of stress, including DNA damage and
oncogene activation followed by apoptosis, cell cycle arrest,
senescence, DNA repair, cell metabolism or autophagy (15). Malfunction and mutations of p53 have
been found in most types of human cancers, leading to deregulated
p53 activity of proliferation and uncontrolled survival.
Most TP53 mutations in human cancers are
missense mutations and focus on exon 5–8. In many cases, the
mutations can either cause a loss of tumor-suppressor function
(LOF) or, in some cases, a gain of oncogenic function (GOF)
(16). A total of 45 mutations in
35 cases were detected in the patients recruited in our study. The
relatively high frequency of point-mutations of p53 protein
including codons 245, 248, 266, 267, 271 and 298 in the present
study was similar with the commonly observed mutation regions of
p53. All of these mutated codons were noted in the above 2 cases.
For codon 245, all mutations, including Gly to Cys, Gly to Ser and
Gly to Asp, led to a change from hydrophobic protein to a polar or
acid one. At codon 271, 4 cases had the same mutation from Glu to
Val, resulting in a change from acid protein to a hydrophobic one.
Moreover, most of the mutations occurred in the L or S motif
structure of p53. Compared with the mutation rates reported in
other countries, the mutation rate in the present study (35/93,
37.6%) was slightly higher than the data in Indian (17,18)
HNSCC tissues which showed various TP53 mutations in 17–21%
of patients, but was slightly lower than the TP53 mutations
reported from USA, Europe and Japan ranging from 39 to 69%
(19–21).
Based on the functional assays of a published
protocol (13) and an online
software, we forecasted that most of the identified point-mutations
in TP53 (25/36) in this study p53 protein will result in
changes of wild-type p53 to a non-functional form. A few
point-mutations (4/36) are a neutral form that do not affect p53
function. Only the mutation at codon 47 in 1 case was predicted by
one technique to be able to induce a change to the supertrans form
that may result in stronger p53 activity. In addition, 5 stop codon
point-mutations and 3 different frameshift insertions, which were
not recognized with the above two methodologies, definitely
interrupt the expression of normal p53 and eventually cause p53
dysfunction. Wide distributions in the non-functional p53-related
point-mutations, as well as stop codon mutations and frameshift
insertions in the TP53 gene in HNSCC cells emphasize again
the essential role of p53 in carcinogenesis.
Normally, the half-life of wild-type p53 protein is
short which makes it difficult to be detected by
immunohistochemistry, whereas the mutated p53 protein is fairly
stable which can easily be identified in tumor cells by
immunohistochemistry (22). In
agreement with previous data, we also determined that p53
overexpression is a frequently observed event in Chinese patients
with HNSCCs (23). However, we did
not find any significant correlation between the p53 expression
profiles in the tumor tissues and a series of parameters, including
gender and age of the patients, various pathological
classifications and clinical stage. Compared with previous studies
that demonstrated that p53 overexpression is more prevalent in
laryngeal tumors than in tumors in other anatomical sites (24), our data found a relatively lower
positive rate of p53 in laryngeal tumors (31.3%) than that in other
HNSCCs (48.3%), although no statistical difference was
achieved.
HPV is another important etiologic factor, in
addition to tobacco and alcohol for HNSCCs, particularly for
oropharyngeal cancer (25). During
the past few decades, HPV DNA has been detected in ~25% of HNSCCs
overall. More importantly, 45–100% of oral SCC cases are reported
to be HPV-positive (25,26). The data in our previous study also
showed that HPV-positive rates had a significant association with
the anatomic sites of tumors (12).
In line with many published data, we also found that the
p53-positive rate of patients with HPV16/18 E6 positivity was lower
than that of the cases with HPV E6 negativity in the group of other
HNSCCs. However, this phenomenon was not observable in the patients
with laryngeal tumors, in which the p53-positive rates of the
patients with HPV16/18 E6 positivity or negativity were comparable.
These data highlight that the contrary correlation between HPV16/18
E6 positivity and p53 positivity was more likely detected in the
SCCs that occurred in the oropharyngeal site. Further statistical
analysis of the presence of p53 mutations and HPV infection or
between HPV DNA and p53 protein expression failed to reveal any
significant relationship in HNSCCs in our study. This possibly
implies that either TP53 mutation or expression of high-risk
HPV E6 may independently lead to p53 inactivation during the
pathogenesis of HNSCCs.
The codon 72 polymorphism of p53 results in a
substitution of Pro for Arg in the amino acid sequence and thus has
an impact on the binding capacity and functional properties of p53
(27). Previous reports suggest
that the p53 codon 72 polymorphism is associated with the
susceptibility to several types of cancers and the survival of
cancer patients (28,29). In the present study, we found that
the p53 Arg/Pro heterozygote was the major phenotype (49.5%) among
the cohort of 92 patients, while the homozygous phenotype Arg/Arg
and Pro/Pro accounted for a lower percentage of 31.2 and 18.3%,
respectively. The SNP72 polymorphism has not been significantly
linked with many factors, such as HPV infection, clinical stage,
tobacco use and alcohol consumption. However, the cases having the
Arg phenotype exhibited an obvious tendency to have an older age of
disease onset and a higher degree of differentiation in pathology
than the cases having Pro72. In addition, in the group of laryngeal
carcinomas, all patients showing strongly positive p53
overexpression had the Pro phenotype. These results are in
accordance with the conclusion that the wild-type Arg allele has a
greater ability to localize to mitochondria, thereby inducing
apoptosis to a greater exent than Pro72 (30,31).
It has been confirmed that Arg/Pro and Prp/Pro phenotypes of p53
codon 72 are significantly associated with an increased risk of
secondary primary malignancy (SPM) in patients with HNSCCs
(32). Whether such an association
exists in our patient cohort deserves long-term follow-up.
Acknowledgements
This study was supported by the China Mega-Project
for Infectious Disease (2011ZX10004-101, 2012ZX10004215), the Young
Scholar Scientific Research Foundation of China CDC (2012A102) and
the SKLID Development Grant (2012SKLID102 and 2011SKLID211).
Abbreviations:
|
HNSCC
|
head and neck squamous cell
carcinoma
|
|
HPV
|
human papillomavirus
|
|
SNP
|
single-nucleotide polymorphism
|
|
SNP72
|
single-nucleotide polymorphisms in
codon 72
|
|
IHC
|
immuno-histochemistry
|
|
PCR
|
polymerase chain reaction
|
|
TA class
|
trans-criptional activity class
|
|
LSH
|
loop-sheet-helix
|
|
LOF
|
loss of tumor-suppressor function
|
|
GOF
|
gain of oncogenic function
|
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Gillison ML, Koch WM, Capone RB, et al:
Evidence for a causal association between human papillomavirus and
a subset of head and neck cancers. J Natl Cancer Inst. 92:709–720.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sturgis EM and Cinciripini PM: Trends in
head and neck cancer incidence in relation to smoking prevalence:
an emerging epidemic of human papillomavirus-associated cancers?
Cancer. 110:1429–1435. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Z, Sturgis EM, Zhang Y, et al:
Combined p53-related genetic variants together with HPV
infection increase oral cancer risk. Int J Cancer. 131:E251–E258.
2012.PubMed/NCBI
|
|
5
|
Rautava J and Syrjänen S: Biology of human
papillomavirus infections in head and neck carcinogenesis. Head
Neck Pathol. 6(Suppl 1): S3–S15. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Braakhuis BJ, Snijders PJ, Keune WJ, et
al: Genetic patterns in head and neck cancers that contain or lack
transcriptionally active human papillomavirus. J Natl Cancer Inst.
96:998–1006. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Smeets SJ, Braakhuis BJ, Abbas S, et al:
Genome-wide DNA copy number alterations in head and neck squamous
cell carcinomas with or without oncogene-expressing human
papillomavirus. Oncogene. 25:2558–2564. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Whibley C, Pharoah PD and Hollstein M: p53
polymorphisms: cancer implications. Nat Rev Cancer. 9:95–107. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dumont P, Leu JI, Della Pietra AC III,
George DL and Murphy M: The codon 72 polymorphic variants of p53
have markedly different apoptotic potential. Nat Genet. 33:357–365.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Buyru N, Altinisik J, Isin M and Dalay N:
p53 codon 72 polymorphism and HPV status in lung cancer. Med Sci
Monit. 14:CR493–CR497. 2008.PubMed/NCBI
|
|
11
|
Storey A, Thomas M, Kalita A, et al: Role
of a p53 polymorphism in the development of human
papillomavirus-associated cancer. Nature. 393:229–234. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wei W, Shi Q, Guo F, et al: The
distribution of human papillomavirus in tissues from patients with
head and neck squamous cell carcinoma. Oncol Rep. 28:1750–1756.
2012.PubMed/NCBI
|
|
13
|
Kato S, Han SY, Liu W, et al:
Understanding the function-structure and function-mutation
relationships of p53 tumor suppressor protein by high-resolution
missense mutation analysis. Proc Natl Acad Sci USA. 100:8424–8429.
2003. View Article : Google Scholar
|
|
14
|
SIFT Journal. http://sift.bii.a-star.edu.sg.
|
|
15
|
Tornesello ML, Buonaguro L and Buonaguro
FM: Mutations of the TP53 gene in adenocarcinoma and
squamous cell carcinoma of the cervix: a systematic review. Gynecol
Oncol. 128:442–448. 2013.
|
|
16
|
Hollstein M, Moriya M, Grollman AP and
Olivier M: Analysis of TP53 mutation spectra reveals the
fingerprint of the potent environmental carcinogen, aristolochic
acid. Mutat Res. Feb 17–2013.(Epub ahead of print). View Article : Google Scholar
|
|
17
|
Kannan K, Munirajan AK, Krishnamurthy J,
et al: Low incidence of p53 mutations in betel quid and
tobacco chewing-associated oral squamous carcinoma from India. Int
J Oncol. 15:1133–1136. 1999.
|
|
18
|
Heinzel PA, Balaram P and Bernard HU:
Mutations and polymorphisms in the p53, p21 and p16
genes in oral carcinomas of Indian betel quid chewers. Int J
Cancer. 68:420–423. 1996.PubMed/NCBI
|
|
19
|
Saranath D, Tandle AT, Teni TR, et al: p53
inactivation in chewing tobacco-induced oral cancers and
leukoplakias from India. Oral Oncol. 35:242–250. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lukas J, Bohr VA and Halazonetis TD:
Cellular responses to DNA damage: current state of the field and
review of the 52nd Benzon Symposium. DNA Repair. 5:591–601. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kilpivaara O and Aaltonen LA: Diagnostic
cancer genome sequencing and the contribution of germline variants.
Science. 339:1559–1562. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bosch FX, Ritter D, Enders C, et al: Head
and neck tumor sites differ in prevalence and spectrum of
p53 alterations but these have limited prognostic value. Int
J Cancer. 111:530–538. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou W, Ma Y, Yang H, Ding Y and Luo X: A
label-free biosensor based on silver nanoparticles array for
clinical detection of serum p53 in head and neck squamous cell
carcinoma. Int J Nanomedicine. 6:381–386. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Peltonen JK, Helppi HM, Pääkkö P,
Turpeenniemi-Hujanen T and Vähäkangas KH: p53 in head and neck
cancer: functional consequences and environmental implications of
TP53 mutations. Head Neck Oncol. 2:362010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Betiol J, Villa LL and Sichero L: Impact
of HPV infection on the development of head and neck cancer. Braz J
Med Biol Res. 46:217–226. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tribius S and Hoffmann M: Human papilloma
virus infection in head and neck cancer. Dtsch Arztebl Int.
110:184–190. 2013.PubMed/NCBI
|
|
27
|
Walter SD, Riddell CA, Rabachini T, Villa
LL and Franco EL: Accuracy of p53 codon 72 polymorphism
status determined by multiple laboratory methods: a latent class
model analysis. PLoS One. 8:e564302013.
|
|
28
|
Chen SP, Hsu NY, Wu JY, et al: Association
of p53 codon 72 genotypes and clinical outcome in human
papillomavirus-infected lung cancer patients. Ann Thorac Surg.
95:1196–1203. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou X, Gu Y and Zhang SL: Association
between p53 codon 72 polymorphism and cervical cancer risk among
Asians: a HuGE review and meta-analysis. Asian Pac J Cancer Prev.
13:4909–4914. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kaghad M, Bonnet H, Yang A, et al:
Monoallelically expressed gene related to p53 at 1p36, a region
frequently deleted in neuroblastoma and other human cancers. Cell.
90:809–819. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Thomas M, Kalita A, Labrecque S, Pim D,
Banks L and Matlashewski G: Two polymorphic variants of wild-type
p53 differ biochemically and biologically. Mol Cell Biol.
19:1092–1100. 1999.PubMed/NCBI
|
|
32
|
Li F, Sturgis EM, Chen X, Zafereo ME, Wei
Q and Li G: Association of p53 codon 72 polymorphism with
risk of second primary malignancy in patients with squamous cell
carcinoma of the head and neck. Cancer. 116:2350–2359.
2010.PubMed/NCBI
|