Introduction
Endometrial carcinoma is the most common malignancy
of the female reproductive tract and is the fourth most common
cancer among women in Europe (1)
and the second most common cancer among women in Slovakia (2). Contrary to its high incidence, the
mortality rate is the lowest of all gynecological malignancies
(19.5–23.7/100,000) indicating a good prognostic outcome when
detected in early stages (1).
Endometrioid carcinoma of the endometrium (EEC),
also known as type I, is the most common histological type of the
disease and accounts for ~80% of cases of endometrial carcinoma
(3). This type of the disease is
associated with an endocrine milieu of estrogen predominance and
develops from endometrial hyperplasia. The majority of cases are
presented in early stages, are usually well differentiated, are
associated with a favorable prognosis (4) compared to type II carcinomas (e.g.
papillary serous and clear cell) and are sensitive to endocrine
treatment (5). The transition from
normal endometrium to a malignant tumor is thought to involve a
stepwise accumulation of alterations in cellular mechanisms leading
to dysfunctional cell growth (6).
Type I and type II carcinoma also present different molecular
pathways in evolution.
The risk factors for EEC include obesity,
anovulatory states, early onset of menarche, late menopause,
nulliparity and exogene exposure of estrogene therapy
(hyperestrogenic status), that promote the development of
endometrial hyperplasia (mainly complex) with or without atypia
which has been proposed as a possible precursor lesion of EEC
(7,8) with progression rates into carcinoma of
up to 40% (9).
Endometrial carcinogenesis is a complex process
requiring the acquisition of genetic abnormalities in oncogenes,
tumor-suppressor genes and genes involved in DNA repair. Among such
genes, GSTP1, CDH1 (E-cadherin) and
RASSF1A are included. The GSTP1 gene is localized on
chromosome 11q13 and encodes production of
glutathion-S-transferases and plays a role in processes such as
cell metabolism, response to stress stimuli, cell proliferation,
apoptosis, carcinogenesis, response to chemotherapy, interaction
with cellular proteins and cellular detoxification (10). The CDH1 (E-cadherin) gene is
localized on chromosome 16q21 and belongs to the calcium-dependent
cell adhesion molecule family. Predominantly, it is found in
epithelial cells where it is responsible for intercellular adhesive
junctions. The loss of CHD1 expression leads to the loss of
tissue homogeneity and predisposes to early invasion and metastatic
spread of malignant cells. Thus, its downregulation is associated
with poor prognosis in many epithelial tumor types (11). The RASSF1A gene is localized
on chromosome 3p21.3 and encodes production of Ras-superfamily
GTP-ases. The Ras superfamily comprises many guanine
nucleotide-binding proteins (G proteins) that are essential to
intracellular signal transduction. These proteins act biologically
as molecular switches, which, cycling between OFF and ON states,
play a fundamental role in cell biological processes, e.g.
regulation of cell proliferation, differentiation, motility and
apoptosis (12,13).
Previously, mainly genetic mutations (PTEN,
p53 and KRAS oncogenes) have been the scope of
molecular studies; yet, recently it has become clear that
epigenetic alterations (e.g. methylation, histone deacetylation or
miRNA expression) may underline the molecular biology of
endometrial lesions (10,14). These changes are defined as
heritable alterations in gene expression without alteration of the
nucleotide sequence (15) and are
the most common molecular alterations in human neoplasias (16). Carcinogens may act by altering the
normal epigenetic control of gene activity in specialized cells,
and thereby produce aberrant heritable phenotypes (17). These phenotypes can be utilized not
only at the level of diagnosis, but also in early prevention
(18). Moreover, epigenetic changes
are dynamic and modifiable upon treatment with pharmacological
agents; thus, potential targets to halt carcinogenesis for example
by using histone deacetylase inhibitors (HDACIs) and/or DNA
methyltransferase inhibitors (DNMTIs) (19,20)
have been studied.
While epigenetics refers to broad changes in several
types of malignancies, including gynecological (21–23),
we focused on the role of DNA methylation in relation to
endometrial carcinogenesis. We aimed to investigate the aberrant
methylation of CpG islands within the promoter regions of three
tumor-suppressor genes, GSTP1, CDH1 and
RASSF1A, in endometrioid endometrial carcinomas and
endometrial complex hyperplasias in order to define the frequency
of the epigenetic alterations in comparison to healthy controls and
to determine the possible impact on the disease histological
pattern.
Materials and methods
Patient population
This was a prospective study enrolling initially a
total of 92 subjects referred to the Department of Obstetrics and
Gynecology, Jessenius Faculty of Medicine, Comenius University,
Bratislava, Slovak Republic for surgical treatment due to uterine
pathology. All participants were of Caucasian race and residents of
the geographic area of Slovakia. After initial consultation, all
subjects signed an informed consent and subsequently underwent
biological sample collection during surgery (hysteroscopy,
hysterectomy, uterine curettage) or by a pathologist during
procurement of a frozen section in case of known endometrial
malignancy. The retrieved tissue samples, sized 3–5 × 5 × 3–5 mm,
and the obtained tissue samples were immediately placed in plastic
tubes with mRNA stable solution and stored frozen at −20°C for
later epigenetic analysis. Exclusion criteria were the history of
previous endometrial surgery, history of a previous gynecological
malignancy, synchronous malignancy and cases with an endometrial
malignancy other than endometrioid adenocarcinoma (e.g. serous or
clear cell type). For healthy controls, we used histologically
negative endometrial samples from paraffin-embedded tissue blocks
from cases operated on for benign uterine fibroids. Of all the
enrolled subjects, only samples with retrieved sufficient mRNA and
later DNA were included in the final analyses: endometroid
adenocarcinoma (EEC, n=41/41), endometrial complex hyperplasia
with/without atypia (EHP, n=19/24), and cases with healthy
endometrium (controls, n=20/27). The stratification into the
observed groups (EEC and EHP) was based retrospectively according
to the histopathological report. The Regional Ethics Committee of
the Jessenius Faculty of Medicine (registered under IRB00005636 at
the Office for Human Research Protection, U.S. Department of Health
and Human Services) approved the study protocol (codes IRB
169/2011). The study was carried out in accordance with the
Declaration of Helsinki for experiments involving humans.
Histopathological analysis
Histological samples were fixed in 10% formol
solution, and assessments were performed using 4- to 5-μm
hematoxylin and eosin-stained sections of formalin-fixed,
paraffin-embedded tumors. Typing was evaluated according to the WHO
Classification of Tumours (24),
and histological grading and staging was carried out according to
the revised pTNM FIGO 2009 classification (25–27).
DNA isolation from formalin-fixed
paraffin-embedded tissue
Formalin-fixed paraffin-embedded tissue was
deparaffinized by organic solvent-xylene and a series of alcohol
solutions, and removed of their water and air-dried at room
temperature. Dry tissue was suspended in 180 μl of lysis buffer and
digested by proteinase K for 3 days at 56°C, following genomic DNA
extraction using the DNeasy blood and tissue kit (Qiagen,
Heidelberg, Germany) according to the manufacturer’s
instructions.
Genomic DNA isolation from fresh
endometrial tissue
Fresh endometrial tissue was cut into 0.3-cm thick
pieces, and each piece was sampled into a 1.5-ml tube containing
RNAlater® protect reagent (Qiagen). Tissue was stored at
4°C for one day until DNA extraction was performed. Protected
tissue was centrifuged 1 min at 14,000 rpm at 4°C. RNAlater was
removed, and one stainless steel bead 5 mm in diameter (stainless
steel beads, 5 mm; Qiagen), 300 μl of RLT buffer (Qiagen) and
β-mercaptoethanol and 1 μl Reagent DX (Qiagen) were added into each
tube. The tissue was homogenized in TissueLyser LT (Qiagen) at 50
Hz for 2 min until the tissue was completely disturbed. Homogenized
tissue was incubated at 56°C for 12 h with addition of 60 μl
proteinase K into each sample. DNA was isolated from the lysed
tissue using the DNeasy blood and tissue kit according to the
manufacturer’s instructions.
DNA quality control
DNA was eluted in 60 μl of elution buffer in both
procedures, and its quality was confirmed by electrophoretic
separation on a 1.5% agarose gel stained by ethidium bromide
(Figs. 1 and 2). DNA concentration was estimated by UV
spectrophotometry at a wavelength of 260 nm.
Bisulfite conversion
Bisulfite conversion of DNA takes advantage of the
bisulfite-mediated chemical conversion of unmethylated cytosine
residues into uracil. Methylated cytosine residues remain
unchanged. DNA methylated and unmethylated genomic regions after
bisulfite conversion can be distinguished by sequence-specific PCR
primers (28). Aliquots of 1 μg of
each DNA sample were subjected to bisulfite treatment using the
EpiTect bisulfite modification kit (Qiagen) following the
manufacturer’s protocol. Briefly, 1 μg of DNA was mixed with 85 μl
of bisulfite mix and 35 μl of DNA protect buffer in a total volume
of 140 μl. Bisulfite conversion was performed using the following
thermal cycling program: denaturation at 95°C for 5 min, incubation
at 60°C for 25 min, denaturation at 95°C for 5 min, incubation at
60°C for 85 min, denaturation at 95°C for 5 min and incubation at
60°C for 175 min. After cycling, converted DNA was purified using
an automatic preparation in QIAcube (Qiagen). Converted DNA was
subsequently eluted in 20 μl of elution buffer and stored at −20°C.
Unmethylated and methylated DNA was included in every bisulfite
treatment as a control sample.
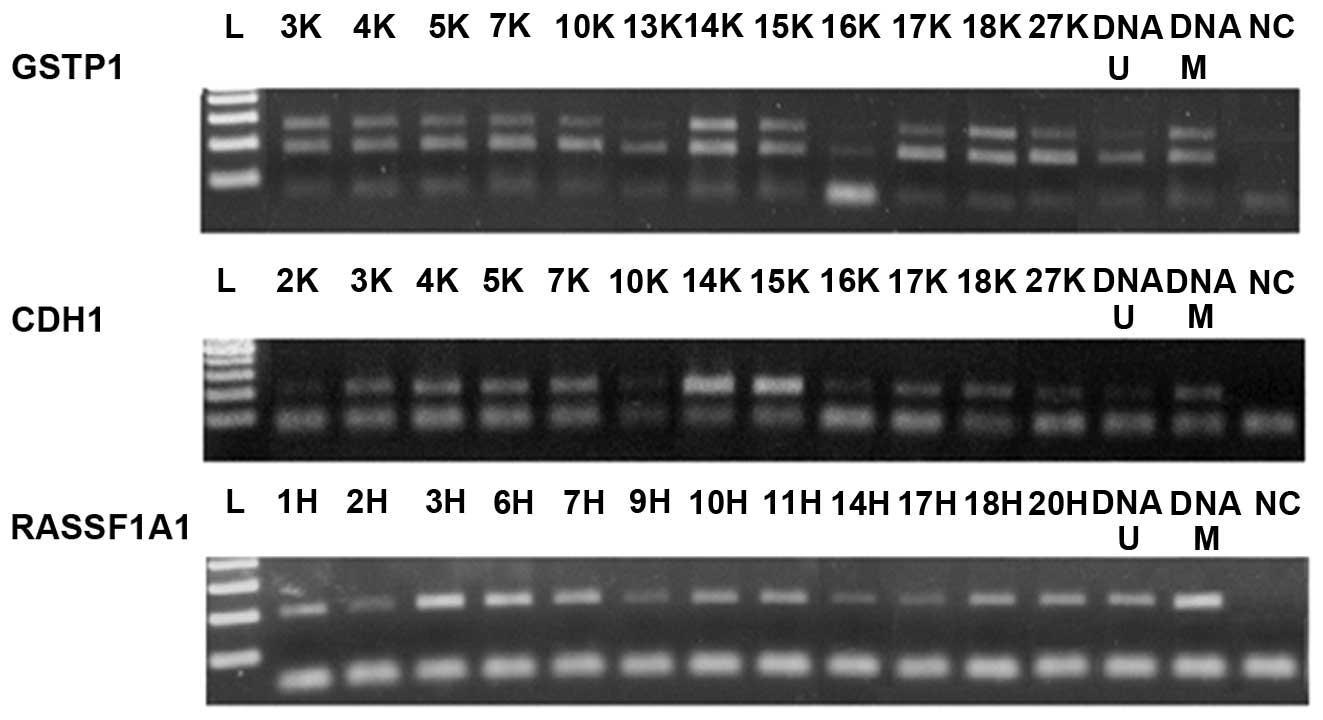
Methylation-specific nested PCR
(MSP)
Nested PCR is a two-step PCR; in the first step
(Fig. 3), PCR initially amplifies
bisulfite-modified DNA with the use of flanking PCR primers. In the
second step (Fig. 4), the amplified
external product is used as the template for the
methylation-specific PCR assay using internal methylated and
unmethylated primers. The modified DNA was subject to
methylation-specific nested PCR (N-MSP) to investigate the
methylation status of the promoter region of the GSTP1,
CDH1 and RASSF1A genes. Primer sequences, annealing
temperatures and the product lengths are listed in Table I.
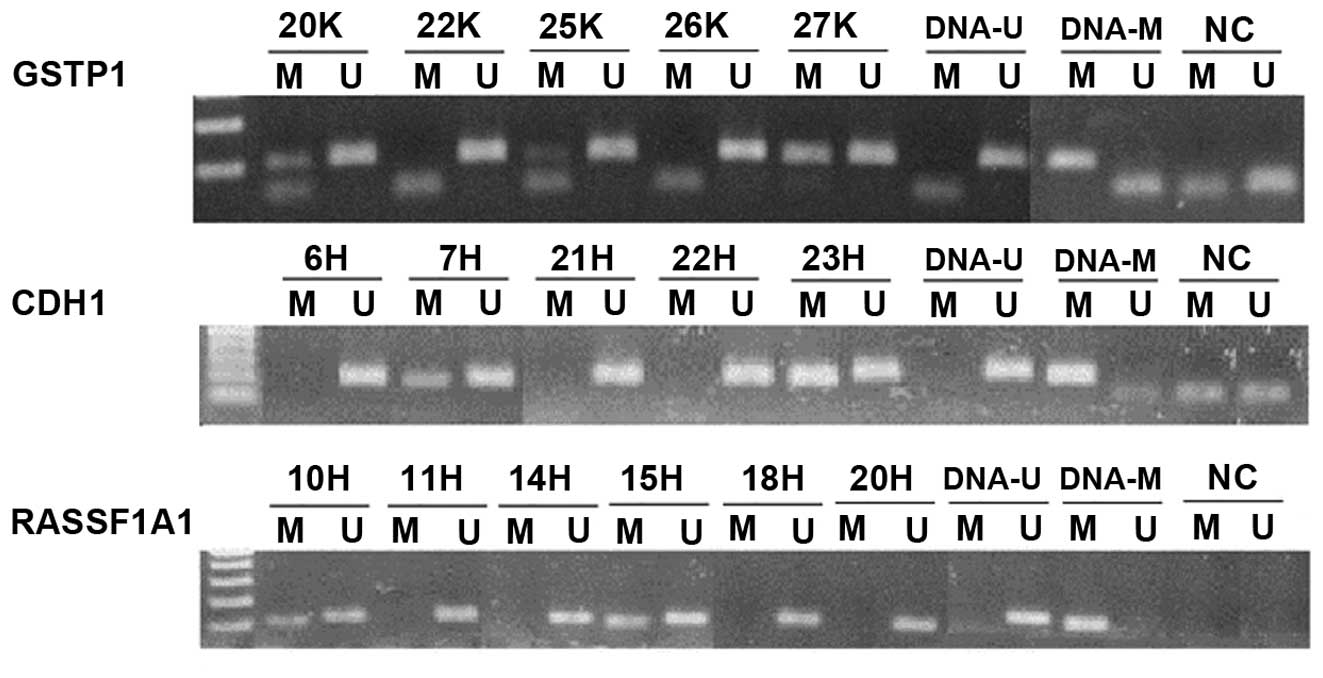 | Figure 4Representative results of the second
step of nested-MSP. In GSTP1, samples 20K, 25K, 27K were
methylated, while 22K and 26K were unmethylated in the promoter
region; in CDH1, samples 7H and 23H were methylated, while
6H, 21H and 22H were unmethylated in the CDH1 promoter
region. On the last RASSF1A1 segment, samples 10H and 15H
were methylated, while 11H, 14H, 18H and 20H were unmethylated in
the RASSF1A1 promoter region. DNA-U, positive control for
umnethylated DNA; DNA-M, positive control for methylated DNA; NC,
negative control. |
 | Table IPrimer sequences of external and
specific internal primers, annealing temperatures, sizes of the PCR
products and studies where the primers were published. |
Table I
Primer sequences of external and
specific internal primers, annealing temperatures, sizes of the PCR
products and studies where the primers were published.
| Gene | Primer type | Sequence | Annealing temp.
(°C) | Size (bp) | Study (ref.) |
|---|
| External
primers |
| GSTP1 | Forward |
5′-GGGATTTTAGGGYGTTTTTTTG-3′ | 56 | 159 | (29) |
| Reverse |
5′-ACCTCCRAACCTTATAAAAATAATCCC-3′ | | | |
| CDH1 | Forward |
5′-GTGTTTTYGGGGTTTATTTGGTTGT-3′ | 60 | 186 | (29) |
| Reverse |
5′-TACRACTCCAAAAACCCATAACTAACC-3′ | | | |
| RASSF1A | Forward |
5′-TTGAGTTGYGGGAGTTGGTATT-3′ | 56 | 210 | (30) |
| Reverse |
5′-CCCAAATAAATCRCCACAAAAAT-3′ | | | |
| Internal
methylated |
| GSTP1 | Forward |
5′-TTCGGGGTGTAGCGGTCGTC-3′ | 62 | 91 | (29) |
| Reverse |
5′-GCCCCAATACTAAATCACGACG-3′ | | | |
| CDH1 | Forward |
5′-TGTAGTTACGTATTTATTTTTAGTGGCGTC-3′ | 62 | 112 | (29) |
| Reverse |
5′-CGAATACGATCGAATCGAACCG-3′ | | | |
| RASSF1A | Forward |
5′-GTGTTAACGCGTTGCGTATC-3′ | 58 | 94 | (31) |
| Reverse |
5′-AACCCCGCGAACTAAAAACGA-3′ | | | |
| Internal
unmethylated |
| GSTP1 | Forward |
5′-GATGTTTGGGGTGTAGTGGTTGTT-3′ | 59 | 97 | (29) |
| Reverse |
5′-CCACCCCAATACTAAATCACAACA-3′ | | | |
| CDH1 | Forward |
5′-TGGTTGTAGTTATGTATTTATTTTTAGTGGTGTT-3′ | 61 | 120 | (29) |
| Reverse |
5′-ACACCAAATACAATCAAATCAAACCAAA-3′ | | | |
| RASSF1A | Forward |
5′-TTTGGTTGGAGTGTGTTAATGTG-3′ | 60 | 108 | (31) |
| Reverse |
5′-CAAACCCCACAAACTAAAAACAA-3′ | | | |
The first step of PCR was carried out in a total
volume of 25 μl per reaction, containing 1 U of FastStart Taq DNA
polymerase (Roche Diagnostics, Indianapolis, IN, USA), 2.5 μl of
10X PCR buffer, 2.5 mmol/l MgCl2, 1.0 mmol/L dNTPs (dNTP
Mix; Applied Biosystems, Foster City, CA, USA) and 10 pmol/l of
each external primer. The first step was run in a thermal cycler
using the following conditions: initial denaturation at 95°C for 5
min; then 35 cycles at 95°C for 30 sec, 56°C (GSTP1 and
RASSF1A) or 60°C (CDH1) for 30 sec and 72°C for 30
sec, and a final extension at 72°C for 5 min. The first step PCR
product (5 μl) was mixed with 2 μl of 6X DNA loading dye
(Fermentas, Germany) and analyzed on 1.75% agarose gel
electrophoresis and visualized by ethidium bromide staining.
First step PCR products were diluted 500-fold, and 2
μl was added to a second PCR in a 25 μl volume, with primers
specific for methylated or unmethylated alleles. The second step
PCR comprised 30 cycles for all genes at 95°C for 30 sec, annealing
at 62°C (GSTP1 and CDH1-methylated), 61°C
(CDH1-unmethylated), 60°C (RASSF1A1-unmethylated),
59°C (GSTP1-unmethylated) or 58°C
(RASSF1A1-methylated) for 30 sec and extension at 72°C for
30 sec. Five microliters and 2 μl of 6X DNA loading dye were loaded
onto 1.75% agarose gel and visualized by ethidium bromide staining
(Fig. 4).
Statistical analysis
We used descriptive statistics expressed as means ±
standard deviation (±SD) or as a number (percentage) to provide a
summary of the data for continuous and categorical variables,
respectively. The homogeneity of the studied groups was assessed
using the Student’s t-test. For statistical analysis between the
CpG methylation and histological findings, the Chi-square
(χ2) test was used based on Pearson’s distribution. The
correlations between tumor-suppressor gene promoter methylation and
histopathological variables were assessed using Pearson’s
correlation coefficient. The trendlines were achieved by applying
linear regression model analysis. The statistical level of
significance was set to P≤0.05. All statistical calculations were
performed by MedCalc 11.1 (MedCalc Software Inc., Mariakerke,
Belgium) software for Windows.
Results
A total of 92 subjects was initially included in the
study of which 41 EEC, 19 EHP and 20 controls were processed for
final analyses due to sufficient DNA extraction and quality. The
mean age of the EEC patients was 63.1 (±9.3) years, the mean age of
the EHP patients was 52.8 (±9.2) years and 48.7 (±11.1) years for
the controls (P<0.0001). There was no significant difference in
onset of menarche, parity, history of oral contraceptives and
hormonal replacement therapy and smoking among the patients and
controls, thus, proving homogeneity of the studied cohort samples
(Table II).
 | Table IIDemographics and clinical features of
the studied groups. |
Table II
Demographics and clinical features of
the studied groups.
| Features | EEC (n=41) | EHP (n=24) | Controls
(n=27) | P-value |
|---|
| Mean age
(years) | 63.1 | 52.8 | 48.7 | 0.0001 |
| Onset of menarche
(years) | 13.5 | 13.3 | 12.9 | NS |
| Parity | 2.3 | 2.0 | 2.5 | NS |
| Smoking (%) | 60.3 | 39.7 | 44.9 | NS |
| History of OC
(%) | 14.6 | 12.5 | 11.1 | NS |
| History of HRT
(%) | 4.8 | 4.1 | 3.7 | NS |
A significant difference was found between the
studied groups and the presence of the promoter CpG hypermethylated
status of the GSTP1 (P<0.05) and RASSF1A
(P<0.0001) genes. Promoter methylation of the RASSF1A,
GSTP1 and CDH1 genes was present in 85.4, 68.3 and
31.4% of EEC samples when compared to the controls with 30.0, 35.0
and 20.0%, respectively (Table
III) and was in line with previously published studies
(Table IV). The CpG methylation
status of all three investigated tumor-suppressor genes was found
in 12.2% of EEC patients, in 4.2% of EHP patients and in 3.7% of
the controls, respectively. The positive findings for promoter
methylation in two investigated genes were revealed in 48.7% of EEC
patients, 26.0% of EHP and in 18.5% controls.
 | Table IIIPresence of CpG promoter
hypermethylation in tumor-suppressor genes according to
histopathology. |
Table III
Presence of CpG promoter
hypermethylation in tumor-suppressor genes according to
histopathology.
| CpG methylation
positivity | EEC (n=41) | EHP (n=19) | Controls
(n=20) | P-value |
|---|
| GSTP1
methylated | 68.3 | 52.6 | 35.0 | |
| GSTP1
unmethylated | 31.7 | 47.4 | 65.0 | <0.05 |
| CDH1
methylated | 31.4 | 21.1 | 20.0 | |
| CDH1
unmethylated | 68.6 | 78.9 | 80.0 | NS |
| RASSF1A
methylated | 85.4 | 36.8 | 30.0 | |
| RASSF1A
unmethylated | 14.6 | 63.2 | 70.0 | <0.0001 |
 | Table IVAnalysis of previously published data
concerning methylation profiles in endometrioid endometrial cancer
(EEC). |
Table IV
Analysis of previously published data
concerning methylation profiles in endometrioid endometrial cancer
(EEC).
| Cases with CpG
island promoter methylation in EEC |
|---|
|
|
|---|
| RASSF1A | GSTP1 | CDH1 |
|---|
| Pallarés et
al(34) | 74.0 | - | - |
| Pijnenborg et
al(41) | 85.0 | - | - |
| Seeber et
al(40) | 79.0 | 15.0 | - |
| Arafa et
al(38) | 74.0 | - | - |
| Liao et
al(39) | 61.5 | - | - |
| Kang et
al(36) | 81.0 | - | 42.9 |
| Nieminen et
al(57) | 54.0 | - | 15.0 |
| Di Domenico et
al(14) | - | - | 61.5 |
| Banno et
al(48) | - | - | 14.0 |
| Saito et
al(47) | - | - | 37.8 |
| Moreno-Bueno et
al(46) | - | - | 21.2 |
| Chan et
al(55) | - | 30.9 | - |
| Fiolka et al
(present study) | 85.4 | 68.3 | 31.4 |
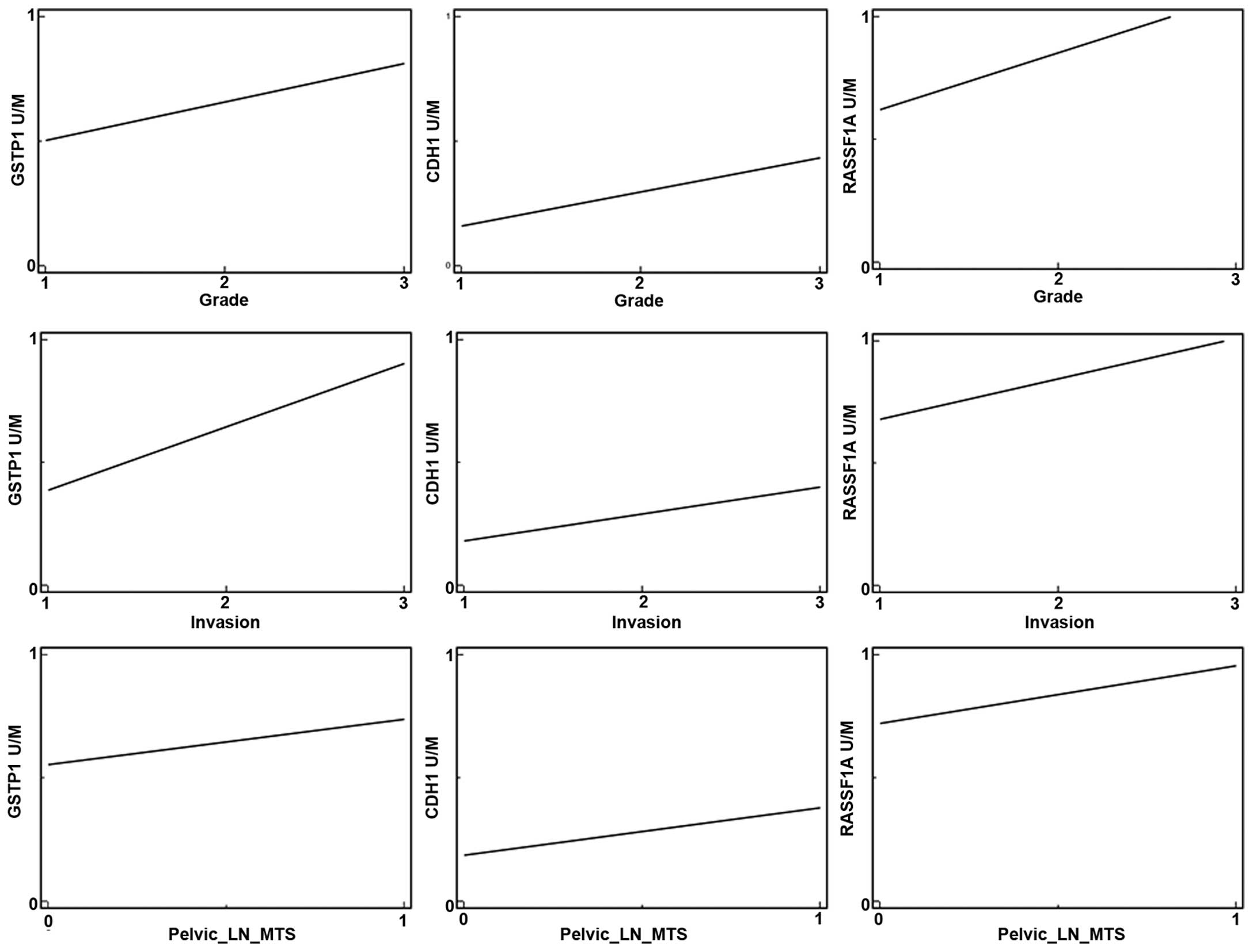
In regards to the histopathological variables and
CpG methylation in the promoter region of the tumor-suppressor
genes we found significant correlations between the RASSF1A
and GSTP1 genes and higher tumor grade, myometrial invasion
and positive metastatic involvement of pelvic lymph nodes (Table V). No associations were noted
between promoter hypermethylation of the CDH1 gene and the
biological features of endometrial cancer; however, a trend of
higher tumor grade, deeper myometrial invasion and metastatic
spread to pelvic lymph nodes was observed (see regression lines of
positive correlations) (Fig.
5).
 | Table VAssociations between biological
features of EEC and promoter CpG methylation of tumor-suppressor
genes. |
Table V
Associations between biological
features of EEC and promoter CpG methylation of tumor-suppressor
genes.
| CpG methylation
positivity | GSTP1 | CDH1 | RASSF1A |
|---|
| Tumor grade | r=0.3875 | r=0.2301 | r=0.5199 |
| p=0.0123 | p=0.1708 | p=0.0005 |
| Myometrial
invasion | r=0.4325 | r=0.1849 | r=0.3727 |
| p=0.0047 | p=0.2732 | p=0.0164 |
| Metastatic pelvic
lymph nodes | r=0.3044 | r=0.1589 | r=0.3211 |
| p=0.0530 | p=0.3477 | p=0.0407 |
Discussion
There is no effective screening or diagnostic tool
for the prevention of endometrial cancer; thus, its incidence is
rising when compared to other gynecological malignancies, e.g.
cervical cancer. In contrast, common risk and predisposing
epidemiological and histopathological factors associated with the
development of uterine carcinomas have been identified. Based on
these factors, we are able to select the women at a higher risk for
disease origin and offer them increased attention. Moreover,
scientists are still in search for new screening methods toward the
aim of detecting premalignant at risk lesions or early stages of
the disease. In the present study, we took advantage of the
techniques of epigenetics and proteomics with their high
sensitivity and specificity in the detection of human malignancies
and their new therapeutic approaches (32,33).
The most common epigenetic alterations analyzed in human cancers
are CpG promoter methylation, histone deacetylation and miRNA
expression. Epigenetic inactivation is defined as a change imposed
onto the functionality of a gene that does not involve alteration
of its coding sequence. Transcriptional silencing by
hypermethylation of CpG islands in the promoter regions of
tumor-suppressor genes has become recognized as a common phenomenon
in carcinogenesis, including endometrial cancer (34).
As endometrial carcinogenesis is a multistep process
involving precursor lesions, the aim of the present study was to
analyze the methylation frequency of three tumor-suppressor genes
(RASSF1A, CDH1/E-cadherin and GSTP1) in
endometrioid endometrial carcinomas, complex endometrial
hyperplasias and in healthy endometrium with an aim to find the
potential value for early cancer diagnosis and the association with
the clinicopathological pattern of the disease. The results
indicated that the pattern of gene promoter methylation was
associated with the biological aggressiveness of the carcinoma and
that the frequency of methylated genes progressively increased with
the type of histological features from normal endometrium to
endometrial hyperplasia and endometrioid carcinomas. Thus, the
panel of examined genes as well as the frequency of the methylation
of these genes may be useful to distinguish between EHP and EEC,
and low vs. high-grade carcinomas in daily pathology practice.
Moreover, it can be useful for oncologists for assessment of the
prognosis of EEC.
In the present study, a significant difference was
found between the studied groups and the presence of promoter CpG
hypermethylation status in the RASSF1A and GSTP1
genes. Promoter methylation of the RASSF1A, GSTP1 and
CDH1 genes was present in 85.4, 68.3 and 31.4% of EEC
samples when compared to the controls with 30.0, 35.0 and 20.0%,
respectively. The CpG methylation in all three investigated
tumor-suppressor genes was noted in 12.2% of EEC patients, in 4.2%
of EHP patients and in 3.7% of the controls, respectively. Positive
findings for promoter methylation in two investigated genes was
noted in 48.7% of EEC patients, 26.0% of EHP patients and in 18.5%
of the controls.
The first report of RASSF1A methylation in
association with endometrial cancer was published in 2004 by Fiegl
et al(35) in a sample of 15
patients aimed for detection of endometrial cancer using epigenetic
markers. Later, several studies focused on this epigenetic event in
endometrial carcinomas as well as in hyperplasias (34,36–40).
Similar to our study, Seeber et al(40) analyzed the promoter hypermethylation
of the RASSF1A and GSTP1 genes in endometrial
carcinomas and detected 79 and 15% methylation positivity for the
observed genes and a significantly higher cumulative methylation
index of tumor-suppressor genes in EC type I compared to type II.
Similarly, RASSF1A was shown to be frequently (74%)
methylated in EEC also in another study where it was a common
finding in advanced-stage disease (34). The high frequency of methylation of
the RASSF1A gene in EEC and atypic EHPs was revealed also by
Arafa et al(38) where a
methylated promoter occurred in 74 and 50% of subjects,
respectively. No significant results were obtained for the other
genes (P16, MGMT and GSTP1). Notably, 36% of
histologically normal endometrial tissues adjacent to EEC showed
RASSF1A gene methylation, indicating that CpG promoter
region methylation is a markedly heterogeneous process, even in the
absence of morphological or other molecular alterations which may
point to the active cancerization process in the surrounding
endometrium region. The 70 and 50% CpG promoter methylation
positivity in the RASSF1A gene in EEC and EHP was detected
also by Pijnenborg et al(41). Collectively, these studies confirmed
a high frequency (33% up to 85%) of CpG promoter methylation of the
RASSF1A gene in endometrial carcinomas. Moreover, this
epigenetic alteration showed different frequencies according to the
type of disease, with a higher incidence in endometrioid compared
to serous or clear cell carcinomas (39,40).
Furthermore, our association analysis demonstrated
that hypermethylation of CpG islands was correlated with
clinicopathological parameters (tumor grade, myometrial invasion
and nodal involvement). There were significant correlations between
the RASSF1A and GSTP1 genes and higher tumor grade,
deeper myometrial invasion and positive metastatic involvement of
pelvic lymph nodes. Similar results were found by Liao et
al(39) who detected higher
RASSF1A hypermethylation in type I endometrioid EC compared
to type II carcinomas, advanced stage and myometrial invasion. The
advanced stage of the disease (FIGO stage III, IV), metastatic
lymph node involvement, and high grade (G3) were more frequent in
patients with RASSF1A hypermethylation than in those without
as revealed in a study by Jo et al(42). Thus, this epigenetic event has the
potential to be used as a molecular marker for cancer diagnosis and
prognosis. Based on these findings, there is increased importance
of CpG promoter methylation of the RASSF1A gene for
clinicians due to its potential association with survival outcome.
There is research where this epigenetic event was reported to be
associated with poor survival showing higher incidence of
recurrences (77.8%) and lower disease-free survival (DFS) (97.0%)
at 5 years for methylated and unmethylated patients (42). However, in another study this
association was controversial (41). Nevertheless, the positive prognostic
outcome is augmented by the findings that EC cells with
RASSF1A promoter hypermethylation treated with
5-aza-2-deoxycytidine demonstrated reexpression and demethylation
of the promoter region of RASSF1A. This suggests that
aberrant hypermethylation of this gene is directly responsible for
transcriptional inactivation of its expression in EC cell lines
(43,44) and its restriction may improve the
survival in EC patients. Moreover, RASSF1A hypermethylation
was found to be significantly associated with microsatellite
instability in endometrial carcinomas and loss of heterozygosity in
cervical cancers; thus, we could block the increasing rate of
genetic abnormalities in uterine carcinogenesis (43,45).
A few studies have evaluated the promoter
methylation of CDH1/E-cadherin, a possible tumor-suppressor
gene, in endometrial cancer (14,46–49).
In the present study, we detected CpG promoter methylation of this
gene in 31.4% of EEC samples, 21.1% of EHP cases and in 20.0% of
cases with healthy endometrium. No associations were found between
promoter hypermethylation of the CDH1 gene and biological
features of endometrial cancer, and there was no difference across
histologic types of the disease (EEC vs. EHP vs. healthy control
endometrium). A higher methylation rate of the CDH1 gene
(42.9% in EEC) was detected by Kang et al(36) who also described the high frequency
of this event in cervical squamous cell carcinoma (80.6%). The
association between CDH1 promoter hypermethylation and
endometrial cancer was also analyzed by Yi et al(49) who found that the hypermethylation of
the CDH1 promoter, which caused low expression of E-cadherin
in endometrial cancer, was associated with not only
clinicopathological progression of endometrial cancer but also with
the overall 5-year clinical survival rate. The findings provide a
potential therapeutic and prognostic target molecule for patients
with endometrial cancer. Furthermore, it has been suggested that
epigenetic change in E-cadherin expression could allow dissociation
of individual cells from the primary tumor mass to facilitate
invasion or metastasis (50). The
positive associations between E-cadherin promoter
methylation, higher tumor grade, myometrial invasion and
involvement of pelvic lymph nodes revealed by Saito et
al(47), suggest a possible
role of CDH1 in endometrial cancer progression. However,
additional studies did not confirm this and found that CDH1
promoter hypermethylation, noted in 21.2% of endometrial
carcinomas, was not associated with clinicopathological or
immunohistochemical variables (46,51).
Moreover, controversial findings of this epigenetic event in
endometrial carcinogenesis were presented by Pijnenborg et
al(37) who did not find
CDH1 gene promoter methylation in the tested endometrial
tumors, although the absence of E-cadherin expression was detected
and found to be associated with the development of distant
metastases. Although the role of CDH1 promoter methylation
in endometrial carcinogenesis must be analyzed in further studies,
similar to hypermethylation of the RASSF1A gene, CHD1
hypermethylation can be modified by DNA methyltransferases. It was
shown that CpG methylation in the promoter region of the
E-cadherin gene and induction of E-cadherin after
treatment with the DNA methyltransferase inhibitor 5-azacytidine in
human cancer cell lines lacked E-cadherin expression (52). In other words, the positive
demethylation effect of DNMTIs and HDACIs on CDH1 promoter
methylation was observed and resulted in the suppression of growth
of endometrial cancer cells (20,53).
CDH1 suppressor gene-mediated invasion in human
cancer is silenced by an epigenic mechanism (54).
The frequency of CpG methylation in the promoter of
the GSTP1 gene in endometrial cancer was reported by Chan
et al(55) who found a
frequency of 30.9% and Seeber et al(40) who found a 15% rate for this
epigenetic alteration. In the present study, we detected a
frequency of 68.3 and 52.6% of CpG methylation in EEC and EHP
samples with significant correlations to tumor aggressiveness.
GSTP1 promoter methylation and linkage to tumor biologic
patterns was the scope of the study of Chan et al(55), who revealed that the extent of
myometrial invasion was significantly correlated with both the
methylation status and the protein expression of the GSTP1
gene. Moreover, they postulated that hypermethylation of the
GSTP1 gene promoter region may act as a dynamic regulation
mechanism contributing to reduced GSTP1 expression, which is
associated with the myometrial invasion potential of endometrial
carcinoma.
As evidenced from our study and the above mentioned
studies, epigenetic molecular changes are commonly present in EEC
and its precursor lesions, and these changes in endometrial tissue
can be detectable several years before endometrial carcinoma in
genetically predisposed individuals. Additionally, the recent data
confirm that the methylation profile of the peritumoral endometrium
is different from the altered molecular background of benign
endometrial polyps and hyperplasias. Therefore, these findings
suggest that the methylation of tumor-suppressor genes may clearly
distinguish between benign and malignant lesions (14) which can be utilized in wide
diagnostics or disease prevention. For example, RASSF1A
promoter methylation in cervical cell smears can predict the
presence of endometrial cancer with a 63.0% sensitivity and 96.3%
specificity (56). The clinical
importance of epigenetic abnormalities is also heightened by its
power to distinguish the biologic risk of a lesion as it has been
proven that abnormal DNA mismatch repair and methylation classify
normal endometrium and simple hyperplasia into one category and
complex hyperplasia without atypia, complex hyperplasia with
atypia, and endometrial carcinoma into another, suggesting that,
contrary to a traditional view, complex hyperplasia without atypia
and complex hyperplasia with atypia are equally important as
precursor lesions of endometrial carcinoma (57). Generally, hypermethylation of
tumor-suppressor genes can be used in the early disease detection
and prediction of the risk of malignant conversion.
In conclusion, promoter methylation of common
tumor-suppressor genes is a frequent epigenetic event in EEC and
EHP indicating their active and flexible role via control of gene
expression in early carcinogenesis. Furthermore, the high
methylation incidence of the RASSP1A and GSTP1 genes
in high-grade and advanced-stage carcinomas emphasizes their
prognostic value in EEC which collectively represents a clinical
tool for the proper management of the disease.
Acknowledgements
The authors thank Dr Pavol Slavik for his valuable
technical assistance and tissue sample collection. The present
study was supported by grant UK 61/2009 and 576/2010 from the
Ministry of Education, Slovak Republic.
References
|
1
|
Ferlay J, Steliarova-Foucher E,
Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D and
Bray F: Cancer incidence and mortality patterns in Europe:
estimates for 40 countries in 2012. Eur J Cancer. 49:1374–1403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lutz JM, Francisci S, Mugno E, Usel M,
Pompe-Kirn V, Coebergh JW and Bieslka-Lasota M; EUROPREVAL Working
Group. Cancer prevalence in Central Europe: the EUROPREVAL Study.
Ann Oncol. 14:313–322. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Emons G, Fleckenstein G, Hinney B,
Huschmand A and Heyl W: Hormonal interactions in endometrial
cancer. Endocr Relat Cancer. 7:227–242. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cirisano FD Jr, Robboy SJ, Dodge RK,
Bentley RC, Krigman HR, Synan IS, Soper JT and Clarke-Pearson DL:
The outcome of stage I–II clinically and surgically staged
papillary serous and clear cell endometrial cancers when compared
with endometrioid carcinoma. Gynecol Oncol. 77:55–65. 2000.
|
|
5
|
Ito K, Utsunomiya H, Yaegashi N and Sasano
H: Biological roles of estrogen and progesterone in human
endometrial carcinoma - new developments in potential endocrine
therapy for endometrial cancer. Endocr J. 54:667–679. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gründker C, Günthert AR and Emons G:
Hormonal heterogeneity of endometrial cancer. Adv Exp Med Biol.
630:166–188. 2008.
|
|
7
|
Armstrong AJ, Hurd WW, Elguero S, Barker
NM and Zanotti KM: Diagnosis and management of endometrial
hyperplasia. J Minim Invasive Gynecol. 19:562–571. 2012. View Article : Google Scholar
|
|
8
|
Sorosky JI: Endometrial cancer. Obstet
Gynecol. 120:383–397. 2012. View Article : Google Scholar
|
|
9
|
Lax S: Precursor lesions of endometrial
carcinoma: diagnostic approach and molecular pathology. Pathologe.
32(Suppl 2): S255–S264. 2011.(In German).
|
|
10
|
Tao MH and Freudenheim JL: DNA methylation
in endometrial cancer. Epigenetics. 5:491–498. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Reinhold WC, Reimers MA, Lorenzi P, Ho J,
Shankavaram UT, Ziegler MS, Bussey KJ, Nishizuka S, Ikediobi O,
Pommier YG and Weinstein JN: Multifactorial regulation of
E-cadherin expression: an integrative study. Mol Cancer Ther.
9:1–16. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fanelli F and Raimondi F: Nucleotide
binding affects intrinsic dynamics and structural communication in
Ras GTPases. Curr Pharm Des. 19:4214–4225. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kang J and Pervaiz S: Crosstalk between
Bcl-2 family and Ras family small GTPases: potential cell fate
regulation? Front Oncol. 2:2062012.PubMed/NCBI
|
|
14
|
Di Domenico M, Santoro A, Ricciardi C,
Iaccarino M, Iaccarino S, Freda M, Feola A, Sanguedolce F, Losito
S, Pasquali D, Di Spiezio Sardo A, Bifulco G, Nappi C, Bufo P,
Guida M, De Rosa G, Abbruzzese A, Caraglia M and Pannone G:
Epigenetic fingerprint in endometrial carcinogenesis: the
hypothesis of a uterine field cancerization. Cancer Biol Ther.
12:447–457. 2011.PubMed/NCBI
|
|
15
|
Holliday R: The inheritance of epigenetic
defects. Science. 238:163–170. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jones PA: DNA methylation errors and
cancer. Cancer Res. 56:2463–2467. 1996.PubMed/NCBI
|
|
17
|
Holliday R: DNA methylation and epigenetic
defects in carcinogenesis. Mutat Res. 181:215–217. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jones A, Lechner M, Fourkala EO,
Kristeleit R and Widschwendter M: Emerging promise of epigenetics
and DNA methylation for the diagnosis and management of women’s
cancers. Epigenomics. 2:9–38. 2010.PubMed/NCBI
|
|
19
|
Banno K, Kisu I, Yanokura M, Masuda K,
Ueki A, Kobayashi Y, Susumu N and Aoki D: Epigenetics and genetics
in endometrial cancer: new carcinogenic mechanisms and relationship
with clinical practice. Epigenomics. 4:147–162. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yi TZ, Li J, Han X, Guo J, Qu Q, Guo L,
Sun HD and Tan WH: DNMT inhibitors and HDAC inhibitors regulate
E-cadherin and Bcl-2 expression in endometrial carcinoma in vitro
and in vivo. Chemotherapy. 58:19–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lasabova Z, Tilandyova P, Kajo K, Zubor P,
Burjanivova T, Danko J and Plank L: Hypermethylation of the GSTP1
promoter region in breast cancer is associated with prognostic
clinicopathological parameters. Neoplasma. 57:35–40. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Culbova M, Lasabova Z, Stanclova A,
Tilandyova P, Zubor P, Fiolka R, Danko J and Visnovsky J:
Methylation of selected tumor-suppressor genes in benign and
malignant ovarian tumors. Ceska Gynekol. 76:274–279. 2011.(In
Slovak).
|
|
23
|
Jha AK, Nikbakht M, Jain V, Capalash N and
Kaur J: p16(INK4a) and p15(INK4b) gene
promoter methylation in cervical cancer patients. Oncol Lett.
3:1331–1335. 2012.
|
|
24
|
Silverberg SG, Kurman RJ, Nogales F,
Mutter GL, Kubik-Huch RA and Tavassoli FA: Epithelial tumours and
related lesions. Pathology and Genetics of Tumours of the Breast
and Female Genital Organs. Tavassoli FA and DeVilee P: IARC Press;
Lyon: pp. 221–232. 2003
|
|
25
|
Zaino RJ, Kurman RJ, Diana KL and Morrow
CP: The utility of the revised International Federation of
Gynecology and Obstetrics histologic grading of endometrial
adenocarcinoma using a defined nuclear grading system. A
Gynecologic Oncology Group study. Cancer. 75:81–86. 1995.
View Article : Google Scholar
|
|
26
|
Zaino RJ: FIGO staging of endometrial
adenocarcinoma: a critical review and proposal. Int J Gynecol
Pathol. 28:1–9. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pecorelli S: Revised FIGO staging for
carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol
Obstet. 105:103–104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Herman JG, Graff JR, Myohanen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: a novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
House MG, Guo MZ, Iacobuzio-Donahue C and
Herman JG: Molecular progression of promoter methylation in
intraductal papillary mucinous neoplasms (IPMN) of the pancreas.
Carcinogenesis. 24:193–198. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leu YW, Rahmatpanah F, Shi H, Wei SH, Liu
JC, Yan PS and Huang TH: Double RNA interference of DNMT3b and
DNMT1 enhances DNA demethylation and gene reactivation. Cancer Res.
63:6110–6115. 2003.PubMed/NCBI
|
|
31
|
Shen WJ, Dai DQ, Teng Y and Liu HB:
Regulation of demethylation and re-expression of RASSF1A gene in
gastric cancer cell lines by combined treatment of 5-Aza-CdR and
NaB. World J Gastroenterol. 14:595–600. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Heichman KA and Warren JD: DNA methylation
biomarkers and their utility for solid cancer diagnostics. Clin
Chem Lab Med. 50:1707–1721. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hatzimichael E and Crook T: Cancer
epigenetics: new therapies and new challenges. J Drug Deliv.
13:5293122013.PubMed/NCBI
|
|
34
|
Pallarés J, Velasco A, Eritja N, Santacana
M, Dolcet X, Cuatrecasas M, Palomar-Asenjo V, Catasús L, Prat J and
Matias-Guiu X: Promoter hypermethylation and reduced expression of
RASSF1A are frequent molecular alterations of endometrial
carcinoma. Mod Pathol. 21:691–699. 2008.
|
|
35
|
Fiegl H, Gattringer C, Widschwendter A,
Schneitter A, Ramoni A, Sarlay D, Gaugg I, Goebel G, Müller HM,
Mueller-Holzner E, Marth C and Widschwendter M: Methylated DNA
collected by tampons - a new tool to detect endometrial cancer.
Cancer Epidemiol Biomarkers Prev. 13:882–888. 2004.PubMed/NCBI
|
|
36
|
Kang S, Kim JW, Kang GH, Lee S, Park NH,
Song YS, Park SY, Kang SB and Lee HP: Comparison of DNA
hypermethylation patterns in different types of uterine cancer:
cervical squamous cell carcinoma, cervical adenocarcinoma and
endometrial adenocarcinoma. Int J Cancer. 118:2168–2171. 2006.
View Article : Google Scholar
|
|
37
|
Pijnenborg JM, Kisters N, van Engeland M,
Dunselman GA, de Haan J, de Goeij AF and Groothuis PG: APC,
beta-catenin, and E-cadherin and the development of recurrent
endometrial carcinoma. Int J Gynecol Cancer. 14:947–956. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Arafa M, Kridelka F, Mathias V,
Vanbellinghen JF, Renard I, Foidart JM, Boniver J and Delvenne P:
High frequency of RASSF1A and RARb2 gene promoter
methylation in morphologically normal endometrium adjacent to
endometrioid adenocarcinoma. Histopathology. 53:525–532. 2008.
|
|
39
|
Liao X, Siu MK, Chan KY, Wong ES, Ngan HY,
Chan QK, Li AS, Khoo US and Cheung AN: Hypermethylation of RAS
effector-related genes and DNA methyltransferase 1 expression in
endometrial carcinogenesis. Int J Cancer. 123:296–302. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Seeber LM, Zweemer RP, Marchionni L,
Massuger LF, Smit VT, van Baal WM, Verheijen RH and van Diest PJ:
Methylation profiles of endometrioid and serous endometrial
cancers. Endocr Relat Cancer. 17:663–673. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pijnenborg JM, Dam-de Veen GC, Kisters N,
Delvoux B, van Engeland M, Herman JG and Groothuis PG:
RASSF1A methylation and K-ras and B-raf
mutations and recurrent endometrial cancer. Ann Oncol. 18:491–497.
2007. View Article : Google Scholar
|
|
42
|
Jo H, Kim JW, Kang GH, Park NH, Song YS,
Kang SB and Lee HP: Association of promoter hypermethylation of the
RASSF1A gene with prognostic parameters in endometrial cancer.
Oncol Res. 16:205–209. 2006.PubMed/NCBI
|
|
43
|
Kang S, Lee JM, Jeon ES, Lee S, Kim H, Kim
HS, Seo SS, Park SY, Sidransky D and Dong SM: RASSF1A
hypermethylation and its inverse correlation with BRAF
and/or KRAS mutations in MSI-associated endometrial
carcinoma. Int J Cancer. 119:1316–1321. 2006. View Article : Google Scholar
|
|
44
|
Zhang YQ, Mao XY, Ma XL, Zhang M and Sheng
N: Effects and mechanisms of 5-aza-2′-deoxycytidine on endometrial
cancer cell. Zhonghua Fu Chan Ke Za Zhi. 44:861–864. 2009.(In
Chinese).
|
|
45
|
Yu MY, Tong JH, Chan PK, Lee TL, Chan MW,
Chan AW, Lo KW and To KF: Hypermethylation of the tumor suppressor
gene RASSFIA and frequent concomitant loss of heterozygosity
at 3p21 in cervical cancers. Int J Cancer. 105:204–209.
2003.PubMed/NCBI
|
|
46
|
Moreno-Bueno G, Hardisson D, Sarrió D,
Sánchez C, Cassia R, Prat J, Herman JG, Esteller M, Matías-Guiu X
and Palacios J: Abnormalities of E- and P-cadherin and catenin (β-,
γ-catenin, and p120ctn) expression in endometrial cancer
and endometrial atypical hyperplasia. J Pathol. 199:471–478.
2003.PubMed/NCBI
|
|
47
|
Saito T, Nishimura M, Yamasaki H and Kudo
R: Hypermethylation in promoter region of E-cadherin gene is
associated with tumor dedifferentiation and myometrial invasion in
endometrial carcinoma. Cancer. 97:1002–1009. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Banno K, Yanokura M, Susumu N, Kawaguchi
M, Hirao N, Hirasawa A, Tsukazaki K and Aoki D: Relationship of the
aberrant DNA hypermethylation of cancer-related genes with
carcinogenesis of endometrial cancer. Oncol Rep. 16:1189–1196.
2006.PubMed/NCBI
|
|
49
|
Yi TZ, Guo J, Zhou L, Chen X, Mi RR, Qu
QX, Zheng JH and Zhai L: Prognostic value of E-cadherin expression
and CDH1 promoter methylation in patients with endometrial
carcinoma. Cancer Invest. 29:86–92. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Graff JR, Gabrielson E, Fujii H, Baylin SB
and Herman JG: Methylation patterns of the E-cadherin 5′ CpG island
are unstable and reflect the dynamic, heterogeneous loss of
E-cadherin expression during metastatic progression. J Biol Chem.
275:2727–2732. 2000.
|
|
51
|
Moreno-Bueno G, Hardisson D, Sánchez C,
Sarrió D, Cassia R, García-Rostán G, Prat J, Guo M, Herman JG,
Matías-Guiu X, Esteller M and Palacios J: Abnormalities of the
APC/β-catenin pathway in endometrial cancer. Oncogene.
21:7981–7990. 2002.
|
|
52
|
Yoshiura K, Kanai Y, Ochiai A, Shimoyama
Y, Sugimura T and Hirohashi S: Silencing of the E-cadherin
invasion-suppressor gene by CpG methylation in human carcinomas.
Proc Natl Acad Sci USA. 92:7416–7419. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Takai N, Desmond JC, Kumagai T, Gui D,
Said JW, Whittaker S, Miyakawa I and Koeffler HP: Histone
deacetylase inhibitors have a profound antigrowth activity in
endometrial cancer cells. Clin Cancer Res. 10:1141–1149. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hirohashi S: Inactivation of the
E-cadherin-mediated cell adhesion system in human cancers. Am J
Pathol. 153:333–339. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chan QK, Khoo US, Chan KY, Ngan HY, Li SS,
Chiu PM, Man LS, Ip PP, Xue WC and Cheung AN: Promoter methylation
and differential expression of π-class glutathione S-transferase in
endometrial carcinoma. J Mol Diagn. 7:8–16. 2005.
|
|
56
|
Kim GE, Kweon SS, Lee JS, Lee JH, Nam JH
and Choi C: Quantitative assessment of DNA methylation for the
detection of cervical and endometrial adenocarcinomas in
liquid-based cytology specimens. Anal Quant Cytol Histol.
34:195–203. 2012.PubMed/NCBI
|
|
57
|
Nieminen TT, Gylling A, Abdel-Rahman WM,
Nuorva K, Aarnio M, Renkonen-Sinisalo L, Järvinen HJ, Mecklin JP,
Bützow R and Peltomäki P: Molecular analysis of endometrial
tumorigenesis: importance of complex hyperplasia regardless of
atypia. Clin Cancer Res. 15:5772–5783. 2009. View Article : Google Scholar : PubMed/NCBI
|