Introduction
Metastasis is the endpoint of tumorigenesis and is a
major cause of cancer-related mortality (1). Prevention or elimination of this level
of disseminated cancer progression is essential to reduce morbidity
and mortality (1,2). Head and neck squamous cell carcinoma
(HNSCC) is the sixth most common cancer in the world (3). Although therapeutic regimens for HNSCC
have been improved, the survival rate of HNSCC patients has not
markedly increased due to locoregional and/or distant
recurrence/metastasis (4).
Therefore, to develop effective therapeutic or preventive
strategies to overcome the progression of this cancer, elucidation
of the mechanisms of metastasis is an urgent issue.
Mounting evidence supports the hypothesis that
tumors contain a small subpopulation of cells, called cancer stem
cells (CSCs) or cancer initiating cells (CICs), which exhibit
self-renewal capacity, and are responsible for tumor growth and
metastasis (5). In addition, we
previously demonstrated the existence of stemness-possessing cells
in HNSCC (6,7). Metastatic potential depends on
multiple factors, including the regulation of cell growth,
survival, angiogenesis and invasion. Furthermore,
epithelial-mesenchymal transition (EMT) is recognized as a crucial
event in the metastatic process (8)
and may enhance stemness and tumor-initiating properties (9–11).
However, modulators of metastatic HNSCC that participate in the EMT
process and stemness remain unclear.
Although tumorigenic and metastatic head and neck
carcinoma cell lines are available, the genetic variation among
these cell lines makes it difficult to compare the observations
derived from these different cell lines. Therefore, developing a
systemic metastatic cellular model derived from the same ancestor
cellular lineage could be an important tool for investigating
metastasis in head and neck cancer. In the present study, we aimed
to develop a cellular model consisting of cell lines possessing
differential malignant potential but with the same genetic
background. Such a system may allow us to avoid the variation found
in cell lines derived from different individuals or types of
tissues.
Previously, we established the tumorigenic SASVO3
cell line, which possesses enhanced in vivo tumorigenicity,
from parental HNSCC SAS cells (12). SASVO3 cells exhibit enhanced cancer
stem cell properties, including an increase in sphere-forming
activity, the number of side population cells and the expression of
the stem cell marker Bmi1. Herein, we successfully established a
metastatic carcinoma cell line from pulmonary metastatic nodules of
SASVO3 cells. Using these cell lines, derived from the same
parental cancer cell line and exhibiting an increase in malignancy
potential, we investigated the underlying mechanisms of
tumorigenesis and metastasis by transcriptome analysis and cellular
assays.
Materials and methods
Cell culture
Human HNSCC SAS cells and the various derived cell
lines, including SASVO3, SASVO3M-1 and SASVO3M-5, were cultured in
Dulbecco’s modified Eagle’s medium (DMEM) (Gibco-BRL). The medium
was supplemented with 10% fetal bovine serum (FBS) (HyClone
Laboratory), 2 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml
streptomycin sulfate. The cells were maintained at 37°C in a
humidified atmosphere containing 5% CO2.
Establishment of the metastatic cell
lines
Female nude mice, aged 6–8 weeks, were obtained from
the National Laboratory Animal Breeding and Research Center,
Taipei, Taiwan. All mice were maintained under standard
pathogen-free laboratory conditions. Animal experiments were
conducted according to the institutional guidelines established by
the Animal Core Facility and IACUC of the Veterans General
Hospital, Taipei, Taiwan. SASVO3 cells at a density of
5×105 cells/100 μl PBS were injected into nude mice via
the tail vein. The mice were euthanized 65 days after injection,
and metastatic nodules were removed from the lungs. Lung tissues
were subjected to pathological examination and in vitro cell
culture studies. For histological analyses, lung tissues were fixed
with buffered-formalin and embedded in paraffin. Sections were
stained using standard hematoxylin and eosin protocol. For cell
line generation, metastatic nodules were torn into tiny pieces and
grown in medium consisting of DMEM and defined keratinocyte
serum-free medium (KSFM) (Gibco-BRL) at a ratio of 1:2. The
outgrowth epithelial tumor cells were cloned and adapted to
complete DMEM. Two clones derived from the metastatic nodules were
designated as SASVO3M-1 and SASVO3M-5. The abbreviations for
SASVO3, SASVO3M-1 and SASVO3M-5 cells are VO3, M-1 and M-5,
respectively.
Global transcription and pathway
analyses
Total RNA was extracted from the parental SAS cells,
as well as the derived cell lines, using TRIzol reagent (Invitrogen
Life Technologies). RNA purity was determined based on the
OD260/280 ratio and electrophoretic analysis. Genome-wide gene
expression profiling was performed using the human HT-12 v4
BeadChip array (Illumina, Inc.). This system utilizes >47,000
probes, which cover 29,000 transcripts. Data were extracted from
GenomeStudio software (Illumina, Inc.). After quantile
normalization, a total of 47,320 probes were included for analysis.
The biological functions and pathways of the selected genes were
interpreted by Ingenuity pathway analysis (Ingenuity®
Systems, Inc.)
Focus formation assay
Parental SAS cells and the derived cell lines were
plated (1.8×106 cells) onto 60-mm dishes and grown in
DMEM containing 2% FBS for 6 days. Cells were subsequently fixed
with methanol and stained with 10% Giemsa solution in PBS.
Fluorescence-activated cell sorting
(FACS)
Primary antibodies against integrin ανβ6
(Millipore), uPAR (American Diagnostica, Inc.) and CD133 (Miltenyi
Biotec) were used to determine the expression of proteins on the
cell surface. APC goat anti-mouse (Ig) (Becton-Dickinson and
Company) was used as a secondary antibody. The isotype antibody was
purified mouse IgG1 (BioLegend). Primary and secondary antibodies
were incubated serially at 4°C for 30 min. Analyses were performed
using a FACScanto flow cytometer (BD Biosciences).
Immunoblotting
The antibodies used in this study included Snail
(3895; Cell Signaling Technology, Inc.), Slug (ab27568; Abcam),
Twist (sc-15393; Santa Cruz Biotechnology, Inc.), vimentin (550513)
and fibronectin (610078; both from BD Biosciences), GAPDH (MAB374;
Millipore), FAK (sc557), Tyr 925-phosphorylated FAK (sc-11766) and
Oct4 (sc-9081) (all from Santa Cruz Biotechnology, Inc.), Nanog
(ab21624; Abcam) and Bmi1 (2830; Cell Signaling Technology, Inc.).
For immunoblotting, the cell pellets were homogenized in 200 ml
lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, 1.5
mM MgCl2, 1% Triton X-100) and incubated on ice for 20
min. After centrifugation at 13,000 × g for 30 min at 4°C, the
supernatant containing the protein extracts was collected. Protein
concentrations were determined using the Bio-Rad protein assay.
Immunoblot analyses were performed as described previously
(12).
Immunofluorescence staining and confocal
microscopy
Parental SAS cells and the derived cell lines were
plated on glass coverslips in 6-well plates. For E-cadherin
immunofluorescence staining, cells were fixed with 4%
paraformaldehyde/phosphate-buffered saline (PBS) and permeabilized
with 0.5% (v/v) Triton X-100 in PBS, followed by incubation with
blocking solution [1% BSA and 0.2% (v/v) Tween-20 in PBS]. Cells
were incubated with primary antibodies against E-cadherin (BD
Transduction Laboratories), and subsequently incubated with the
secondary antibody conjugated with FITC (Sigma-Aldrich). Nuclei
were counterstained with DAPI, and the cells were mounted using
fluorescence mounting medium (Dako). Fluorescence images were
captured using a Zeiss Axio Observer A1 microscope. For F-actin
staining, cells were fixed with 4% paraformaldehyde, incubated in
blocking solution and stained with TRITC-conjugated phalloidin
(Sigma-Aldrich). The cells were subsequently counterstained with
DAPI and examined using an Olympus FV1000 confocal microscope.
Cell migration assay
Cell migration assays were performed using an Oris™
cell migration assembly kit (Platypus Technologies) according to
the manufacturer’s instructions. In brief, the assay utilizes Oris™
cell seeding stoppers to restrict cell seeding to the outer annular
region of the wells. SAS cells and the derived cell lines were
plated onto each well at the indicated density in serum-free DMEM
and allowed to attach for 6 h at 37°C. The stopper was subsequently
removed to form an unseeded region at the center of each well. An
image of the clear detection zone was captured as a control for
cell migration. The cells were then incubated at 37°C for 16 h to
permit cell migration into the detection zone. The cells were
stained with Calcein AM fluorescent dye (Sigma-Aldrich) for 1 h
before images were captured using a Zeiss Axio Observer A1
microscope.
Cell invasion assay
Cell invasion assays were performed using an Oris™
96-well cell migration assay kit following the manufacturer’s
instructions. In brief, a thin layer of basal membrane was coated
by rinsing each well with 3.5 mg/ml BME coating solution, after
which the seeding stopper was pressed down into each well. The
cells were starved in medium containing 1% FBS for 18 h prior to
plating. The cells were incubated for 6 h to permit cell
attachment. The stopper was then removed, and 40 μl BME coating
solution was added to create a 3-D BME layer. After incubating for
48 h, the cells were stained for the cell migration assay.
Sphere formation assay
SAS cells and the derived cell lines were cultured
in tumor sphere medium consisting of serum-free DMEM/F12 medium, N2
supplement (both from Gibco-BRL), 10 ng/ml human recombinant basic
fibroblast growth factor-basic (FGF) and 10 ng/ml epidermal growth
factor (EGF) (R&D Systems). Cells were plated at a density of
7.5×104 to 1×105 live cells/100-mm dish, and
the medium was replaced every other day until tumor sphere
formation was observed (~4 weeks) (7).
In vivo pulmonary metastasis assay
Nude mice were randomly grouped into sets of 5 mice
each. Cells at the indicated cell number were individually injected
into mice via the tail vein. The date of animal death or euthanasia
was recorded, and the lung tissue from these mice was fixed with
Bouin’s solution.
Statistical analyses
Kaplan-Meier survival was analyzed with the
Statistical Package for the Social Sciences (SPSS) statistical
program version 17.0 (SPSS, Inc.).
Results
Establishment of the metastatic HNSCC
cell lines
To establish a cellular model for investigating the
progression of metastasis, we attempted to generate metastatic cell
lines from the highly tumorigenic SASVO3 (VO3) cell line, which was
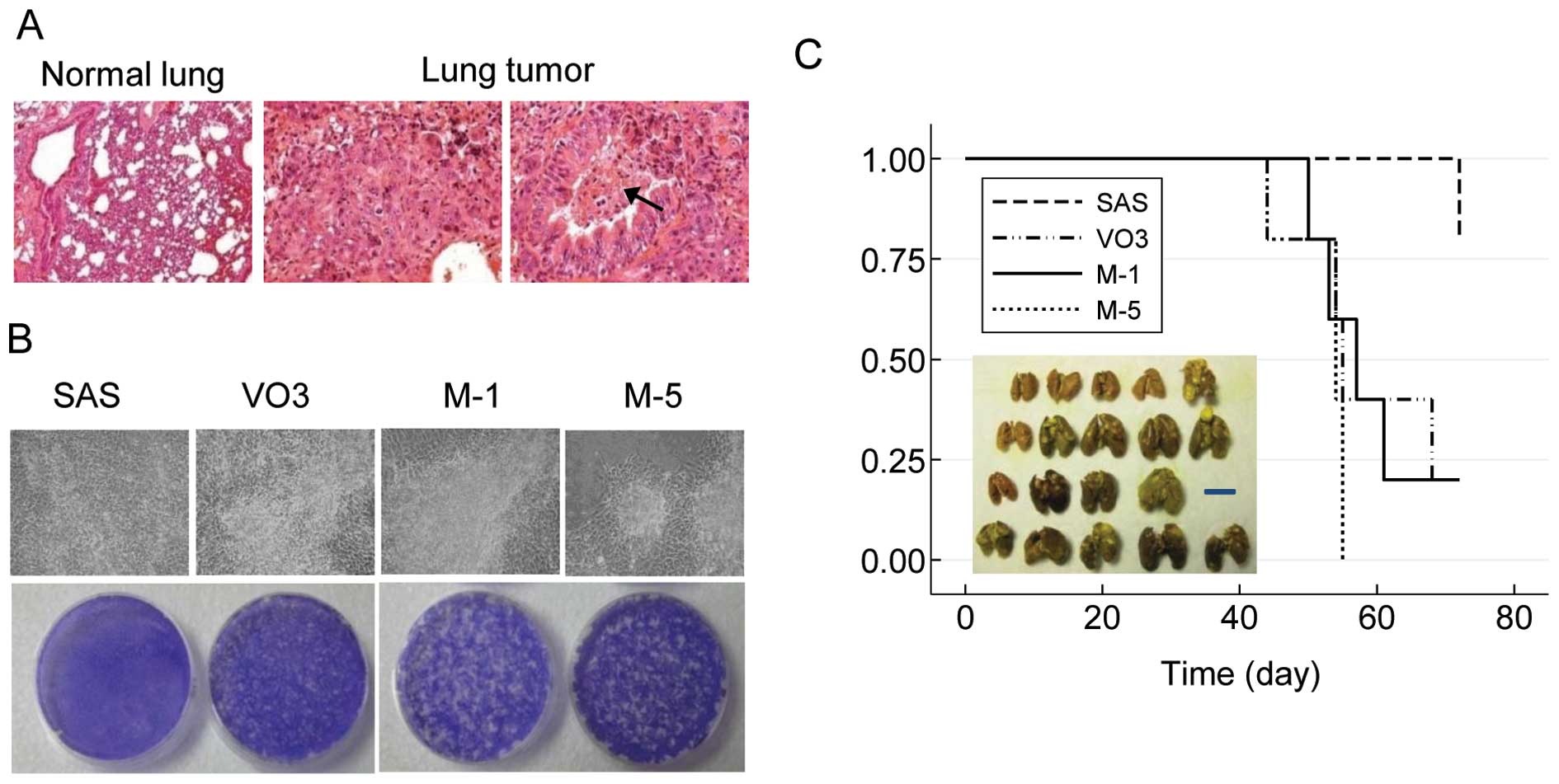
previously developed from parental SAS cells (12). Empirically, we induced pulmonary
metastatic tumors by injecting VO3 cells into the tail vein of nude
mice. Pathological analysis revealed that the tumor cells
subsequently found in the lung were capable of invading into the
trachea (Fig. 1A). Two metastatic
cell line clones were generated and designated as M-1 and M-5. The
in vitro malignancy of VO3, M-1 and M5 were further examined
by a focus formation assay. As shown in Fig. 1B, these metastatic cell lines
exhibited significantly enhanced focus formation when compared with
the original VO3 cells, even when the cells were not confluent. In
contrast, the SAS cells with reduced tumorigenicity rarely formed
foci, even at full confluence (Fig.
1B).
Enhanced pulmonary metastatic activity of
metastatic HNSCC cells
To compare the lung colonization activity of cell
lines with different malignant potential, SAS cells and cells from
the 3 derived cell lines were collected and injected into nude mice
via the tail vein. The results showed that the metastatic M-1 and
M-5 cells, even when injected at a 10-fold lower cell number than
SAS or VO3 cells, were capable of inducing more pulmonary
metastatic colonies and shortening the overall survival of the
injected mice (Fig. 1C). In
contrast, non-metastatic SAS cells rarely formed pulmonary
metastatic colonies. The data thus revealed that the newly
established metastatic cells possessed an enhanced ability to
extravasate and proliferate in the lung parenchyma as compared to
the ancestor VO3 cells.
Distinct transcriptome signatures of the
metastatic HNSCC cells
To understand the transcriptional changes in the
cell lines with different malignant potential, genome-wide gene
expression profiling of SAS, VO3, M-1 and M-5 cells was carried
out. Scatter plot analysis revealed a high correlation of
metastatic M-1 and M-5 clones (R2=0.9941). After
quantile normalization, 47,320 probes were subjected to 2 systemic
analyses to measure the correlation of cells with differing
malignant potential (heatmap) and to select the genes that
displayed a marked alteration in parallel with malignancy
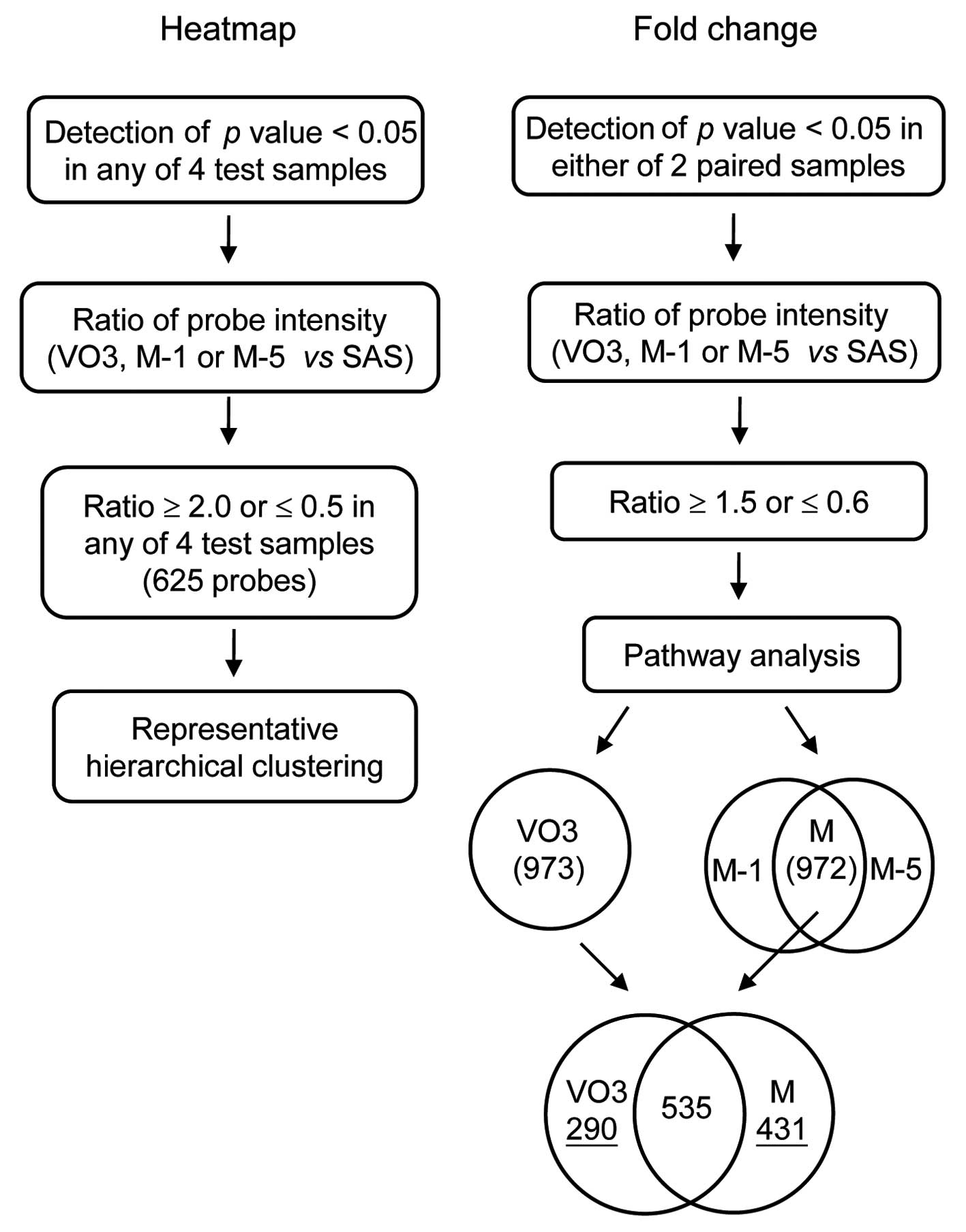
(fold-change). A schematic of the transcriptome data analysis is
shown in Fig. 2. The criteria of
selection were: i) to limit false results arising from
low-expression genes. Probes with detection of a p-value ≥0.05 in
any of the 4 test samples (heatmap), or in either of the paired
samples (fold-change) were excluded. ii) The ratio of probe
intensity of VO3, M-1 or M-5 vs. that of SAS was calculated. iii)
Cutoff values of the ratio were as indicated in Fig. 2.
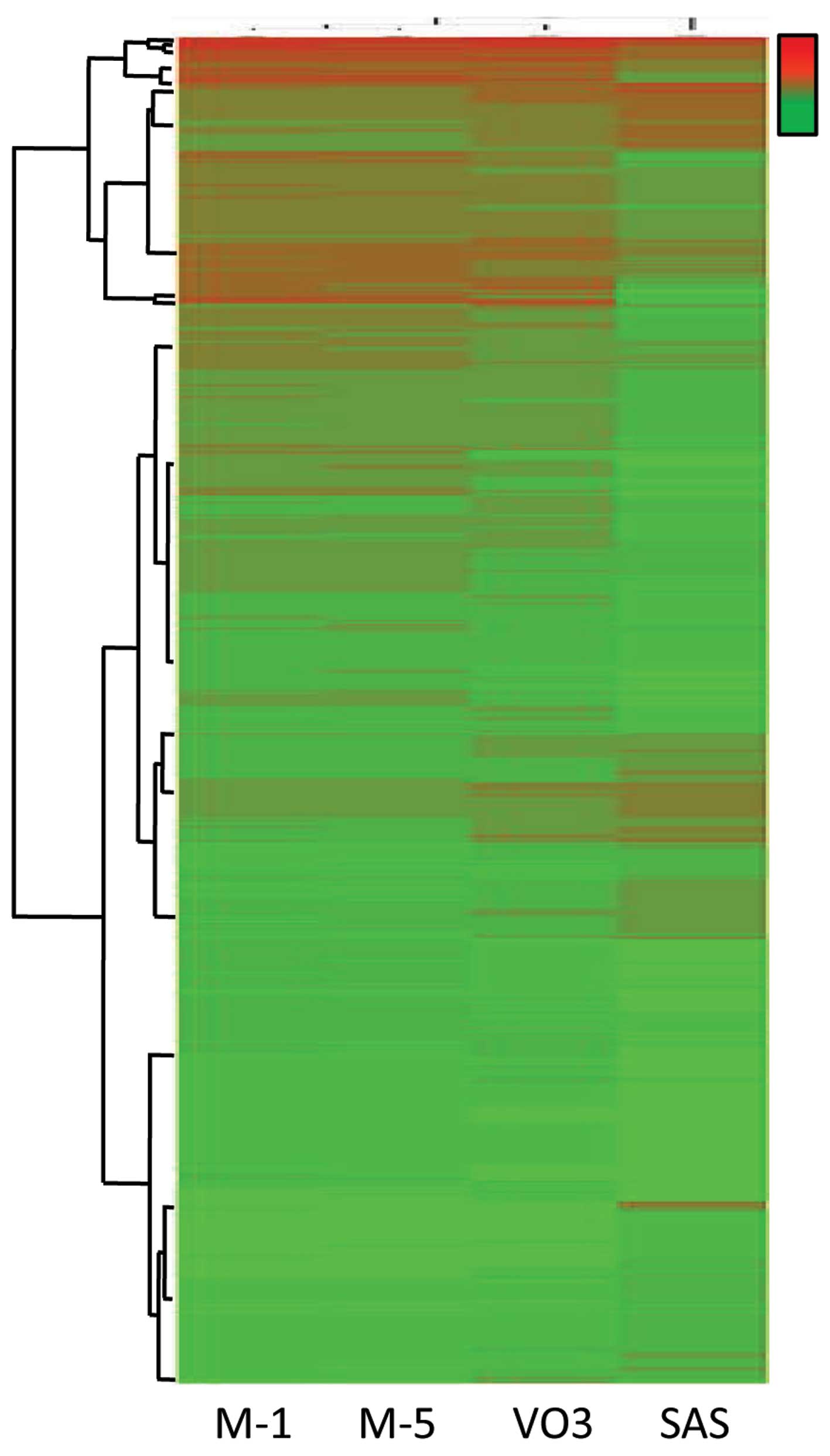
As shown in Fig. 3,
hierarchical clustering revealed a differential signature in cells
with varying malignant potential. The signatures of the 2
metastatic cell lines were the most closely related to each other,
but were most distinct from the non-metastatic SAS cells. However,
the signature of highly tumorigenic VO3 cells was in-between the
aforementioned cell lines and contained certain uniquely expressed
genes. To transcriptionally profile these changes between the
highly tumorigenic cells and metastatic cells, genes with a
statistically significant difference in expression in the VO3, M-1
and M-5 cells were selected and subjected to pathway analysis.
There were 825 and 972 ingenuity pathway analysis (IPA)-identified
genes displaying significant changes in VO3 cells and with common
preservation in M-1 and M-5 cells (designate M), respectively.
Furthermore, a total of 535 overlapping genes were observed in both
tumorigenic and metastatic cells, while 209 and 431 genes were
uniquely found in VO3 cells and commonly reserved in M cells,
respectively (Fig. 2, right).
Biological pathways involved in VO3 (825) and common M (972) gene
sets were then analyzed. Several canonical pathways were identified
in both the VO3 and the common M groups, including the integrin
signaling pathway, VDR/RXR activation pathway and IL-17A, which has
a role in psoriasis. Of note, Notch signaling was found exclusively
in the highly tumorigenic cells, while interferon signaling, RhoA
signaling and Oct4, which plays a role in mammalian embryonic stem
cell pluripotency, were significantly altered in the metastatic
cells. Table I depicts the top 10
canonical pathways identified by IPA. Table II shows the top 5 upregulated and
downregulated genes uniquely identified in the VO3 or metastatic
cells. Collectively, the transcriptome analysis suggests that
metastatic HNSCC cells may have profound changes associated with
cell movement and stemness activity.
 | Table IIngenuity pathway analysis of top
canonical pathways in the highly tumorigenic or metastatic cell
lines. |
Table I
Ingenuity pathway analysis of top
canonical pathways in the highly tumorigenic or metastatic cell
lines.
| Pathway name | P-value | Ratio |
|---|
| Highly tumorigenic
cells (VO3) |
| VDR/RXR
activationa | 1.65E-05 | 13/81 (0.160) |
| Integrin
signaling | 4.17E-04 | 19/207 (0.092) |
| Notch signaling | 7.5E-04 | 7/43 (0.163) |
| Cyclins and cell
cycle regulation | 1.33E-03 | 10/89 (0.112) |
| Role of IL-17A in
psoriasis | 1.45E-03 | 4/13 (0.308) |
| ILK
signalingb | 1.67E-03 | 17/192 (0.089) |
| Role of IL-17F in
allergic inflammatory airway diseases | 2.11E-03 | 7/48 (0.146) |
| Interferon
signaling | 2.25E-03 | 6/36 (0.167) |
| Glioma
invasiveness signaling | 2.29E-03 | 8/60 (0.133) |
| Neurotrophin/TRK
signaling | 2.5E-03 | 9/75 (0.120) |
| Metastatic cells
(M-1 and M-5) |
| Interferon
signaling | 6.39E-06 | 10/36 (0.278) |
| Role of IL-17A in
psoriasis | 2.61E-05 | 6/13 (0.462) |
| VDR/RXR
activationa | 4.95E-05 | 14/81 (0.173) |
| RhoA
signaling | 2.24E-04 | 16/114 (0.140) |
| Antigen
presentation pathway | 5.63E-04 | 8/40 (0.20) |
| Role of tissue
factor in cancer | 5.78E-04 | 15/114 (0.132) |
| Integrin
signaling | 6.61E-04 | 22/207 (0.106) |
| Clathrin-mediated
endocytosis signaling | 1.8E-03 | 20/196 (0.102) |
| Role of Oct4 in
mammalian embryonic stem cell pluripotency | 2.17E-03 | 8/45 (0.178) |
| Virus entry via
endocytic pathways | 2.37E-03 | 12/99 (0.121) |
 | Table IIIngenuity pathway analysis of the
most significant 5 genes uniquely altered in the highly tumorigenic
or metastatic cells. |
Table II
Ingenuity pathway analysis of the
most significant 5 genes uniquely altered in the highly tumorigenic
or metastatic cells.
| Gene symbol
(name) | Log ratioa |
|---|
| Highly tumorigenic
cells |
| Upregulated
genes |
| BGN
(biglycan) | 2.121 |
| HEG1 (HEG homolog
1) | 2.114 |
| SERPINB3 [serpin
peptidase inhibitor, clade B (ovalbumin), member 3] | 2.003 |
| SERPINE1 [serpin
peptidase inhibitor, clade E (nexin, plasminogen activator
inhibitor type 1), member 1] | 1.847 |
| EFEMP1 (EGF
containing fibulin-like extracellular matrix protein 1) | 1.840 |
| Downregulated
genes |
| HCLS1
(hematopoietic cell-specific Lyn substrate 1) | −2.082 |
| UST
(uronyl-2-sulfotransferase) | −1.207 |
| H2AFY2 (H2A
histone family, member Y2) | −1.158 |
| CADM4 (cell
adhesion molecule 4) | −1.112 |
| ENO2 (enolase
2) | −1.084 |
| Metastatic
cells |
| Upregulated
genes |
| GJB2 (gap
junction protein, β) | 3.183 |
| PPP1R14C [protein
phosphatase 1, regulatory (inhibitor) subunit 14C] | 2.566 |
| S100A7 (S100
calcium binding protein A7) | 2.123 |
| SAA1 (serum
amyloid A1) | 1.808 |
| SGPP2
(sphingosine-1-phosphate phosphatase 2) | 1.607 |
| Downregulated
genes |
| MT1G
(metallothionein 1G) | −1.991 |
| BLMH (bleomycin
hydrolase) | −1.824 |
| CENPV (centromere
protein V) | −1.798 |
| OVOL2 (ovo-like
2) | −1.780 |
| FOXA2 (forkhead
box A2) | −1.679 |
Acquirement of EMT in tumorigenic cells
and increased expression of focal adhesion-related genes and
increased filopodium formation in metastatic HNSCC cells
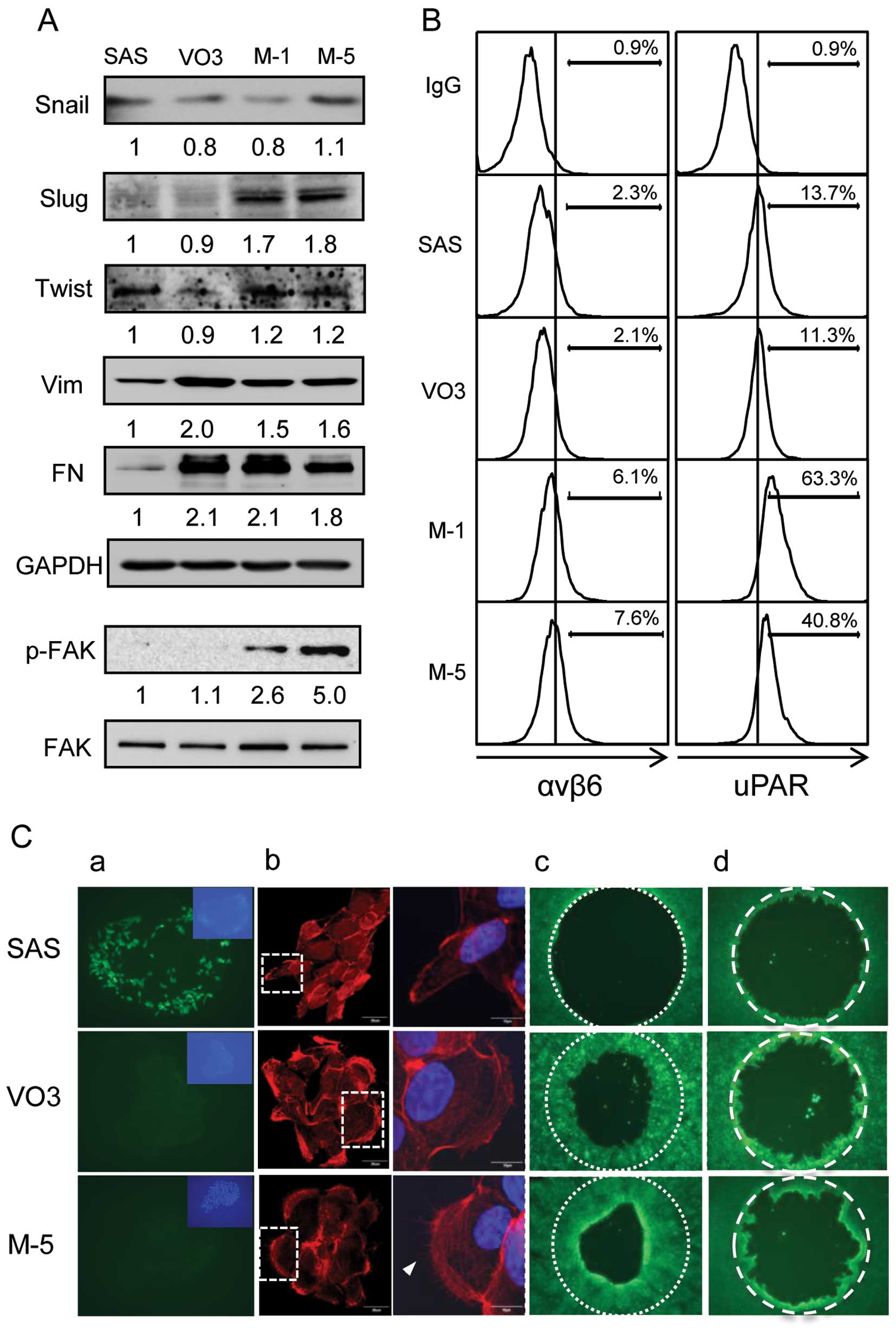
To verify the differential expression of cell
movement-related genes identified from transcriptome analysis, the
protein levels in the distinct cell lines of the above-mentioned
genes were determined. Highly tumorigenic VO3 cells displayed
upregulation of vimentin and fibronectin (Fig. 4A) and downregulation of the
epithelial marker E-cadherin (Fig.
4C-a), suggesting that a gain of EMT may have occurred in VO3
cells. In the 2 metastatic clones, the expression patterns of the 3
genes listed above remained similar, without any significant
change. Moreover, the protein levels of genes related to the EMT
transcription factor Slug, focal adhesion
(αvβ6 and p-FAK), and proteolytic
degradation, urokinase-type plasminogen activator receptor (uPAR)
were increased in the metastatic cells (Fig. 4A and B). However, no obvious changes
in Snail or Twist proteins (EMT-related transcription factors) in
the 4 tested cell lines were observed (Fig. 4A).
IPA analysis identified the upregulation of small
GTPase RhoA signaling in the metastatic cell lines (Table I). To further verify downstream
activation of RhoA signaling, the polymerization of F-actin was
characterized using phalloidin staining. The metastatic M-5 clone
displayed significant actin reorganization in lamellipodia, as well
as filopodium formation, when compared to the parental SAS cells
(Fig. 4C-b). The highly tumorigenic
VO3 cells also showed augmentation of lamellipodia, but less of
filopodia (Fig. 4C-b). The mobility
assays revealed that the migratory ability of VO3 and M-5 cells was
higher than that of the parental SAS cells (Fig. 4C-c). Additionally, the invasive
ability of M-5 cells was moderately higher than that of VO3 cells
(Fig. 4C-d). Taken together, our
data revealed that actin-polymerization, cell migration activity
and partial EMT changes occurred in the highly tumorigenic VO3
cells, while focal adhesion, filopodium formation and invasive
activity were enhanced in the metastatic cells.
Increased stemness-associated
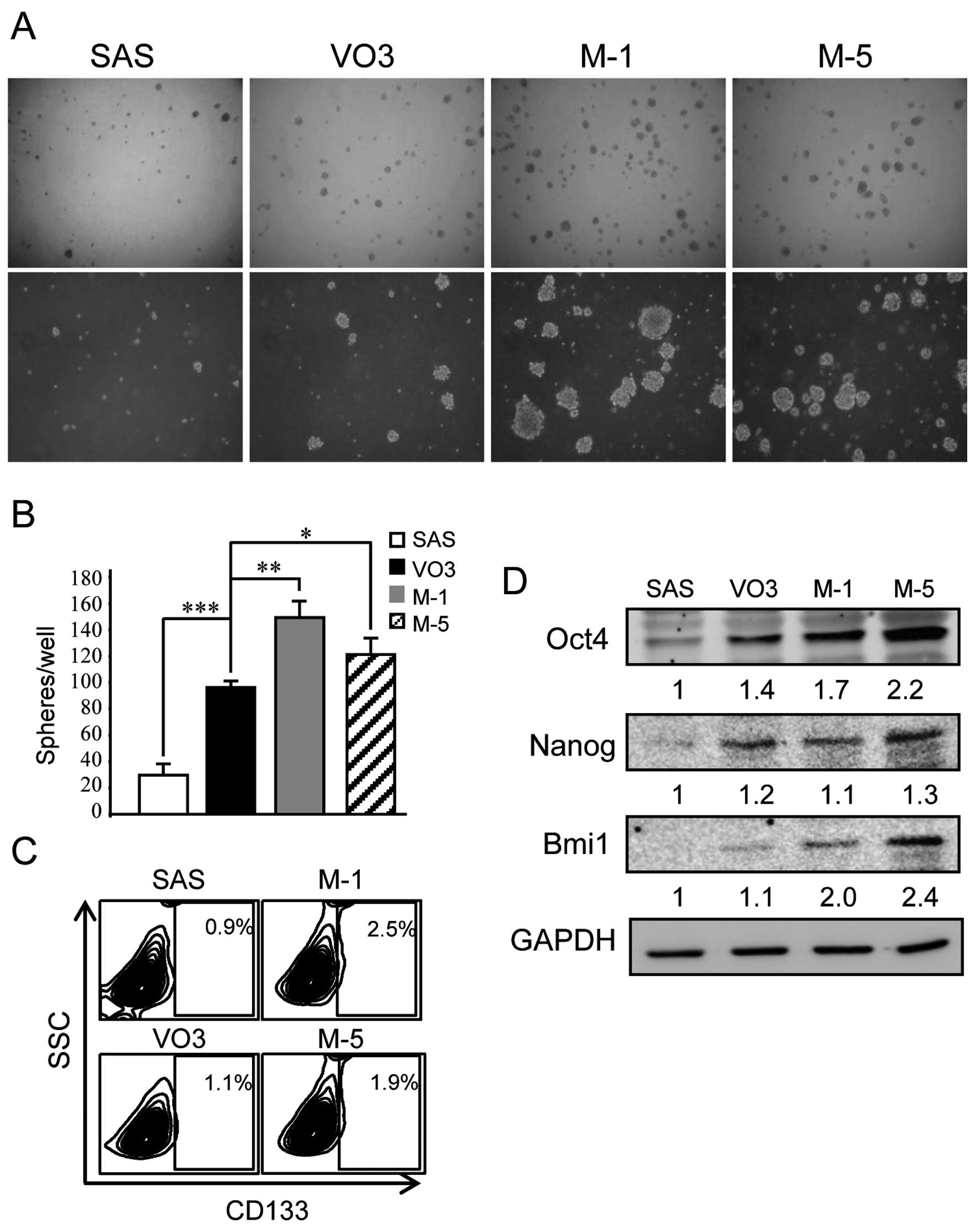
characteristics in metastatic HNSCC cells
Transcriptome analysis revealed activation of the
canonical Oct4 pathway, which plays a role in mammalian embryonic
stem cell pluripotency, in the metastatic M-1 and M-5 cells
(Table I). Hence, we examined the
acquisition of stem cell properties in the metastatic cells by
sphere formation which is one of the characteristics of stemness
(7). The metastatic cells showed
increased sphere formation ability in regards to both sphere size
and number as compared to the parental SAS cells that rarely form
spheres and VO3 cells that displayed moderate sphere formation
ability (Fig. 5A and B).
Furthermore, the expression of the stem cell markers, CD133, Oct4
and Bmi1, was also augmented in the metastatic cells, along with a
moderate increase in Nanog protein levels (Fig. 5C and D).
Discussion
Metastasis is an end stage of cancer and is the
major cause of cancer-related death. Investigation into the
mechanisms underlying metastasis may lead to improvements in
diagnosis and therapy. Our previous study generated the highly
tumorigenic cell line SASVO3, which acquired increased stem-like
properties in comparison to the parental SAS cell line (12). Here, we showed that EMT-related
genes (vimentin, fibronectin and E-cadherin) and cell migration
activity also increased in the highly tumorigenic cells. Moreover,
we demonstrated that the derived metastatic cell lines (SASVO3-M-1
and SASVO3-M-5) acquired further alterations in canonical pathways,
such as small GTPase signaling and Oct4-mediated pluripotency
signaling. Expression of genes involved in focal adhesion (integrin
αvβ6 and p-FAK), EMT (Slug), ECM degradation
(uPAR) and stemness (Oct4, Bmi1 and CD133) was also augmented. The
metastatic cells also displayed significant filopodium formation
and increased cell invasion, sphere formation and pulmonary
metastatic colony formation ability compared to the SASVO3 cells.
Together, our findings suggest that in the current system, partial
EMT changes may occur during tumor formation, while further changes
in cell invasiveness and survival may be obtained in parallel with
the acquisition of metastatic properties.
Lamellipodia and filopodia play pivotal roles in
tumor metastasis and invasion (13)
and are known to be controlled by small GTPases. The small GTPase
family has been reported to be involved in multiple aspects of
cancer development including cell proliferation, cell polarity,
adhesion, migration and invasion (14,15).
Small Rho GTPases act as molecular switches to translate
extracellular signals into intracellular events that induce changes
in actin organization (16). Rho,
Rac and Cdc42 are the most well-characterized family members and
contribute to the formation of stress fibers, lamellipodia and
filopodia, respectively (17).
Among them, Cdc42 is specifically involved in the generation of
cell polarity and in the orientation of migrating cells (18,19).
Integrins are cell surface adhesion receptors mediating adhesion
stabilization of filopodia (20).
The integrin-FAK axis has been shown to direct the proliferation of
disseminated metastatic cells (21). A recent study (22) demonstrated that when using
extracellular matrix culture conditions, the formation of
filopodium-like protrusions is a prerequisite for integrin-adhesion
plaque formation, which then triggers FAK/ERK signaling and
ultimately permits cell proliferation and the establishment of
metastatic colonies. This finding shows that integrin, FAK
activation and F-actin reorganization by small GTPases modulate the
dissemination and proliferation of metastatic cells. Our present
findings agree with these studies and suggest that our established
cellular system may be a useful model for investigating the
mechanisms underlying cellular dissemination, cell polarity and
cell survival in tumor metastasis.
Slug is one of the transcription factors of EMT and
has been shown to regulate cell invasion and metastasis (23). However, in concert with Sox9, these
transcription factors can convert differentiated murine luminal
cells into mammary stem cells (24). These studies demonstrate that the
EMT transcription factor slug may play a role in stem cell
maintenance, which supports our finding that the metastatic HNSCC
clones possessed both pulmonary metastatic and stem-like
activity.
Multiple lines of evidence reveal that Oct4 plays
fundamental roles in stem cell self-renewal and pluripotency, as
well as somatic cell reprogramming of murine embryonic stem (ES)
cells by connecting epigenetic pathways (25,26).
In conjunction with Cdk1, Oct4 maintains stemness by inhibiting
cellular differentiation (27). In
human ES cells, Oct4, Nanog and SOX2 play distinct roles in lineage
specification (28). In combination
with Bmi1, a member of polycomb repressor complex 1 (PRC1), Oct4 is
sufficient to reprogram mouse embryonic and adult fibroblasts into
induced pluripotent stem (iPS) cells (29). Our pathway analysis demonstrated
that Oct4-mediated stem cell pluripotency signaling was
significantly altered in the metastatic cells and that Oct4 and
Bmi1 protein levels were increased in metastatic cells. Therefore,
the roles of Oct4 and Bmi1 in stemness and differentiation, or even
metastasis, may be explored using our established metastatic
cells.
Overall, our findings suggest that the gain of EMT
properties, migratory ability, invasiveness and stem cell-like
properties are the major signatures of metastatic cells, along with
enhanced malignancy. Furthermore, we established an important
cellular system for the future study of metastasis, which may
benefit the future treatment of head and neck cancer.
Acknowledgements
This study was supported by grants from the National
Science Council (NSC 99-2314-B-075-041-MY3 and NSC
101-2320-B-010-050), the Taipei Veterans General Hospital
(V99ER2-004 and V102E2-003) and the Ministry of Education, Aim for
the Top University Plan, National Yang-Ming University
(101AC-T513).
References
|
1
|
Valastyan S and Weinberg R: Tumor
metastasis: molecular insights and evolving paradigms. Cell.
147:275–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Eccles SA and Welch DR: Metastasis: recent
discoveries and novel treatment strategies. Lancet. 369:1742–1757.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics, 2007. CA Cancer J Clin. 57:43–66.
2007. View Article : Google Scholar
|
|
4
|
Hinerman RW, Mendenhall WM, Morris CG,
Amdur RJ, Werning JW and Villaret DB: Postoperative irradiation for
squamous cell carcinoma of the oral cavity: 35-year experience.
Head Neck. 26:984–994. 2004.PubMed/NCBI
|
|
5
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu MJ, Jan CI, Tsay YG, et al: Elimination
of head and neck cancer initiating cells through targeting glucose
regulated protein 78 signaling. Mol Cancer. 9:2832010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chiou SH, Yu CC, Huang CY, et al: Positive
correlations of Oct-4 and Nanog in oral cancer stem-like cells and
high-grade oral squamous cell carcinoma. Clin Cancer Res.
14:4085–4095. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Loboda A, Nebozhyn M, Watters J, et al:
EMT is the dominant program in human colon cancer. BMC Med
Genomics. 4:92011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mani SA, Guo W, Liao MJ, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen C, Wei Y, Hummel M, et al: Evidence
for epithelial-mesenchymal transition in cancer stem cells of head
and neck squamous cell carcinoma. PLoS One. 6:e164662011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lo JF, Yu CC, Chiou SH, et al: The
epithelial-mesenchymal transition mediator S100A4 maintains
cancer-initiating cells in head and neck cancers. Cancer Res.
71:1912–1923. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen CY, Chiou SH, Huang CY, et al:
Distinct population of highly malignant cells in a head and neck
squamous cell carcinoma cell line established by xenograft model. J
Biomed Sci. 16:1002009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Machesky LM: Lamellipodia and filopodia in
metastasis and invasion. FEBS Lett. 582:2102–2111. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Leve F and Morgado-Díaz JA: Rho GTPase
signaling in the development of colorectal cancer. J Cell Biochem.
113:2549–2559. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Khalil BD and El-Sibai M: Rho GTPases in
primary brain tumor malignancy and invasion. J Neurooncol.
108:333–339. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hall A: Rho GTPases and the actin
cytoskeleton. Science. 279:509–514. 1998. View Article : Google Scholar
|
|
17
|
Etienne-Manneville S and Hall A: Rho
GTPases in cell biology. Nature. 420:629–635. 2002. View Article : Google Scholar
|
|
18
|
Etienne-Manneville S: Polarity proteins in
migration and invasion. Oncogene. 27:6970–6980. 2008. View Article : Google Scholar
|
|
19
|
Gupton SL and Gertler FB: Filopodia: the
fingers that do the walking. Sci STKE. 2007:re52007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Arjonen A, Kaukonen R and Ivaska J:
Filopodia and adhesion in cancer cell motility. Cell Adh Migr.
5:421–430. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shibue T and Weinberg RA: Integrin
β1-focal adhesion kinase signaling directs the
proliferation of metastatic cancer cells disseminated in the lungs.
Proc Natl Acad Sci USA. 106:10290–10295. 2009.
|
|
22
|
Shibue T, Brooks MW, Inan MF, Reinhardt F
and Weinberg RA: The outgrowth of micrometastases is enabled by the
formation of filopodium-like protrusions. Cancer Discov. 2:706–721.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang YJ, Wang QY, Zhou CX, et al: MiR-124
targets Slug to regulate epithelial-mesenchymal transition and
metastasis of breast cancer. Carcinogenesis. 34:713–722. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo W, Keckesova Z, Donaher JL, et al:
Slug and Sox9 cooperatively determine the mammary stem cell state.
Cell. 148:1015–1028. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pardo M, Lang B, Yu L, et al: An expanded
Oct4 interaction network: implications for stem cell biology,
development, and disease. Cell Stem Cell. 6:382–395. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ding J, Xu H, Faiola F, Ma’ayan A and Wang
J: Oct4 links multiple epigenetic pathways to the pluripotency
network. Cell Res. 22:155–167. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li L, Wang J, Hou J, et al: Cdk1
interplays with Oct4 to repress differentiation of embryonic stem
cells into trophectoderm. FEBS Lett. 586:4100–4107. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Z, Oron E, Nelson B, Razis S and
Ivanova N: Distinct lineage specification roles for NANOG, OCT4,
and SOX2 in human embryonic stem cells. Cell Stem Cell. 10:440–454.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Moon JH, Heo JS, Kim JS, et al:
Reprogramming fibroblasts into induced pluripotent stem cells with
Bmi1. Cell Res. 21:1305–1315. 2011. View Article : Google Scholar : PubMed/NCBI
|