Introduction
The tumor necrosis factor receptor (TNFR)-associated
factors (TRAFs) family of proteins consists of six members (TRAF1,
TRAF2, TRAF3, TRAF4, TRAF5 and TRAF6) which act as major signal
transducers for the TNFR superfamily as well as the interleukin-1
receptor/Toll-like receptor (IL-1R/TLR) superfamily (1). A common structural feature of TRAF
proteins is a C-terminal TRAF domain (2) and TRAFs are associated with various
biological functions including adaptive and innate immunity,
embryonic development, stress response and bone metabolism
(1,3).
TRAF4 was originally identified as cysteine-rich
motif associated with RING and TRAF domains (CART1) by differential
screening of a cDNA library from lymph node metastasis of breast
cancer (4). Unlike the other TRAF
family proteins, the biological role of TRAF4 remains elusive,
TRAF4 has been shown to be upregulated in ovarian, bladder, lung
adenocarcinoma, small cell lung carcinoma, colon, breast cancers
and prostate carcinomas by immunohistochemistry and TRAF4 gene
amplification followed by TRAF4 overexpression was detected in ~20%
of the cases from six different types of carcinomas (5). However, the exact mechanism of TRAF4
overexpression in human cancers is still under investigation.
microRNAs (miRNAs) are small non-coding RNAs which
negatively regulate gene expression at the post-transcriptional
level and are involved in essential cellular functions such as
proliferation, differentiation, cell cycle and apoptosis (6). Differential expression of miRNAs has
been observed in several types of cancers and is shown to play an
important role in cancer either as oncogenes or tumor suppressors
(7–10). More specifically, it has been shown
that microRNA-29a (miR-29a) is involved in apoptosis (11,12),
suggesting that its aberrant expression is closely related to
cancer. Altered expression of miR-29 family has been observed in
multiple cancers including cholangiocarcinoma, non-small lung
cancer, nasopharyngeal cancer and acute myeloid leukemia (13–16).
Furthermore, miR-29a has been reported to be downregulated in
hepatocellular carcinoma and acute myeloid leukemia (14,17).
In prostate cancer, miR-29a has been shown to be downregulated only
in castration-resistant prostate cancer compared to benign
prostatic hyperplasia (18),
indicating the possibility that miR-29a has a tumor suppressive
function in prostate cancer.
Using the algorithm, TargetScan (19), we found that miR-29a is a putative
miRNA that binds to the TRAF4 3′ untranslated region (UTR). The aim
of the present study was to investigate the expression of TRAF4 and
miR-29a in localized and metastatic prostate cancer and examine
whether TRAF4 expression is regulated by miR-29a in prostate
cancer.
Materials and methods
Cells
The human prostate cancer cell lines (LNCaP, DU145
and PC3) were maintained in RPMI-1640 medium with 10% fetal bovine
serum (10% FBS) at 37ºC in a humidified atmosphere containing 5%
CO2.
RNA extraction and quantitative real-time
PCR for mRNA and miRNA
Total RNA was isolated using the miRNeasy kit
(Qiagen, Valencia, CA, USA). For mRNA detection, first strand cDNA
was made from 1 μg RNA using iScript cDNA synthesis kit (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) following the manufacturer’s
protocol in a total volume of 20 μl. Quantitative real-time PCR
(Q-PCR) were carried out as previously described (20). In brief, the PCR reactions were
performed with 0.2 μl of cDNA template in 25 μl of reaction mixture
containing 12.5 μl of iQ SYBR-Green Supermix (Bio-Rad Laboratories)
and 0.25 μmol/l each primer. PCR reactions were subjected to hot
start at 95ºC for 3 min followed by 45 cycles of denaturation at
95ºC for 10 sec, annealing at 60ºC for 30 sec and extension at 72ºC
for 30 sec using the CFX96 Real-Time PCR Detection System (Bio-Rad
Laboratories). PCR primers were 5′-CCTGGTGCCTTTGAC AATCT-3′
(forward) and 5′-CTCAGTGACGTGCTGTGG TT-3′ (reverse) for TRAF4 and
5′-GAATATAATCCCAAGC GGTTTG-3′ (forward) and 5′-ACTTCACATCACAGCTC
CCC-3′ (reverse) for TATA binding protein (TBP). TBP was used as an
internal control.
For miRNA expression analysis, cDNA was synthesized
from 10 ng total RNA using TaqMan MicroRNA reverse transcription
kit with miRNA specific RT primer (Applied Biosystems, Foster City,
CA, USA) according to the manufacturer’s protocol in a total volume
of 15 μl. Q-PCR were carried out following the manufacturer’s
protocol. In brief, the PCR reactions were performed with 1.33 μl
of cDNA template in 20 μl of reaction mixture containing 10 μl of
TaqMan 2X Universal PCR Master Mix, No AmpErase UNG (Applied
Biosystems) and 1 μl of TaqMan MicroRNA assays (20X). PCR reactions
were subjected to hot start at 95ºC for 10 min followed by 40
cycles of denaturation at 95ºC for 15 sec and annealing and
extension at 60ºC for 60 sec using the CFX96 Real-Time PCR
Detection System (Bio-Rad Laboratories). The assay names used in
the present study were hsa-miR-29a (#000412) and RNU24 (#001001)
(Applied Biosystems). RNU24 was used as an internal control.
Analysis and fold- differences were determined using the
comparative threshold cycle method. All experiments were performed
in triplicate and the data presented represents mean ± SD.
Western blot analysis
Western blot analysis was performed as previously
described (21). Briefly, cell
lysates were prepared in whole cell lysis buffer (50 mM Tris-HCl pH
7.5, 1% SDS) with 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl
fluoride and 1X Halt Protease Inhibitor Cocktail (Thermo Fisher
Scientific, Rockford, IL, USA) followed by sonication and
centrifugation (14,000 rpm). Extracts were quantified using the
Bio-Rad protein assay (Bio-Rad Laboratories). Lysates were
subjected to SDS-PAGE and transferred to polyvinylidene difluoride
membranes (Millipore, Bedford, MA, USA). Membranes were incubated
with primary antibodies followed by horseradish
peroxidase-conjugated secondary antibodies, and developed with the
Super Signal West Dura Extended Duration Substrate kit (Thermo
Fisher Scientific). Mouse monoclonal TRAF4 antibody (Origene,
Rockville, MD, USA) was used as a primary antibody.
Mimic/inhibitor miRNA treatment
miRIDIAN microRNA mimics and hairpin inhibitors for
hsa-miR-29a and controls were purchased from Thermo Fisher
Scientific. One day prior to the transfection, cells were seeded
without antibiotics at a density of 30–40%. Cells were transiently
transfected with each microRNA mimics and hairpin inhibitors (50
nM) using DharmaFECT1 transfection reagent (Thermo Fisher
Scientific) according to the manufacturer’s protocol with some
modification, in which the volume of DharmaFECT1 transfection
reagent was reduced to half of the recommended volume to limit
toxic effects.
Plasmids
To generate TRAF4 3′UTR reporter construct, the
TRAF4 3′UTR region (1319 bp) was generated by PCR using PrimeSTAR
MAX DNA polymerase (Takara, Shiga, Japan) from LNCaP genomic DNA.
The primers used for amplification were the following:
5′-AATGCTAGCCATGACCTCAGT CAGGCACT-3′
(forward) and 5′-ATACTCGAGAACAAATC TGGGAGGTGAGC-3′
(reverse). The PCR product was digested with
NheI/XhoI and ligated into the same sites of pmirGLO
Dual-luciferase miRNA target expression vector (Promega Corp.,
Madison, WI, USA) to produce pmir-TR4_WT.
The construct, pmir-TR4_MT, which has mutations at
the predicted miR-29a binding sites, were generated by
site-directed mutagenesis using the Quick Change XL Site-Directed
Mutagenesis kit (Stratagene, La Jolla, CA, USA) with the following
oligonucleotides: sense (M1), 5′-CCTCAGGTGC CTCCAATTATGATTTCAGCCCTGGCCCCTG-3′;
and antisense (M1), 5′-CAGGGGCCAGGGCTGAAATCATAA
TTGGAGGCACCTGAGG-3′. Each sequence is identical to that of
pmir-TR4_WT, except for the sequence in bold letters. All
constructs were confirmed by sequencing.
Luciferase assay
The pmir-TR4_WT and the pmir-TR4_MT (0.5 μg) were
transfected into cells (1×105 cells/well in 24-well
plates) using Lipofectamine LTX reagent (Invitrogen, Grand Island,
NY, USA) with miRIDIAN microRNA mimics for hsa-miR-29a or
non-targeting control (final 50 nM). After 48 h, the cells were
harvested using Reporter Lysis Buffer (Promega). The luciferase
activity of the cell lysate was measured using the Dual-Glo
luciferase reporter assay system (Promega) with FLUOstar Omega
microplate reader (BMG Labtech, Ortenberg, Germany).
Clinical samples
Surgical specimens from 23 patients with clinically
localized prostate cancer, ranging in age from 43 to 70 years
(median 59 years) were collected and frozen at the time of radical
prostatectomy at the Johns Hopkins Hospital. A preoperative serum
PSA was a median of 7.59 (ng/ml) (range, 1.89–29.0). The Gleason
score sum (GS) was: 6 (n=15), 7 (n=7) and 9 (n=1), respectively.
The use of surgical specimens for molecular analysis was approved
by the Johns Hopkins Medicine Institutional Review Boards.
In other experiments, samples from clinically
localized prostate cancer (n=20) and soft tissue metastasis (n=20)
were obtained at University of Washington. The age range of the
patients with clinically localized prostate cancer was 48–75 years
(median, 58 years) and a preoperative serum PSA was a median of
7.54 (ng/ml) (range, 2.4–64.0). The GS was: 6 (n=3), 7 (n=14), 8
(n=1) and 9 (n=2), respectively. Soft tissue metastasis was
obtained from lymph node (n=8), liver (n=5), adrenal (n=1), bladder
(n=1), kidney (n=1), lung (n=1) and pancreas (n=1), respectively.
The specimens were used with the approval of the University of
Washington Institutional Review Boards.
Statistical analysis
Data are presented as the mean ± SD and statistical
differences between two groups of data were analyzed using the
Student’s t-test. TRAF4 expression levels in each patient group
were compared by the Mann-Whitney U test. Correlation between TRAF4
and miR-29a expression were analyzed using the Pearson’s
correlation coefficient test. Statistical significance was applied
to P-values of <0.05.
Results
Expression of TRAF4 and miR-29a in
prostate cancer cells
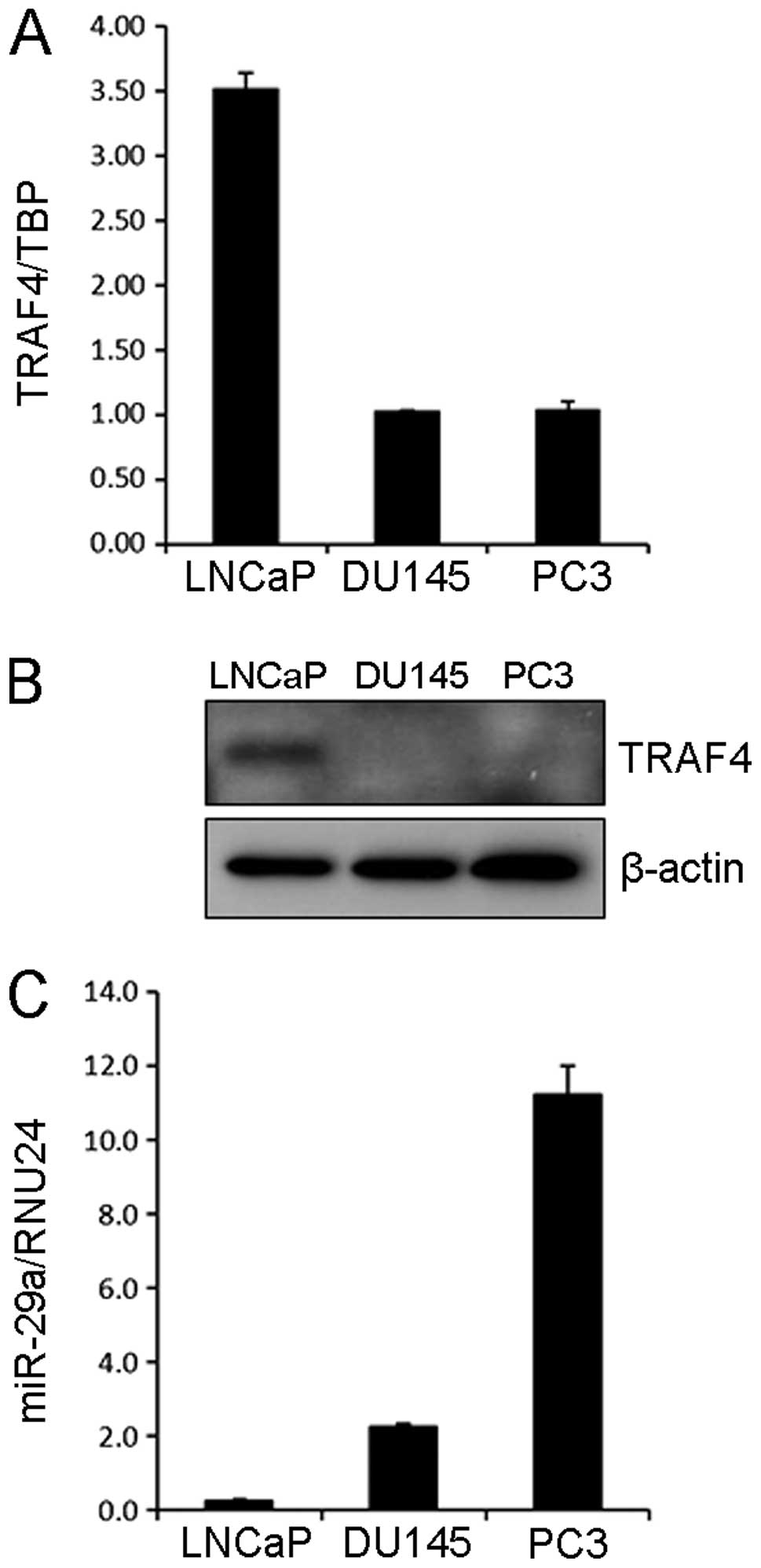
We first analyzed the TRAF4 expression levels in
three different PCa cell lines (LNCaP, DU145 and PC3 cells). TRAF4
was highly expressed in LNCaP cells compared to DU145 and PC3 cells
at both mRNA and protein levels (Fig.
1A and B). Since miR-29a was predicted to bind TRAF4 3′UTR by
the algorithm, TargetScan (19), we
examined the expression of miR-29a in these cell lines. As shown in
Fig. 1C, high and moderate
expression levels of miR-29a were observed in PC3 and DU145 cells,
respectively while LNCaP cells exhibited the lowest expression of
miR-29a among these cell lines.
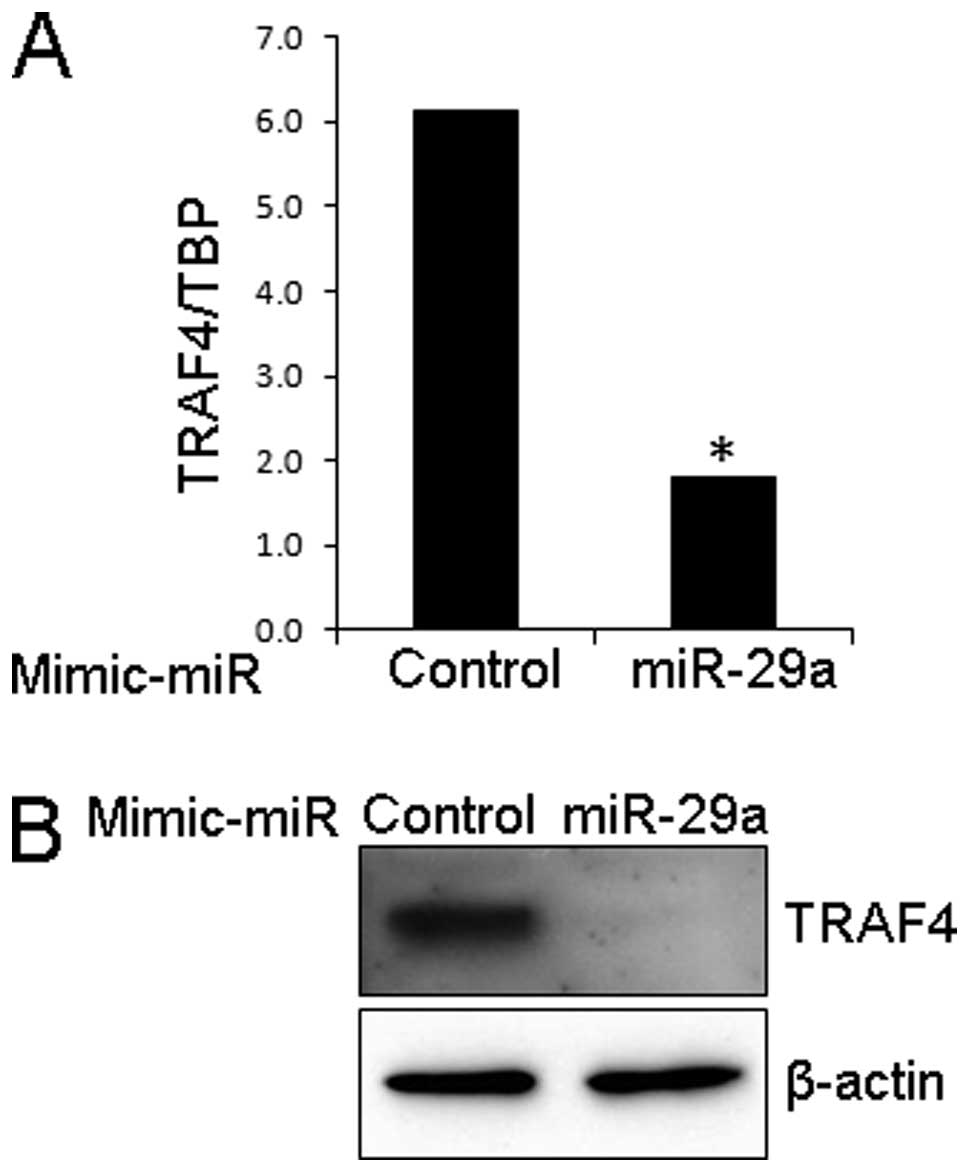
Effect of mimic miR-29a on TRAF4
expression in LNCaP cells
Since we observed an inverse association between
TRAF4 and miR-29a expression in PCa cell lines, we next
investigated the effect of mimic miR-29a on TRAF4 expression in
LNCaP cells in which miR-29a was downregulated and endogenous TRAF4
was expressed. When cells were treated with mimic miR-29a, the
expression of TRAF4 was significantly reduced at both mRNA and
protein levels in LNCaP cells (Fig.
2).
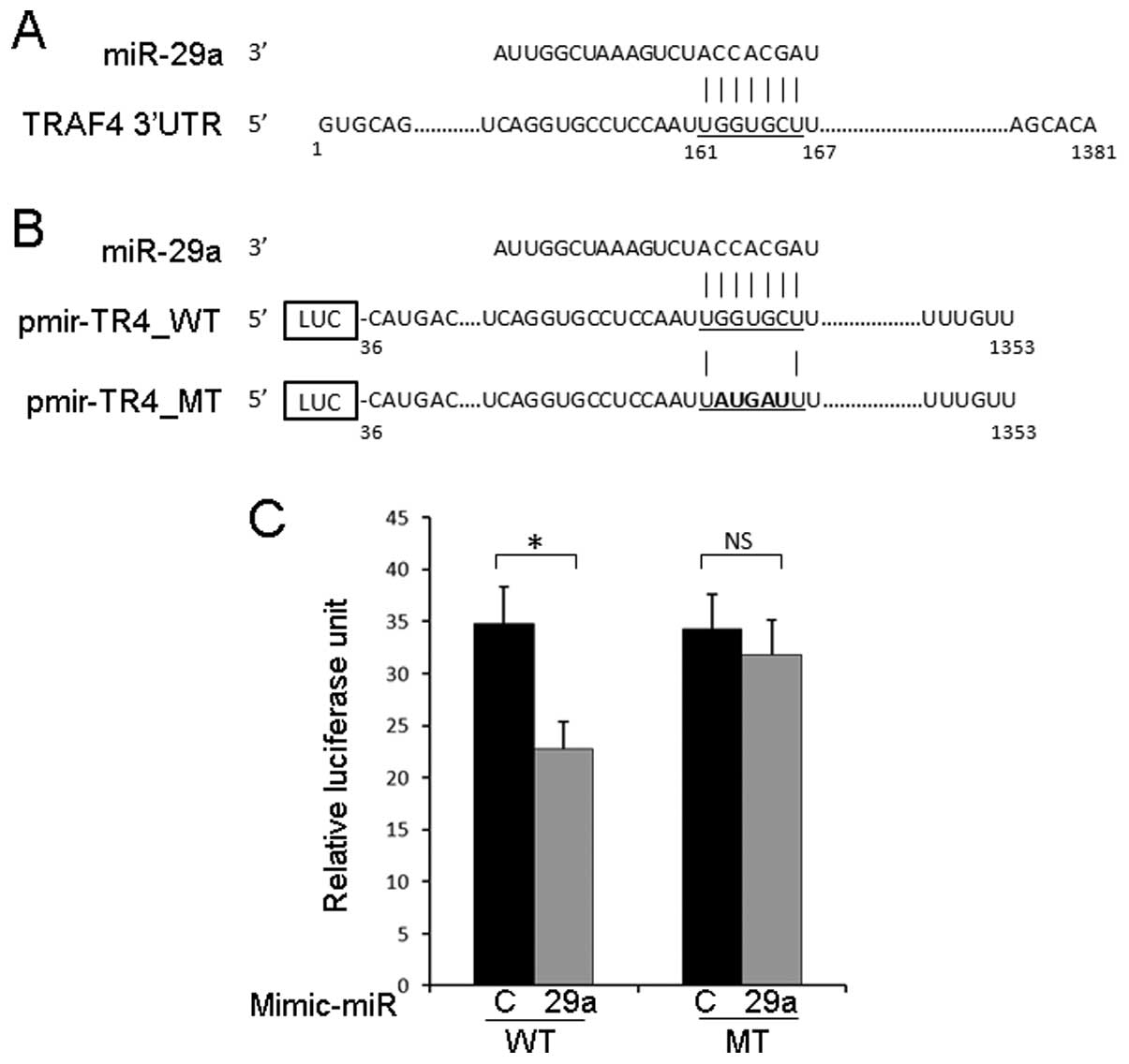
TRAF4 3′UTR is a direct target of
miR-29a
To investigate whether miR-29a binds to the TRAF4
3′UTR, we generated a reporter construct harboring a 1319-bp
fragment of the TRAF4 3′UTR downstream of the firefly luciferase
gene (Fig. 3A and B). As shown in
Fig. 3C, luciferase activity from
pmir-TR4_WT was significantly reduced when mimic miR-29a was
co-transfected, and this suppressive effect of miR-29a was
attenuated by the introduction of mutation at the predicted miR-29a
binding site. These results suggested that miR-29a inhibits the
expression of TRAF4 through the direct binding to the TRAF4
3′UTR.
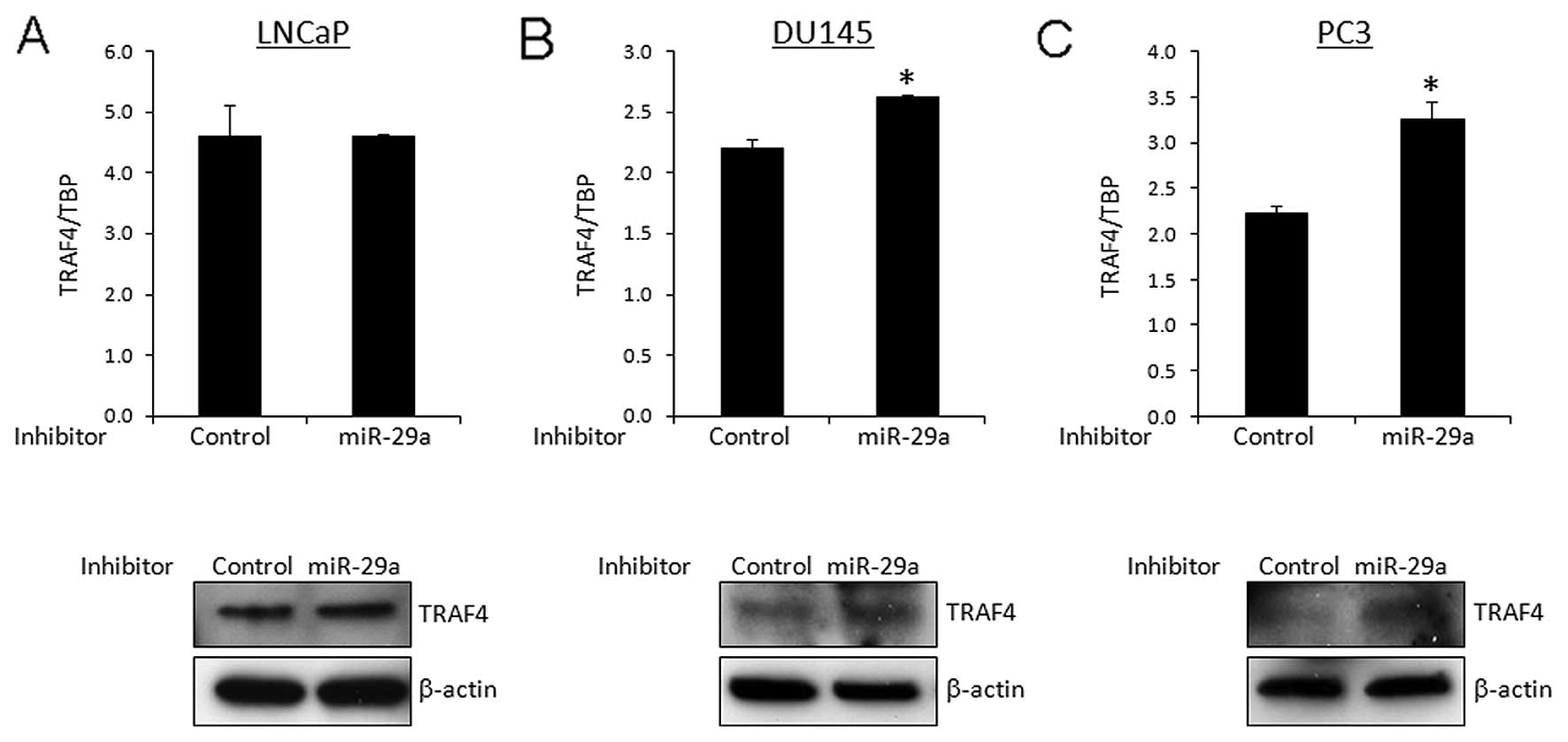
Effect of inhibitor miR-29a on TRAF4
expression in prostate cancer cells
To confirm the suppressive effect of TRAF4
expression by miR-29a, we treated prostate cancer cells with
inhibitor miR-29a. In DU145 and PC3 cells expressing moderate to
high levels of miR-29a, the treatment with inhibitor miR-29a
increased the expression of TRAF4 at both mRNA and protein levels
(Fig. 4B and C). On the other hand,
no obvious effect was observed by the introduction of inhibitor
miR-29a in LNCaP cells in which the expression level of miR-29a was
low compared to DU145 and PC3 cells (Fig. 4A).
Inverse correlation between TRAF4 and
miR-29a expression in patients with prostate cancer
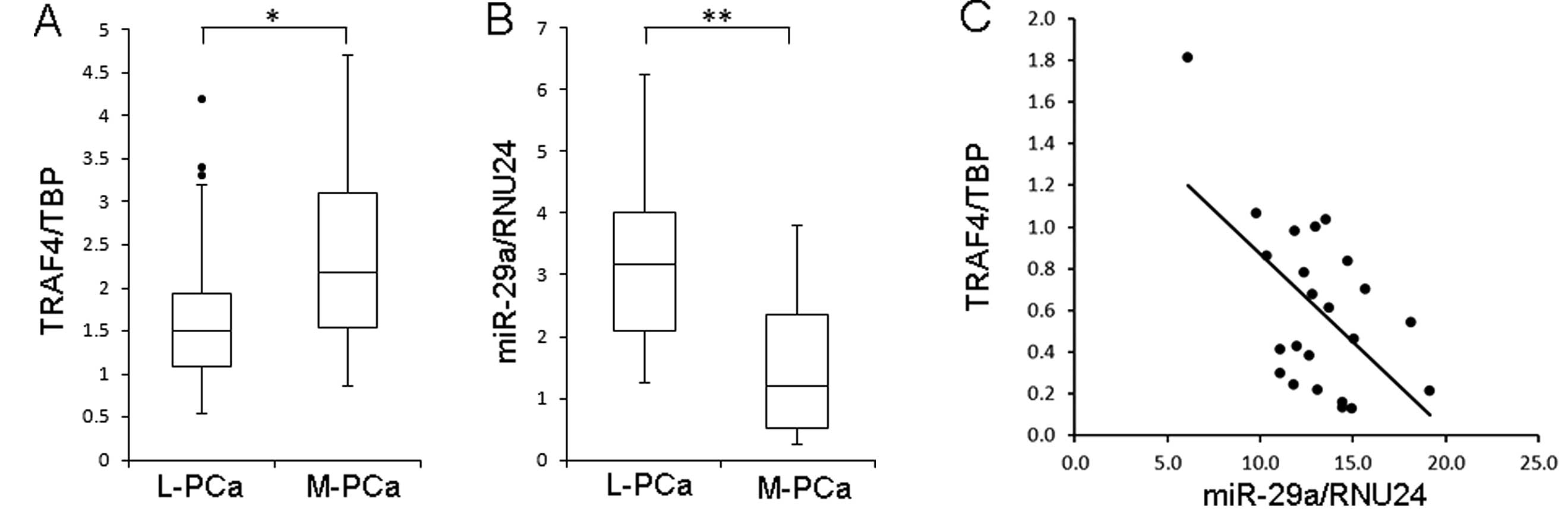
We next examined the expression of TRAF4 and miR-29a
in prostate tumor samples from radical prostatectomy (n=20) and
soft tissue metastasis (n=20). As shown in Fig. 5A, the expression of TRAF4 was
significantly higher in samples from metastatic prostate cancer
compared to localized prostate cancer. On the other hand, miR-29a
expression was significantly lower in metastatic prostate cancer
than in localized prostate cancer (Fig.
5B). Furthermore, there was a significant inverse correlation
between TRAF4 and miR-29a expression in localized prostate cancer
(n=23).
Discussion
In the present study, we demonstrated that TRAF4 is
upregulated and miR-29a is downregulated in metastatic prostate
cancer and TRAF4 is a direct target of miR-29a. These results might
provide a clue to elucidate the molecular mechanism of prostate
cancer progression.
TRAF4 is a unique member of TRAF family proteins
which are known to be involved in immunity, inflammation and
apoptosis (1). However, TRAF4
appears to be a distinct member of the TRAF family, and its
functional role, especially in cancer, remains under investigation
(22). It was reported that TRAF4
is a positive regulator of bone morphogenetic proteins (BMP)
(23). BMPs are members of the
transforming growth factor-β (TGF-β) superfamily and play important
roles in embryogenesis and organogenesis. Since BMPs are well known
to be involved in the development of bone metastasis in prostate
cancer (24), overexpression of
TRAF4 might play an important role in the bone metastasis by
activating BMP signaling in prostate cancer. Bone is the most
common metastatic site in patients with prostate cancer (25) and the complications from bone
metastasis are a major problem when treating patients with prostate
cancer. Thus, TRAF4 may represent a novel therapeutic target to
treat patients with bone metastasis, and further experiments are
required to investigate the role of TRAF4 in the development of
bone metastasis.
The exact role of miR-29a in cancer still remains
elusive. However, accumulating evidence suggests that miR-29a
mainly acts as a tumor suppressor by modulating apoptosis.
Interestingly, it has been shown that the expression of miR-29a is
repressed by c-Myc, Hedgehog and NF-κB at the transcriptional level
(15,26). The oncogenic role of c-Myc in
prostate cancer has been well studied (27) and it is known that c-Myc is highly
upregulated in castration-resistant prostate cancer (28,29).
Thus, overexpression of TRAF4 through the downregulation of miR-29a
by c-Myc might be one of the mechanisms in prostate cancer
progression and contribute to the development of bone
metastasis.
Nuclear factor-κB (NF-κB) proteins are an important
class of transcriptional regulators that sustain the malignant
phenotype in inflammation-related cancer (30). In prostate cancer, the NF-κB
activity is reported to be higher in metastatic prostate cancer
than in localized disease (31),
and elevated activity of NF-κB is correlated with a poor prognosis
in primary prostate cancer (32,33).
Furthermore, significant deregulation of the NF-κB pathway was
identified in metastatic prostate cancer (34). It has been shown that TRAF4
activates NF-κB through the glucocorticoid-induced TNF-R (GITR)
(35). Considering the suppressive
effect of NF-κB on miR-29a transcription (15), there might be a positive feedback
loop from NF-κB to TRAF4 via miR-29a, leading to the progression of
prostate cancer.
In summary, we have demonstrated that TRAF4 is
overexpressed and miR-29a is downregulated in metastatic prostate
cancer. Furthermore, we clearly showed that miR-29a is a negative
regulator of TRAF4 through the direct binding at the TRAF4 3′UTR.
Although further studies are required to elucidate the functional
role of TRAF4, these findings will shed new light on the molecular
mechanism of prostate cancer progression and may provide additional
insight to develop a novel therapeutic strategy to manage patients
with metastatic prostate cancer.
Acknowledgements
The authors wish to thank the members of the
Kulkarni Laboratory for many helpful discussions.
References
|
1
|
Chung JY, Park YC, Ye H and Wu H: All
TRAFs are not created equal: common and distinct molecular
mechanisms of TRAF-mediated signal transduction. J Cell Sci.
115:679–688. 2002.PubMed/NCBI
|
|
2
|
Rothe M, Wong SC, Henzel WJ and Goeddel
DV: A novel family of putative signal transducers associated with
the cytoplasmic domain of the 75 kDa tumor necrosis factor
receptor. Cell. 78:681–692. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xie P: TRAF molecules in cell signaling
and in human diseases. J Mol Signal. 8:72013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Regnier CH, Tomasetto C, Moog-Lutz C, et
al: Presence of a new conserved domain in CART1, a novel member of
the tumor necrosis factor receptor-associated protein family, which
is expressed in breast carcinoma. J Biol Chem. 270:25715–25721.
1995. View Article : Google Scholar
|
|
5
|
Camilleri-Broet S, Cremer I, Marmey B, et
al: TRAF4 overexpression is a common characteristic of human
carcinomas. Oncogene. 26:142–147. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dews M, Homayouni A, Yu D, et al:
Augmentation of tumor angiogenesis by a Myc-activated microRNA
cluster. Nat Genet. 38:1060–1065. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
El Baroudi M, Cora D, Bosia C, Osella M
and Caselle M: A curated database of miRNA mediated feed-forward
loops involving MYC as master regulator. PLoS One.
6:e147422011.PubMed/NCBI
|
|
9
|
Hermeking H: p53 enters the microRNA
world. Cancer Cell. 12:414–418. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hermeking H: The miR-34 family in cancer
and apoptosis. Cell Death Differ. 17:193–199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kole AJ, Swahari V, Hammond SM and
Deshmukh M: miR-29b is activated during neuronal maturation and
targets BH3-only genes to restrict apoptosis. Genes Dev.
25:125–130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Park SY, Lee JH, Ha M, Nam JW and Kim VN:
miR-29 miRNAs activate p53 by targeting p85 α and CDC42. Nat Struct
Mol Biol. 16:23–29. 2009.PubMed/NCBI
|
|
13
|
Fabbri M, Garzon R, Cimmino A, et al:
MicroRNA-29 family reverts aberrant methylation in lung cancer by
targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA.
104:15805–15810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Garzon R, Heaphy CE, Havelange V, et al:
MicroRNA 29b functions in acute myeloid leukemia. Blood.
114:5331–5341. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mott JL, Kobayashi S, Bronk SF and Gores
GJ: mir-29 regulates Mcl-1 protein expression and apoptosis.
Oncogene. 26:6133–6140. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sengupta S, den Boon JA, Chen IH, et al:
MicroRNA 29c is down-regulated in nasopharyngeal carcinomas,
up-regulating mRNAs encoding extracellular matrix proteins. Proc
Natl Acad Sci USA. 105:5874–5878. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiong Y, Fang JH, Yun JP, et al: Effects
of microRNA-29 on apoptosis, tumorigenicity, and prognosis of
hepatocellular carcinoma. Hepatology. 51:836–845. 2010.PubMed/NCBI
|
|
18
|
Porkka KP, Pfeiffer MJ, Waltering KK,
Vessella RL, Tammela TL and Visakorpi T: MicroRNA expression
profiling in prostate cancer. Cancer Res. 67:6130–6135. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shiraishi T, Terada N, Zeng Y, et al:
Cancer/Testis Antigens as potential predictors of biochemical
recurrence of prostate cancer following radical prostatectomy. J
Transl Med. 9:1532011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yoshida T, Shiraishi T, Horinaka M, et al:
Lipoxygenase inhibitors induce death receptor 5/TRAIL-R2 expression
and sensitize malignant tumor cells to TRAIL-induced apoptosis.
Cancer Sci. 98:1417–1423. 2007. View Article : Google Scholar
|
|
22
|
Kedinger V and Rio MC: TRAF4, the unique
family member. Adv Exp Med Biol. 597:60–71. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kalkan T, Iwasaki Y, Park CY and Thomsen
GH: Tumor necrosis factor-receptor-associated factor-4 is a
positive regulator of transforming growth factor-β signaling that
affects neural crest formation. Mol Biol Cell. 20:3436–3450.
2009.PubMed/NCBI
|
|
24
|
Ye L, Lewis-Russell JM, Kyanaston HG and
Jiang WG: Bone morphogenetic proteins and their receptor signaling
in prostate cancer. Histol Histopathol. 22:1129–1147.
2007.PubMed/NCBI
|
|
25
|
Bubendorf L, Schopfer A, Wagner U, et al:
Metastatic patterns of prostate cancer: an autopsy study of 1,589
patients. Hum Pathol. 31:578–583. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chang TC, Yu D, Lee YS, et al: Widespread
microRNA repression by Myc contributes to tumorigenesis. Nat Genet.
40:43–50. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ellwood-Yen K, Graeber TG, Wongvipat J, et
al: Myc-driven murine prostate cancer shares molecular features
with human prostate tumors. Cancer Cell. 4:223–238. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hawksworth D, Ravindranath L, Chen Y, et
al: Overexpression of C-MYC oncogene in prostate cancer predicts
biochemical recurrence. Prostate Cancer Prostatic Dis. 13:311–315.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jenkins RB, Qian J, Lieber MM and Bostwick
DG: Detection of c-myc oncogene amplification and chromosomal
anomalies in metastatic prostatic carcinoma by fluorescence in situ
hybridization. Cancer Res. 57:524–531. 1997.PubMed/NCBI
|
|
30
|
Karin M: Nuclear factor-κB in cancer
development and progression. Nature. 441:431–436. 2006.
|
|
31
|
Ismail HA, Lessard L, Mes-Masson AM and
Saad F: Expression of NF-κB in prostate cancer lymph node
metastases. Prostate. 58:308–313. 2004.
|
|
32
|
Lessard L, Begin LR, Gleave ME, Mes-Masson
AM and Saad F: Nuclear localisation of nuclear factor-kappaB
transcription factors in prostate cancer: an immunohistochemical
study. Br J Cancer. 93:1019–1023. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lessard L, Karakiewicz PI, Bellon-Gagnon
P, et al: Nuclear localization of nuclear factor-kappaB p65 in
primary prostate tumors is highly predictive of pelvic lymph node
metastases. Clin Cancer Res. 12:5741–5745. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Setlur SR, Royce TE, Sboner A, et al:
Integrative microarray analysis of pathways dysregulated in
metastatic prostate cancer. Cancer Res. 67:10296–10303. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Esparza EM and Arch RH: TRAF4 functions as
an intermediate of GITR-induced NF-kappaB activation. Cell Mol Life
Sci. 61:3087–3092. 2004. View Article : Google Scholar : PubMed/NCBI
|