Introduction
The RON receptor tyrosine kinase along with c-Sea,
c-Met and Stk are members of the MET proto-oncogene family
(1). The RON gene consists of 20
exons (2). RON protein is a 180-kDa
heterodimeric protein composed of a 40-kDa α chain and a 150-kDa β
chain linked by disulfide bonds (3). While the α chain contains the
extracellular domain for ligand binding, the β chain includes the
intracellular part that contains a kinase domain and a
transmembrane domain (4). These two
chains are derived from the 180-kDa precursor protein by a
proteolytic cleavage (5). The
macrophage stimulating protein (MSP) was first identified as a
ligand for Ron protein (6). MSP
binds to the RON receptor to upregulate RON kinase activity, which
leads to autophosphorylation on the tyrosine residues in the kinase
domain and the C-terminal docking site (7–9).
Activation of the RON receptor by MSP stimulates a large number of
downstream intracellular pathways (10). Tumor formation and progression
occurs when the accumulation and activation of receptor tyrosine
kinases are abnormal (11). RON
overexpression and activation induce tumor progression and invasive
growth of certain types of epithelial tumor cells (12,13).
Alternative splicing of Ron pre-mRNA produces various protein
isoforms (14). RONΔ165 protein,
identified in gastric cancer cell line KATOIII, is generated by
skipping of exon 11 (15). RONΔ165
does not undergo proteolytic processing and is retained
intracellularly. Furthermore, uneven numbers of cysteine residues
in RONΔ165 produces the RON oligomer. Therefore, RONΔ165 is
constitutively activated without the binding of the MSP ligand.
Abnormal accumulation of this isoform was found in some types of
breast and colon cancer cell lines (16). Furthermore, overexpression of this
splice variant can induce invasive growth and metastasis (15). Apart for the fact that ASF/SF2
induces skipping of exon 11 to control cell motility (16), the splicing mechanism of Ron exon 11
is not yet well understood.
Pre-mRNA splicing is a process in which introns are
removed and then exons are ligated (17–19).
The RNA sequences required for splicing are called splicing signals
that include the 5′ splice site, 3′ splice site, branch point and
polypyrimidine tracts (PPT) (20).
In the alternative splicing procedure, different splicing signals
are selected to produce multiple mRNA isoforms from a single gene
through the numerous combinations of multiple exons (21). Alternative splicing is one of the
critical mechanisms for gene regulation that generates proteomic
diversity (22,23). Abnormal regulation of alternative
splicing causes a variety of human diseases including cancer
(24). Alternative splicing is
finely regulated by several cis-acting elements and
trans-acting elements (25,26).
cis-acting elements are RNA sequences on pre-mRNA that
function as either enhancers or inhibitors to regulate exon
inclusion or skipping. Some cis-elements provide binding
sites for SR proteins and hnRNP proteins to regulate splicing.
Juxtaposed enhancers and inhibitors functionally antagonize each
other (27,28). Exon 11 inclusion of Ron pre-mRNA is
regulated by a juxtaposed enhancer and inhibitor on exon 12
(16). In the present study, we
showed that exon 11 of Ron pre-mRNA also contains various
cis-regulating elements for exon 11 inclusion. Specifically,
a 2-nt RNA, located at 74 nt upstream from the 5′ splice site of
exon 11, functions as an enhancer for exon 11 inclusion. Through
double base and single base substitution analysis on the 2-nt RNA,
we demonstrated that the GA, CC, UG and AC dinucleotides on exon
11, in addition to the wild-type AG sequence, function as enhancers
for exon 11 inclusion of Ron pre-mRNA.
Materials and methods
Construction of plasmids
The wild-type RON exon 10–12 sequences were
amplified from human genomic DNA using RON10-HindIII-for and
RON12-XhoI-rev primers (Table
I) and cloned into HindIII and XhoI restriction
enzyme sites of the pCDNA3.1 (+) vector. Every deletion and
mutation construct was produced with overlapping PCR. All primers
used for minigene constructs are listed in Table I.
 | Table IThe primers used. |
Table I
The primers used.
| Name | Sequences
(5′-3′) |
|---|
|
RON10-HindIII-for |
ATGTTAAGCTTCCTGAATATGTGGTCCGAGAC |
|
RON12-XhoI-rev |
CTTACCTCGAGCTAGCTGCTTCCTCCGCCACC |
| Δ11-1-for |
TATATTGGGCTGGGCTATCAACGTGACCGT |
| Δ11-1-rev |
ACGGTCACGTTGATAGCCCAGCCCAATATA |
| Δ11-2-for |
GGCTGACTGTGTGGGGTGAGAGCTGCCAGC |
| Δ11-2-rev |
GCTGGCAGCTCTCACCCCACACAGTCAGCC |
| Δ11-3-for |
ACGTGACCGTGGGTGTTCCGGGGGGACATG |
| Δ11-3-rev |
CATGTCCCCCCGGAACACCCACGGTCACGT |
| Δ11-4-for |
AGCTGCCAGCACGAGCTGCCCCCTGCCCCC |
| Δ11-4-rev |
GGGGGCAGGGGGCAGCTCGTGCTGGCAGCT |
| Δ11-5-for |
GGGGGACATGGTTGTTGCAGCTTGGCCAGG |
| Δ11-5-rev |
CCTGGCCAAGCTGCAACAACCATGTCCCCC |
| Δ11-6-for |
CCCTGCCCCCATCCCGGTGCCCCATTGCAG |
| Δ11-6-rev |
CTGCAATGGGGCACCGGGATGGGGGCAGGG |
| Δ11-3-1-for |
ACGTGACCGTGGGTGCCAGCACGAGTTCCG |
| Δ11-3-1-rev |
CGGAACTCGTGCTGGCACCCACGGTCACGT |
| Δ11-3-2-for |
GGGTGGTGAGAGCTGTTCCGGGGGGACATG |
| Δ11-3-2-rev |
CATGTCCCCCCGGAACAGCTCTCACCACCC |
| Δ11-3-2(R2)-for |
CTGCCAGCACGTTCCGGGGGGA |
| Δ11-3-2(R2)-rev |
TCCCCCCGGAACGTGCTGGCAG |
| UU-for |
CTGCCAGCACGTTTTCCGGGGGGA |
| UU-rev |
TCCCCCCGGAAAACGTGCTGGCAG |
| CA-for |
CTGCCAGCACGCATTCCGGGGGGA |
| CA-rev |
TCCCCCCGGAATGCGTGCTGGCAG |
| CU-for |
CTGCCAGCACGCTTTCCGGGGGGA |
| CU-rev |
TCCCCCCGGAAAGCGTGCTGGCAG |
| CC-for |
CTGCCAGCACGCCTTCCGGGGGGA |
| CC-rev |
TCCCCCCGGAAGGCGTGCTGGCAG |
| GA-for |
CTGCCAGCACGGATTCCGGGGGGA |
| GA-rev |
TCCCCCCGGAATCCGTGCTGGCAG |
| GU-for |
CTGCCAGCACGGTTTCCGGGGGGA |
| GU-rev |
TCCCCCCGGAAACCGTGCTGGCAG |
| GC-for |
CTGCCAGCACGGCTTCCGGGGGGA |
| GC-rev |
TCCCCCCGGAAGCCGTGCTGGCAG |
| UA-for |
CTGCCAGCACGTATTCCGGGGGGA |
| UA-rev |
TCCCCCCGGAATACGTGCTGGCAG |
| UC-for |
CTGCCAGCACGTCTTCCGGGGGGA |
| UC-rev |
TCCCCCCGGAAGACGTGCTGGCAG |
| AA-for |
CTGCCAGCACGAATTCCGGGGGGA |
| AA-rev |
TCCCCCCGGAATTCGTGCTGGCAG |
| AU-for |
CTGCCAGCACGATTTCCGGGGGGA |
| AU-rev |
TCCCCCCGGAAATCGTGCTGGCAG |
| AC-for |
CTGCCAGCACGACTTCCGGGGGGA |
| AC-rev |
TCCCCCCGGAAGTCGTGCTGGCAG |
| CG-for |
CTGCCAGCACGCGTTCCGGGGGGA |
| CG-rev |
TCCCCCCGGAACGCGTGCTGGCAG |
| GG-for |
CTGCCAGCACGGGTTCCGGGGGGA |
| GG-rev |
TCCCCCCGGAACCCGTGCTGGCAG |
| UG-for |
CTGCCAGCACGTGTTCCGGGGGGA |
| UG-rev |
TCCCCCCGGAACACGTGCTGGCAG |
Cell culture and transfection
MDA-MB-231 cells were grown in RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS) at 37°C in a
humidified 5% CO2 atmosphere. Ron minigene transfection
into MDA-MB-231 cells was carried out with polyethyleneimide (PEI)
according to the manufacturer’s protocol.
RT-PCR
Total RNA was extracted from the MDA-MB-231
transfected cells using RiboEx reagent (GeneAll, Korea) following
the manufacturer’s protocol. Total RNA (1 μg) was reverse
transcribed using oligo dT18 using ImProm-II™ reverse
transcriptase (Promega, Madison, WI, USA) following the
manufacturer’s protocol. cDNA (1 μl) was amplified by PCR using
G-Taq polymerase (Cosmo Genetech, Seoul, Korea). RON minigenes were
as following: RON10-forward (5′-CCTGGCTTTCGCTTCCTACC-3′) and
pCDNA-reverse (5′-CTAGAAGGCACAGTCGAGGCT-3′). GAPDH primer sequences
were as following: GADPH-forward (5′-ACCACAG TCCATGCCATCA-3′) and
GAPDH-reverse (5′-TCCACC ACCCTGTTGCTGTA-3′).
Results
Exon 11 contains various regulatory
elements for exon 11 inclusion of Ron pre-mRNA
In order to identify the enhancer on exon 11 for
exon 11 inclusion of Ron pre-mRNA, we performed mutagenesis
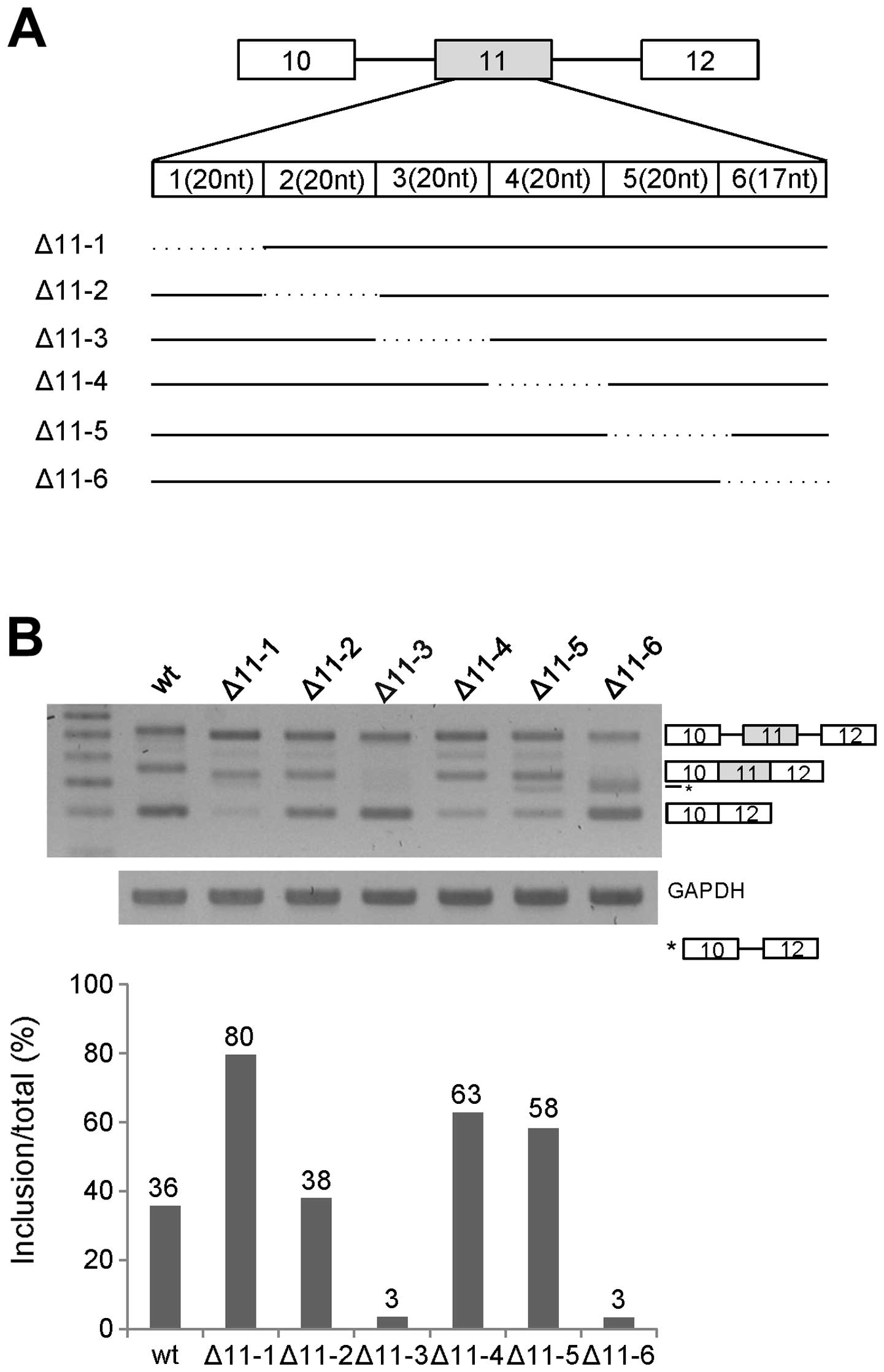
analysis on exon 11. In the first step, we divided exon 11 into six
parts (11-1, 11-2, 11-3, 11-4, 11-5 and 11-6) with the first five
parts containing 20-nt RNA in each and the last part containing
17-nt RNA. The first and last parts are located 15 nt apart from
the 3′ and 5′ splicing sites of exon 11 (Fig. 1A). We produced deletion mutants for
each part of RNA named as Δ11-1, Δ11-2, Δ11-3, Δ11-4, Δ11-5 and
Δ11-6. We extracted RNA from the mutant minigene-transfected cells,
and then performed RT-PCR analysis for Ron exon 11 splicing on each
mutant. As shown in Fig. 1B, exon
11 splicing of Δ11-2 had the similar level of exon 11 inclusion as
that of the wild-type. However, exon 11 inclusion was increased
significantly in the Δ11-1 Δ11-4 and Δ11-5 mutants (~44, ~27 and
~22% each). In addition, the Δ11-3 and Δ11-6 mutants showed
decreased exon 11 inclusion (~33 and ~33% each). Thus, we concluded
that the RNA length (20 nt) and the sequences of 11-3 and 11-6
contained an enhancer for exon 11 inclusion, whereas 11-1, 11-4 and
11-5 RNA contained an inhibitor for exon 11 inclusion of Ron
pre-mRNA. Since the Δ11-6 mutant produced a product caused by
partial splicing (shown as *), the 11-6 RNA part probably also
regulated the partial splicing. To further identify the enhancer
for exon 11 inclusion, we selected the 11-3 RNA part for further
study.
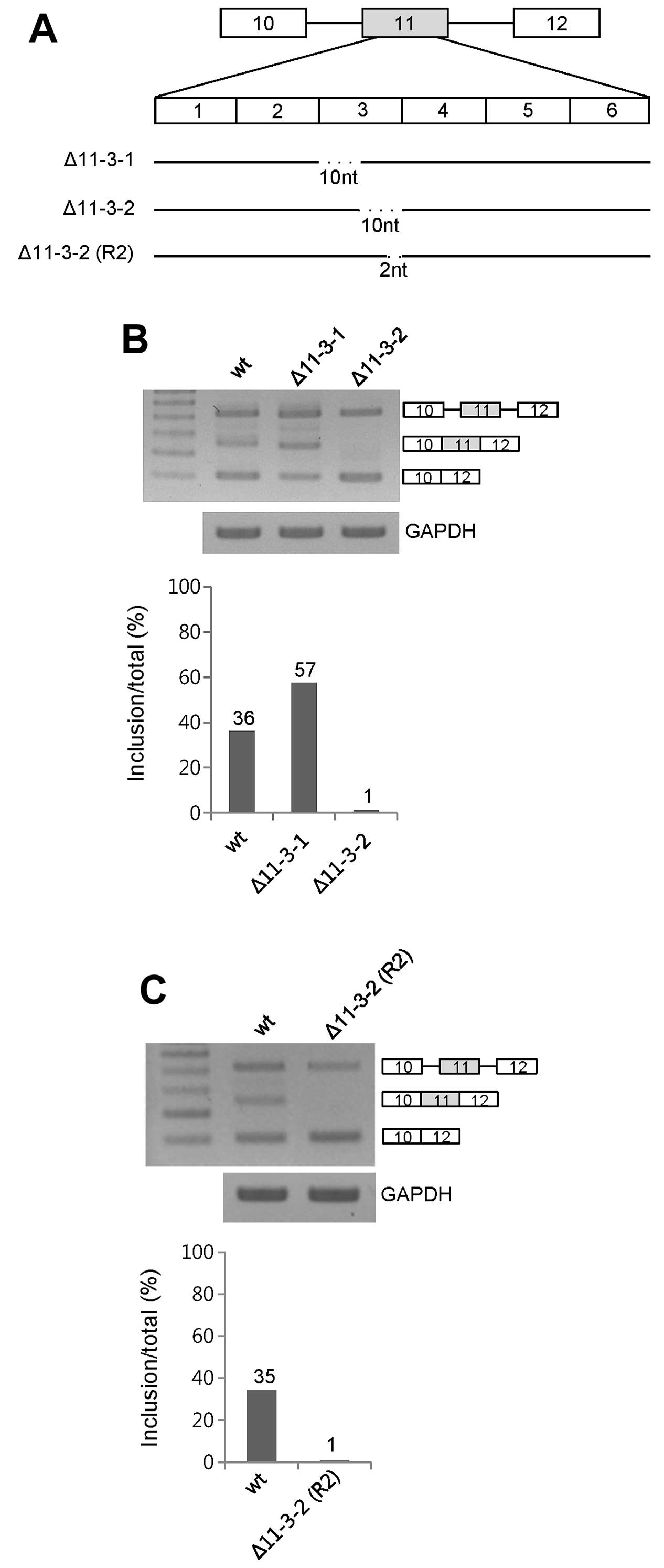
The 2nd 10 nt but not the 1st 10 nt in
11-3 functions as an enhancer for exon 11 inclusion
To further identify the enhancer for exon 11
inclusion in 11-3 RNA of exon 11, we dissected 20 nt of 11-3 RNA
into two 10-nt RNA sections. The two 10-nt deleted mutants were
produced as shown in Fig. 2A,
labeled Δ11-3-1 and Δ11-3-2. RT-PCR analysis showed that only the
Δ11-3-2 mutant decreased exon 11 inclusion (~35%) whereas Δ11-3-1
increased exon 11 inclusion (~21%) (Fig. 2B). Therefore, we conclude that 10 nt
of the 11-3-2 RNA includes the enhancer for exon 11 inclusion.
The 2-nt RNA at the 3′ end of the 11-3-2
RNA functions as an enhancer for exon 11 inclusion
To further understand the enhancer for exon 11
inclusion, we dissected the 2nd 10-nt RNA of 11-3. We deleted 2 nt
from the 3′ end of the 10-nt RNA, labeled Δ11-3-2 (R2) (Fig. 2A). After extraction of RNA from the
minigene-transfected cells, we performed RT-PCR. The results in
Fig. 3B demonstrated that the
Δ11-3-2 (R2) mutant expressed the exon 11 skipped form exclusively
(Fig. 2C). Therefore, we concluded
that the 2-nt RNA at the 3′ end of the 11-3-2 RNA contains
enhancers for exon 11 inclusion.
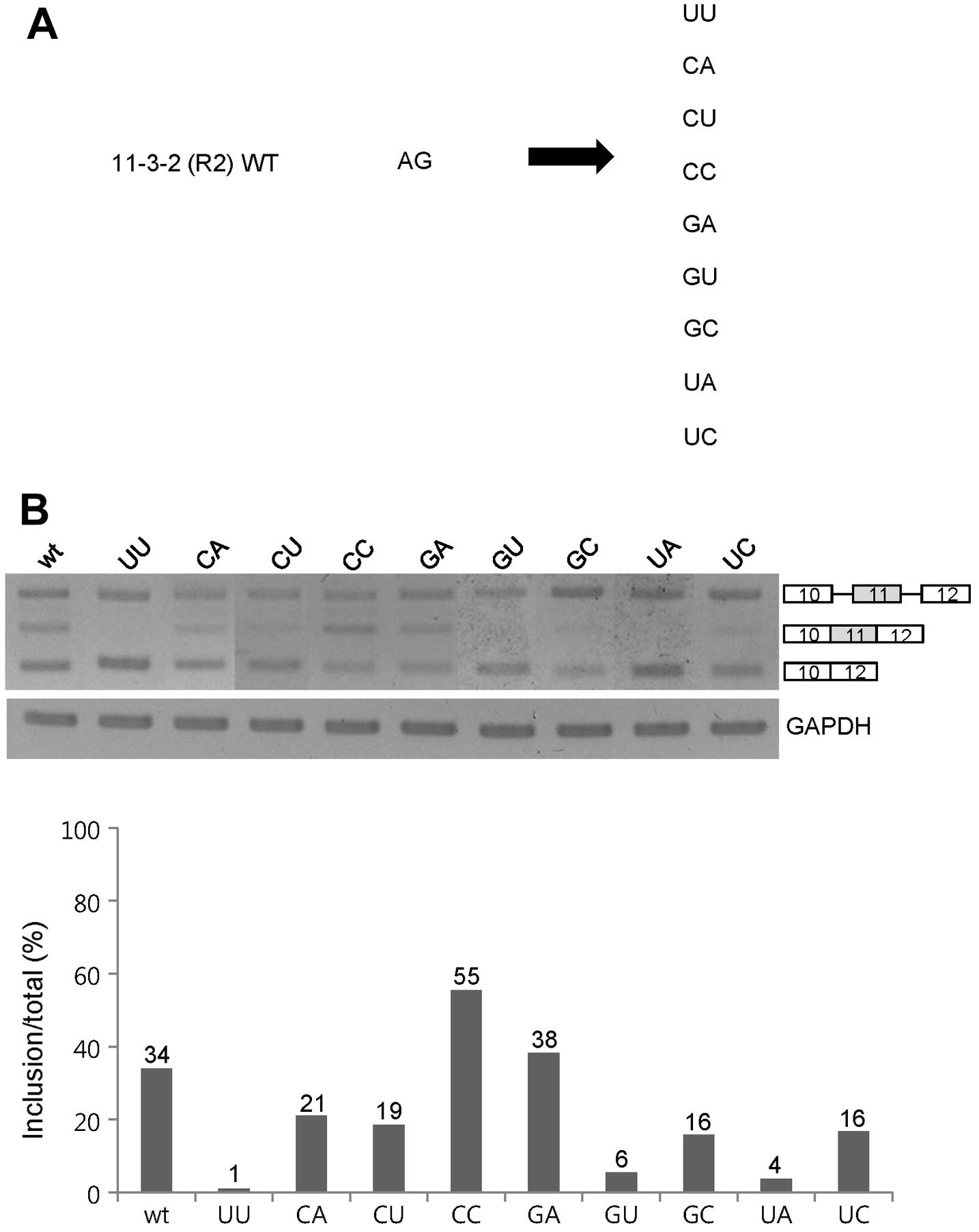
GA, CC and the wild-type AG sequences
function as enhancers for exon 11 inclusion of Ron pre-mRNA
Since the 2-nt RNA at the 11-3-2 section on exon 11
acts as a strong enhancer for exon 11 inclusion, we decided to
pinpoint the sequence requirements for the 2-nt RNA. As the first
approach, we performed substitution mutagenesis analysis on both
nucleotides of the 2-nt RNA. We mutated the AG sequence into
various sequences that cover all of the four base combinations
(Fig. 3A). Among the mutants, as
shown in Fig. 3B, exon 11 inclusion
was completely compromised in the UU, GU and UA mutants. The CA,
CU, GC and UC mutants showed a significant decrease in exon 11
inclusions (~13, ~15, ~18 and ~18%). However, exon 11 inclusion was
increased in the CC mutant, whereas the GA mutant showed a
comparable level of exon 11 inclusion as the wild-type minigene.
The substitution mutant results indicate that most 2-nt sequences
promoted exon 11 skipping of Ron pre-mRNA. One opposite case was
the CC RNA sequence that promoted exon 11 inclusion (~21%).
Therefore, we concluded that the GA, CC and AG sequences at the 3′
end of 11-3-2 RNA are required for the function of the 2-nt RNA as
an enhancer for exon 11 inclusion of Ron pre-mRNA.
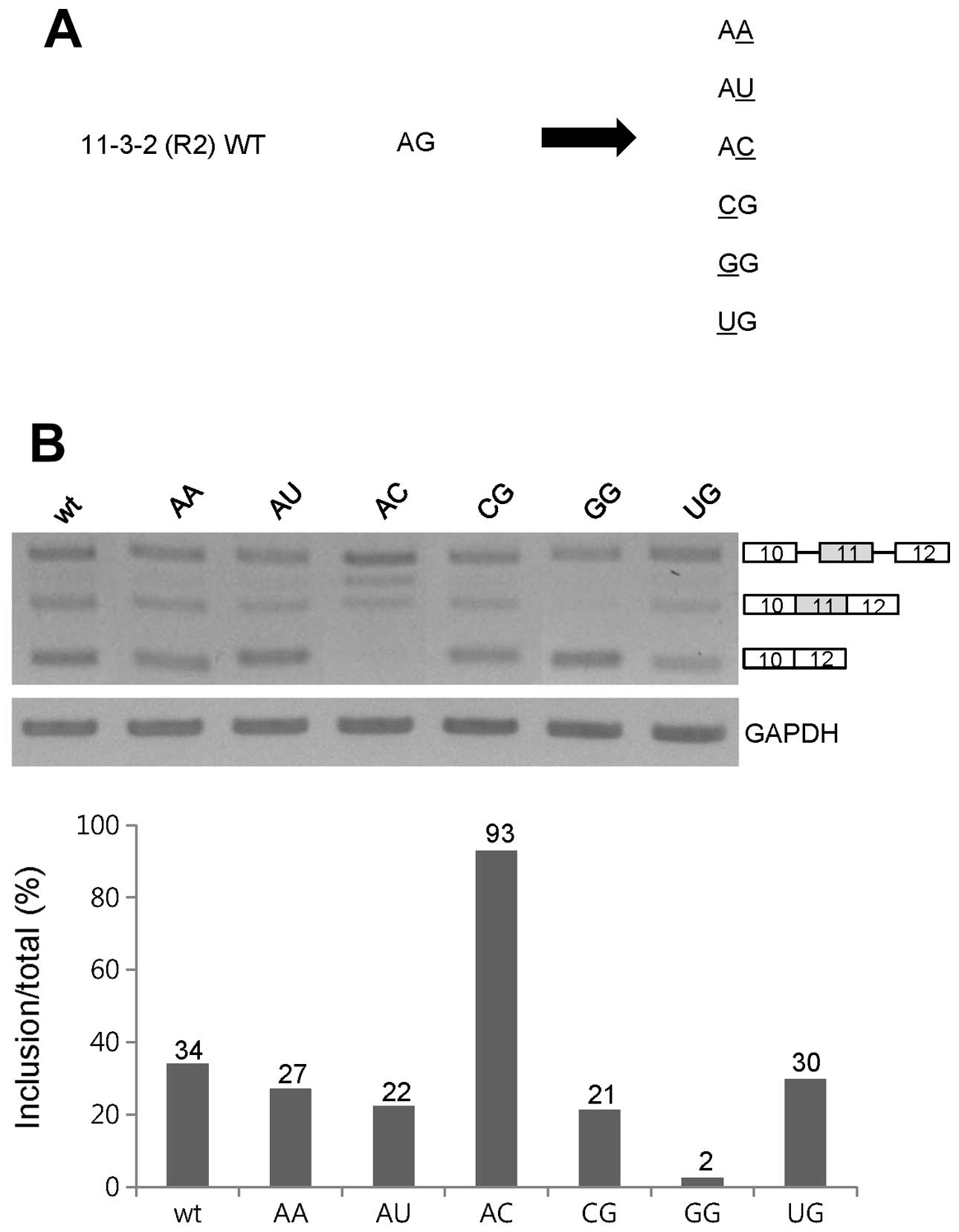
UG and AC also function as enhancers for
exon 11 inclusion of Ron pre-mRNA
In order to further understand the sequence
requirement of the 2-nt enhancer at the 3′ end of the 11-3-2 RNA,
as the second approach, we performed single nucleotide substitution
mutagenesis analysis. In the first set of single nucleotide
mutagenesis assay, we mutated the A nucleotide of wild-type AG at
the 11-3-2 (R2) RNA into the C, G or U nucleotide, whereas the G
residue at the wild-type AG remain unchanged, and were named as CG,
1GG and UG. In the second set of mutagenesis, we mutated the G
nucleotide of the AG dinucleotide into A, U and C nucleotides
separately, whereas the A residue of AG remained unchanged, and
were named as AA, AU and AC (Fig.
4A). RT-PCR analysis in Fig. 4B
shows that the GG mutant demonstrated a completely compromised exon
11 inclusion. In addition, exon 11 inclusion was significantly
reduced in the CG and UG mutant (~13%). In contrast, exon 11
skipping was not significantly decreased in the UG mutant (~3%).
Thus, to maintain the enhancer function of the wild-type AG
dinucleotide for exon 11 inclusion, the first position should be A
and U but not C and G residue. In the second residue substitution
mutants, as shown in Fig. 4B, the
AC mutant, in which the G nucleotide of the AG dinucleotide was
substituted by the C residue, showed the exon 11 inclusion form
exclusively (~93%). Thus, the AC sequence functions as a stronger
enhancer. However, exon 11 inclusion was reduced in the AU and AA
mutants (~12 and ~7%). Therefore, we conclude that the G or C
nucleotide at the second position of the AG dinucleotide maintains
or increases its enhancer function for exon 11 inclusion.
Collectively, the A or U residue at the first position, in
combination with G or C at the second position of the wild-type AG
dinucleotide are required for the enhancer function for exon 11
inclusion. We summarize that UG and AC function as enhancers for
exon 11 inclusion of Ron pre-mRNA.
Discussion
Ron proto-oncogene, a receptor tyrosine kinase,
produces the Δ165 isoform through exon 11 skipping. The Δ165
isoform is a constitutively active isoform without the binding of
the MSP ligand. Exon 11 inclusion is regulated by the juxtaposed
enhancers and inhibitors which are located at exon 12. We
identified a 2-nt enhancer for exon 11 inclusion at exon 11,
located at 74 nt upstream from the 5′ splice site of exon 11,
through serial deletion analysis. Through double base substitution
analysis, we demonstrated that, in addition to the AG sequence, GA
and CC also maintained their enhancer function. Furthermore,
through the single base substitution analysis, we found that UG and
AC function as enhancers for exon 11 inclusion of Ron pre-mRNA.
Exon 11 inclusion/skipping is regulated
by multiple cis-acting elements
Previously, it was shown that an enhancer is located
at exon 12 to promote exon 11 skipping. It was also shown that the
inhibitor RNA, which is located next to the enhancer, promotes exon
11 inclusion. Most importantly, the antagonistic effects of the
enhancer and inhibitor regulate exon 11 inclusion and skipping. Our
results here demonstrated that exon 11 inclusion/skipping is
regulated by, in addition to the enhancer and inhibitor on exon 12,
the enhancer on exon 11. Through multiple 20-nt deletion analyses,
we found that different 20-nt deletions had different effects on
exon 11 splicing.
Exon 11 inclusion was increased significantly in the
Δ11-1, Δ11-4 and Δ11-5 mutants, was decreased significantly in the
Δ11-3 and Δ11-6 mutants, and remained at a similar level for the
wild-type minigene in the Δ11-2 mutant. Our results indicate that
most deletion mutations of exon 11 showed the alteration of exon 11
inclusion. Thus, we concluded that exon inclusion/skipping is
regulated by multiple cis-elements.
Simple determination of the exon enhancer
by deletion mutagenesis is not always correct
Our deletion mutagenesis analysis showed that the
20-nt RNA (11-3) had the enhancer function (Fig. 1B). However, through further deletion
we found that the upstream 10 nt had an inhibitor function, whereas
the downstream 10 nt functioned as an enhancer (Fig. 2B). Thus, it is not correct to
determine the splicing enhancer by deletion mutagenesis, although
deletion mutagenesis definitely provides important information.
cis-acting elements of pre-mRNA splicing are composed of
different combinations of four nucleotides, and usually provide the
functional targets for trans-acting elements. It is not
surprising that each 10-nt RNA had the opposite functions on exon
11 inclusion. One possibility is that one 10-nt RNA section
provided the contact for activator proteins, and the other one
provided the contact for the inhibitory proteins. Another
possibility is that the deletion of 10 nt made the flanking
sequences to be connected to produce another enhancer sequence.
Therefore, determination of splicing enhancer by deletion
mutagenesis is not always correct.
Length and sequence of RNA play roles in
exon 11 inclusion/skipping
Our substitution analysis of the AG dinucleotides
demonstrated that different bases had different effects on exon 11
inclusion of Ron pre-mRNA. By double nucleotide mutagenesis, we
found that the GA, CC as well as AG sequences at the 3′ end of the
11-3-2 RNA were required for the function of the 2-nt RNA as an
enhancer for exon 11 inclusion of Ron pre-mRNA. By single base
substitution analysis, we found that the A or U residue at the
first position, and the G or C at the second position of the AG
dinucleotide were required for the enhancer function for exon 11
inclusion. Surprisingly, we found that several mutants (UA, GC, UU
and GG) completely destroyed exon 11 inclusion, whereas the AC
mutant completely destroyed exon 11 skipping. Our results indicate
the high sequence requirement of the enhancer for exon 11 inclusion
of Ron pre-mRNA.
Acknowledgements
The present study was supported by the Mid-Career
Researcher Program through a National Research Foundation (NRF)
grant (2013029711) funded by the Ministry of Education, Science,
and Technology (MEST), Korea; and a Systems Biology Infrastructure
Establishment grant provided by the Gwangju Institute of Science
and Technology (GIST) in 2013.
References
|
1
|
Ronsin C, Muscatelli F, Mattei MG and
Breathnach R: A novel putative receptor protein tyrosine kinase of
the met family. Oncogene. 8:1195–1202. 1993.PubMed/NCBI
|
|
2
|
Camp ER, Liu W, Fan F, Yang A, Somcio R
and Ellis LM: RON, a tyrosine kinase receptor involved in tumor
progression and metastasis. Ann Surg Oncol. 12:273–281. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gaudino G, Follenzi A, Naldini L, et al:
RON is a heterodimeric tyrosine kinase receptor activated by the
HGF homologue MSP. EMBO J. 13:3524–3532. 1994.PubMed/NCBI
|
|
4
|
Angeloni D, Danilkovitch-Miagkova A,
Ivanov SV, et al: Gene structure of the human receptor tyrosine
kinase RON and mutation analysis in lung cancer samples. Genes
Chromosomes Cancer. 29:147–156. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Follenzi A, Bakovic S, Gual P, Stella MC,
Longati P and Comoglio PM: Cross-talk between the proto-oncogenes
Met and Ron. Oncogene. 19:3041–3049. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang MH, Ronsin C, Gesnel MC, et al:
Identification of the ron gene product as the receptor for the
human macrophage stimulating protein. Science. 266:117–119. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Danilkovitch A and Leonard EJ: Kinases
involved in MSP/RON signaling. J Leukoc Biol. 65:345–348.
1999.PubMed/NCBI
|
|
8
|
Longati P, Bardelli A, Ponzetto C, Naldini
L and Comoglio PM: Tyrosines1234–1235 are critical for activation
of the tyrosine kinase encoded by the MET proto-oncogene (HGF
receptor). Oncogene. 9:49–57. 1994.PubMed/NCBI
|
|
9
|
Iwama A, Yamaguchi N and Suda T: STK/RON
receptor tyrosine kinase mediates both apoptotic and growth signals
via the multifunctional docking site conserved among the HGF
receptor family. EMBO J. 15:5866–5875. 1996.PubMed/NCBI
|
|
10
|
Danilkovitch-Miagkova A: Oncogenic
signaling pathways activated by RON receptor tyrosine kinase. Curr
Cancer Drug Targets. 3:31–40. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Q, Seol DW, Carr B and Zarnegar R:
Co-expression and regulation of Met and Ron proto-oncogenes in
human hepatocellular carcinoma tissues and cell lines. Hepatology.
26:59–66. 1997.PubMed/NCBI
|
|
12
|
Maggiora P, Marchio S, Stella MC, et al:
Overexpression of the RON gene in human breast carcinoma.
Oncogene. 16:2927–2933. 1998.
|
|
13
|
Wang MH, Yao HP and Zhou YQ: Oncogenesis
of RON receptor tyrosine kinase: a molecular target for malignant
epithelial cancers. Acta Pharmacol Sin. 27:641–650. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lu Y, Yao HP and Wang MH: Multiple
variants of the RON receptor tyrosine kinase: biochemical
properties, tumorigenic activities, and potential drug targets.
Cancer Lett. 257:157–164. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Collesi C, Santoro MM, Gaudino G and
Comoglio PM: A splicing variant of the RON transcript induces
constitutive tyrosine kinase activity and an invasive phenotype.
Mol Cell Biol. 16:5518–5526. 1996.PubMed/NCBI
|
|
16
|
Ghigna C, Giordano S, Shen H, et al: Cell
motility is controlled by SF2/ASF through alternative splicing of
the Ron protooncogene. Mol Cell. 20:881–890. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wahl MC, Will CL and Luhrmann R: The
spliceosome: design principles of a dynamic RNP machine. Cell.
136:701–718. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moore MJ and Proudfoot NJ: Pre-mRNA
processing reaches back to transcription and ahead to translation.
Cell. 136:688–700. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Han J, Xiong J, Wang D and Fu XD: Pre-mRNA
splicing: where and when in the nucleus. Trends Cell Biol.
21:336–343. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Black DL: Mechanisms of alternative
pre-messenger RNA splicing. Annu Rev Biochem. 72:291–336. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Blencowe BJ: Alternative splicing: new
insights from global analyses. Cell. 126:37–47. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Keren H, Lev-Maor G and Ast G: Alternative
splicing and evolution: diversification, exon definition and
function. Nat Rev Genet. 11:345–355. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Grabowski PJ and Black DL: Alternative RNA
splicing in the nervous system. Prog Neurobiol. 65:289–308. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
David CJ and Manley JL: Alternative
pre-mRNA splicing regulation in cancer: pathways and programs
unhinged. Genes Dev. 24:2343–2364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee J, Zhou J, Zheng X, et al:
Identification of a novel cis-element that regulates
alternative splicing of Bcl-x pre-mRNA. Biochem Biophys Res Commun.
420:467–472. 2012.
|
|
26
|
Cho S, Moon H, Yang X, et al: Validation
of trans-acting elements that promote exon 7 skipping of SMN2 in
SMN2-GFP stable cell line. Biochem Biophys Res Commun. 423:531–535.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kan JL and Green MR: Pre-mRNA splicing of
IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer
and inhibitor. Genes Dev. 13:462–471. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen H, Kan JL, Ghigna C, Biamonti G and
Green MR: A single polypyrimidine tract binding protein (PTB)
binding site mediates splicing inhibition at mouse IgM exons M1 and
M2. RNA. 10:787–794. 2004. View Article : Google Scholar : PubMed/NCBI
|