Introduction
Cis-diamminedichloroplatinum II (cisplatin)
is one of the most potent antitumor agents and has clinical
activity against a wide variety of solid tumors, such as ovary,
lung, head and neck, and bladder cancer (1–5). It is
generally accepted that the cytotoxic activity of cisplatin results
from its interactions with DNA, inhibition of DNA replication and
DNA repair, disturbance of the cell cycle and the beneficial
process of apoptosis in cancer therapy (6–8).
However, resistance to cisplatin sometimes becomes a limiting
factor in cisplatin-based chemotherapy (9–12). The
mechanisms of resistance include accelerated DNA repair,
inactivation of cisplatin by glutathione, altered apoptosis-related
signals, activation of signaling pathways and declined accumulation
of cisplatin due to decreased uptake and/or increased efflux
(11–14).
With regard to the decline in the accumulation of
cisplatin, it is known that the copper transporter 1 (CTR1)
contributes to cisplatin uptake and regulates sensitivity to
cisplatin (15,16). The copper efflux transporter ATP7B
has been reported to export cisplatin and its overexpression
contributes to clinical cisplatin resistance (17,18).
Additionally, it has been suggested that the ATP-binding cassette
(ABC) transporters MDR1, MRP1 and MRP2 may play a role in enhanced
cisplatin efflux and cisplatin resistance (14,19,20).
KCP-4 is a highly cisplatin-resistant cell line
derived from the human epidermoid carcinoma cell line KB-3-1
(21,22). We previously investigated the
resistance mechanisms of KCP-4 cells and reported that one of the
mechanisms underlying cisplatin resistance in KCP-4 cells involves
activation of NF-κB (23). In
contrast, the accumulation of cisplatin was markedly reduced in
KCP-4 cells when compared with the parent KB-3-1 cells. The
time-dependent cisplatin accumulation in KCP-4 cells in response to
the addition of cisplatin to the culture medium decreased rapidly,
after an initial transient increase. This accumulation was enhanced
by 2,4-dinitrophenol, an inhibitor of phosphorylation of ADP to ATP
(21,22,24).
Therefore, it has been proposed that an ATP-dependent cisplatin
efflux system exists in KCP-4 cells. However, ABC transporters,
namely, MDR1, MRP1 and MRP2, were not expressed in KCP-4 cells
(24,25). Furthermore, ATP-dependent transport
of leukotriene C4 (LTC4), an endogenous
substrate for the glutathione S-conjugate export pump (GS-X pump),
has been found in membrane vesicles prepared from KCP-4 cells
(21,25). LTC4 transport was
inhibited by a GS-platinum complex and by cisplatin or glutathione,
but it was not significant. These results suggested that the GS-X
pump is involved in reducing the accumulation of cisplatin in KCP-4
cells (25), but, to date, no GS-X
pump has been found to be expressed in KCP-4 cells.
The tumor suppressor p53, a transcription factor,
inhibits tumor growth through the induction of apoptosis by
activation of its target genes (26,27).
The p53 mutation has been found in approximately half of all types
of cancer from a variety of tissues (28) and p53 is thought to be an important
factor in the initiation and promotion of various types of cancer.
It is known that p53 mutation enhances cisplatin resistance
(9,14,27).
Exposure of cells to cisplatin activates several genes that mediate
the activation of wild-type p53 and induces cell cycle arrest, DNA
repair and apoptosis. When mutated, the apoptotic function of p53
is abrogated; the mutated protein cannot activate the cell death
program and the sensitivity to cisplatin is reduced by disruption
of the normal signal transduction pathways (9). Thus, the presence of mutations in p53
is an important factor for the development of cisplatin resistance.
However, the characteristics of p53 in KCP-4 cells are also
unclear.
The aim of the present study was to identify the
factors involved in the cisplatin resistance of KCP-4 cells. We
demonstrated that mRNA expressions of ABCA1, ABCA3,
ABCA7 and ABCB10 were increased in KCP-4 cells when
compared with those in KB-3-1 cells; moreover, we found that there
was a heterozygous missense mutation in the p53-encoding gene of
KB-3-1 and KCP-4 cells.
Materials and methods
Cell culture
Dulbecco's modified Eagle's medium (DMEM) was
purchased from Invitrogen Life Technologies (Carlsbad, CA, USA).
KB-3-1, KCP-4 and cisplatin-sensitive revertant KCP-4R cells were
established by Fujii et al (22) and Akiyama et al (29). All cells were cultured in DMEM
containing 10% fetal bovine serum and 100 U/ml of penicillin
(Invitrogen Life Technologies) at 37°C in a 5% CO2
humidified atmosphere.
MTT assay
The
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
was purchased from Wako (Osaka, Japan). Cisplatin was purchased
from Sigma (St. Louis, MO, USA). The MTT colorimetric assay was
used to determine the relative sensitivity of cell lines to
cisplatin, as previously reported (23). Briefly, KB-3-1, KCP-4 and KCP-4R
cells were seeded in each well of 96-well plates at
3×103, 1×104 and 3×103
cells/200-μl/well, respectively, and cultured for 24 h. After a
further 48 h in culture with cisplatin, 50 μl of MTT solution (1
mg/ml in PBS) was added to each well and culturing continued for 4
h. The resultant formazan was dissolved with 100 μl of dimethyl
sulfoxide after aspiration of the culture medium; its absorbance at
595 nm was determined using a microplate reader.
Cisplatin accumulation
Cisplatin accumulation was assessed by the
intracellular concentrations of platinum determined by inductive
coupled plasma spectrometry (ICP). KB-3-1, KCP-4 and KCP-4R cells
were cultured with cisplatin (300 μmol/l) for 2 h at 37°C. Cells
were washed 3 times with cold phosphate-buffered saline (PBS) and
immediately harvested. The harvested cells were further washed with
cold PBS and cell numbers were counted with a hemocytometer before
the aspiration of PBS. Cell pellets were lysed in nitric acid and
the concentrations of platinum were determined by ICP (23).
Reverse transcription-polymerase chain
reaction (RT-PCR) analysis
Total RNA was extracted from KB-3-1, KCP-4 and
KCP-4R cells using TRIzol (Invitrogen Life Technologies). Synthesis
of first-strand cDNA was performed using the SuperScript III
First-Strand Synthesis System (Invitrogen Life Technologies). PCR
was performed using KOD-Plus- (Toyobo, Tokyo, Japan). The reaction
solutions were prepared in a final volume of 50 μl, containing 1 μl
of first-strand cDNA and 0.3 μmol/l sense and antisense primers.
The PCR conditions included an initial denaturation step of 2 min
at 94°C, which was followed by 35 cycles of denaturation for 15 sec
at 94°C, annealing for 30 sec at 55–60°C and extension for 1 min at
68°C.
Quantitative real-time RT-PCR
(qRT-PCR)
The relative mRNA expression levels of ABCA1,
ABCA3, ABCA7 and ABCB10 in the KB-3-1, KCP-4
and KCP-4R cells were evaluated by qRT-PCR using Fast SYBR-Green
Master Mix (Life Technologies, Carlsbad, CA, USA) according to the
manufacturer's instructions. To prepare the standard curve, 1 μg of
total RNA from KB-3-1, KCP-4, or KCP-4R cells was reverse
transcribed with SuperScript III First-Strand Synthesis System,
followed by the preparation of various cDNA dilutions. PCR, using
an Applied Biosystems 7900HT Fast Real-Time PCR System (Life
Technologies) apparatus, was performed in a final volume of 20 μl
of a reaction mixture composed of 10 μl of 2X SYBR-Green PCR Master
Mix, 0.4 pmol of the primers, and 2 μl of diluted cDNA. The
reaction mixture was then loaded onto a 386-well plate and
subjected to an initial denaturation at 95°C for 20 sec, followed
by 40 cycles of amplification at 95°C (2 sec) for denaturation,
60°C (20 sec) for annealing and extension. Primers used for qRT-PCR
were: ABCA1 sense, 5′-GGACCACTGCCCCAGTTCCC-3′ and antisense,
5′-GGGGGACACACAGGCAGCAT-3′; ABCA3 sense, 5′-GC
TGGTGGACAGCAGTATGG-3′ and antisense, 5′-CTCCTC GATGAGGGCTCCAA-3′;
ABCA7 sense, 5′-TACGGCAGAC GTCTTCAGCC-3′ and antisense,
5′-TACTGGCCTGGGCA CACAGC-3′; and ABCB10 sense,
5′-TTGAGCGTGGTGCC TCCAGT-3′ and antisense, 5′-GCTGAGTGGCTTGTGCCA
GG-3′. The transcript amounts were estimated from the respective
standard curves and normalized to 18S ribosomal RNA (sense,
5′-GTAACCCGTTGAACCCCATT-3′ and antisense,
5′-CCATCCAATCGGTAGTAGCG-3′).
Immunoblot analysis
An antibody against p53 (DO-1) was obtained from
Millipore (Billerica, MA, USA). Whole-cell lysates were prepared by
lysing KB-3-1 and KCP-4 cells with detergent buffer [10 mmol/l
Tris-HCl, pH 7.5, 5 mmol/l EDTA, 150 mmol/l sodium chloride, 1%
Triton X-100, 10% glycerol, 1X complete EDTA-free protease
inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA) and 1
mmol/l benzylsulfonyl fluoride]. Insoluble fractions were removed
by centrifugation at 16,000 × g for 10 min at 4°C. Whole-cell
lysates were then boiled in a quarter-volume of sample buffer (125
mmol/l Tris-HCl, pH 7.5, 25% glycerol, 5% sodium dodecyl sulfate,
0.2% bromophenol blue and 25% 2-mercaptoethanol). Proteins in these
samples were separated by SDS-PAGE (10%) and transferred to a PVDF
membrane (Bio-Rad Laboratories, Hercules, CA, USA). The membrane
was incubated for 1 h in Tris-buffered saline (TBS) containing 5%
non-fat milk as blocking buffer and then treated overnight with
1:1,000 anti-p53 antibodies in blocking buffer at 4°C. The membrane
was washed in TBS and then incubated with 1:3,000 HRP-conjugated
goat anti-mouse IgG antibody (Nacalai Tesque, Inc., Kyoto, Japan)
in blocking buffer at room temperature for 1 h. It was then washed
again in TBS. Antibody binding was visualized using the ECL Plus
Western Blotting Detection System (GE Healthcare Bio-Sciences,
Buckingham, UK).
Sequence analysis of p53 gene in KB-3-1
and KCP-4
Complementary DNA samples from total RNAs of KB-3-1
and KCP-4 amplified by RT-PCR were used as templates in the cycle
sequence reaction. Primers used for amplification by RT-PCR were:
sense, 5′-GTGACACGCTTCCCTGGATT-3′ and antisense,
5′-GCTGTCAGTGGGGAACAAGA-3′. Cycle sequence reactions were performed
using a BigDye Terminator v3.1 Cycle Sequencing kit (Life
Technologies). Sequence primers were: 5′-GTGACACGCTTCCCTGGATT-3′
and 5′-AGTTCCTGCATGGGCGGCAT-3′. The reactions were cycled at 96°C
for 60 sec, followed by 25 cycles at 96°C for 10 sec, 50°C for 5
sec and 60°C for 4 min. After purification, products were subjected
to automated sequencing by capillary electrophoresis on an ABI3130
Genetic Analyzer (Life Technologies).
Statistical analysis
Differences between groups were tested by one-way
ANOVA followed by Tukey's test for multiple comparisons. Data are
presented as the means ± SD. Differences were considered
statistically significant at P<0.05.
Results
Comparison of cisplatin resistance among
KB-3-1, KCP-4 and KCP-4R cells
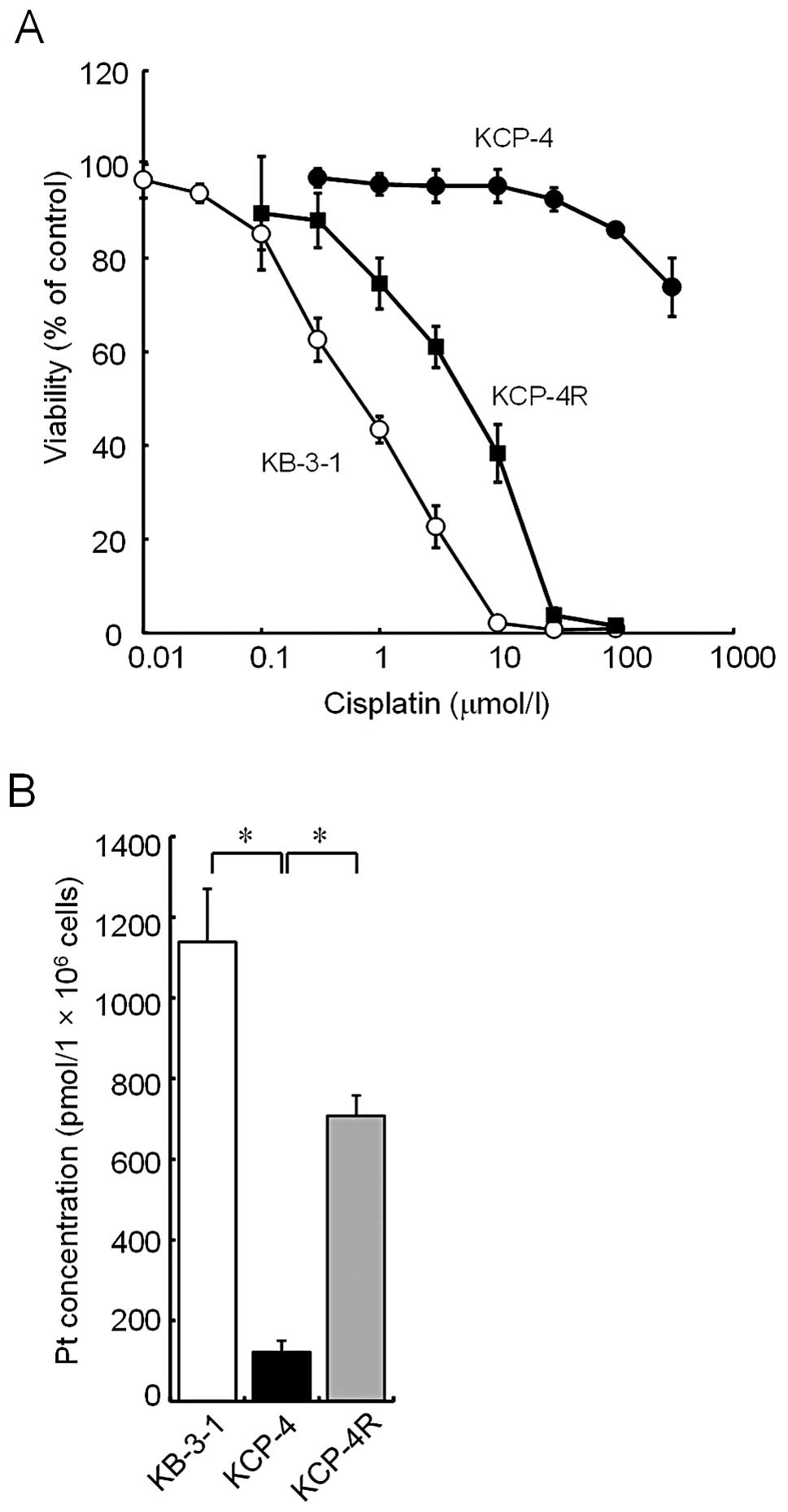
The cell viability of KCP-4 cells in
cisplatin-containing medium, as determined by the MTT assay, was
compared with that of KB-3-1 and KCP-4R, which are a parental cell
line and a cisplatin-sensitive revertant cell line of KCP-4,
respectively (Fig. 1A). KCP-4 cells
were considerably more resistant to cisplatin than the parental
KB-3-1 cells. Sensitivity of KCP-4R cells to cisplatin was
intermediate between that of KCP-4 and KB-3-1 cells.
EC50 values of KB-3-1, KCP-4R and KCP-4 cells were ~0.3,
3 and >300 μmol/l, respectively. Subsequently, the intracellular
accumulation level of platinum in each cell line was measured.
Platinum levels of KCP-4 cells were much lower than those of KB-3-1
cells (122±27 and 1,138±132 pmol/106 cells,
respectively; Fig. 1B). The levels
of KCP-4R cells were 708±50 pmol/106 cells.
Messenger RNA expression of ABC protein
in KB-3-1 and KCP-4 cells
Previous studies suggested that an ATP-dependent
cisplatin efflux system exists in KCP-4 cells (21,22,24).
It is well known that several ABC proteins function as an
ATP-dependent efflux pump. We therefore investigated the mRNA
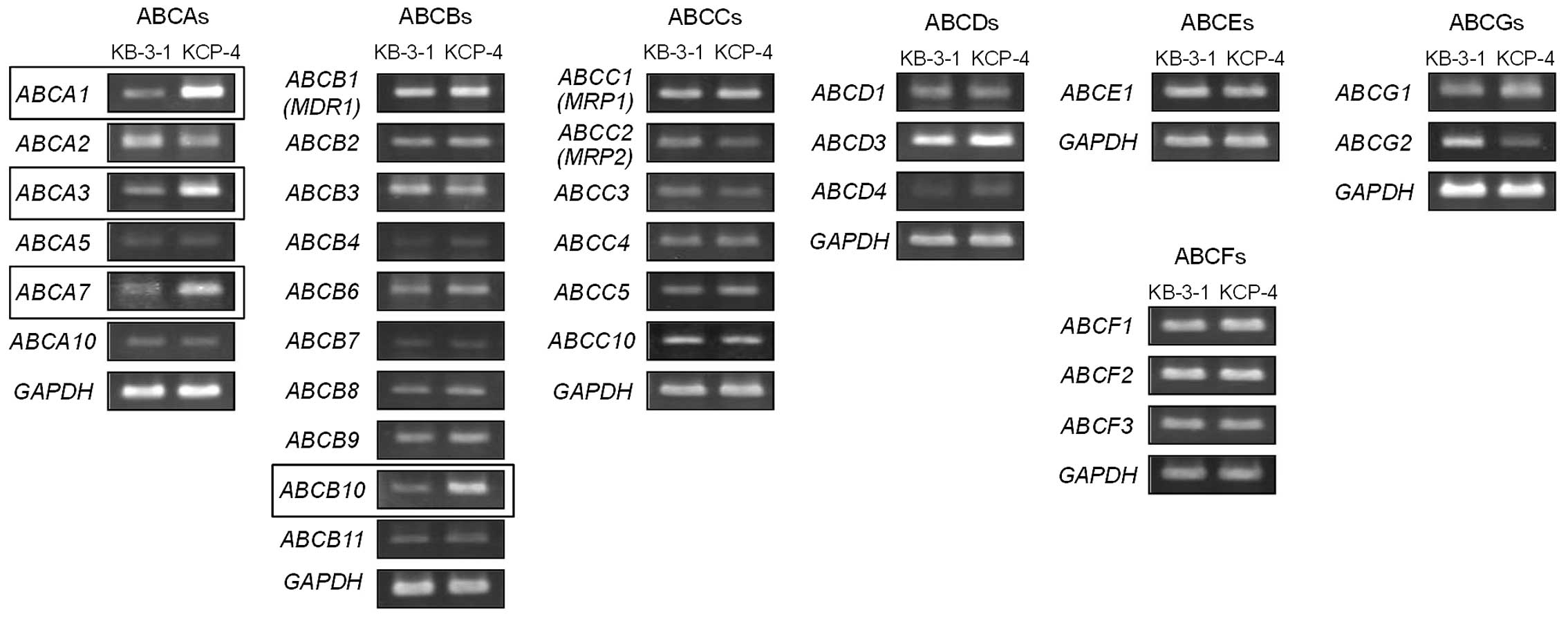
expression of all 48 ABC protein-encoding genes, including
ABCAs, ABCBs, ABCCs, ABCDs,
ABCEs, ABCFs, and ABCGs in KCP-4 and KB-3-1
cells (Table I). RT-PCR analysis
indicated increased mRNA expression of ABCA1, ABCA3,
ABCA7 and ABCB10 in KCP-4 cells (Fig. 2).
 | Table IRT-PCR primer sequences for
amplifying ABC protein-encoding genes. |
Table I
RT-PCR primer sequences for
amplifying ABC protein-encoding genes.
| No. | Gene name | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| 1 | ABCA1 |
gtcattatcatcttcatctgcttc |
cctcacatcttcatcttcatcatt |
| 2 | ABCA2 |
gcagccagagtgtgaaggacgtgg |
gcagagcgtgtccgtgttgaagga |
| 3 | ABCA3 |
gtgcaggccaagcatgtgcag |
cagcaccaggaacacgtgatc |
| 4 | ABCA4 |
aacgtcatcgtgagcatcatcaga |
gaggtcatgactttcagtctgctg |
| 5 | ABCA5 |
ggactggatagaaaacctagaagt |
tactactctatcttcttgtgttcg |
| 6 | ABCA6 |
ggcaaggattacattctagag |
ggaggagtttccatctcattg |
| 7 | ABCA7 |
atcgtggtgctcatctttctg |
tggttccgcaccatgtcaatg |
| 8 | ABCA8 |
ctcctaaccacccactacatg |
tactcctctaggtcaaagctc |
| 9 | ABCA9 |
acccttctgcatactggtttg |
tccatggaatcaggagaaatc |
| 10 | ABCA10 |
aggaacgctaaggtgtattgg |
ctcctgctctttacagagttc |
| 11 | ABCA12 |
ctcacagcatggaagaatgtg |
agagtggtctgactcactaag |
| 12 | ABCA13 |
actgtggactggagacaatac |
ggtacatccatggaagagttg |
| 13 | ABCB1
(MDR1) |
tacagcacggaaggcctaatg |
tgttctcagcaatgctgcagt |
| 14 | ABCB2 |
tggtctgttgactcccttaca |
aaatacctgtggctcttgtcc |
| 15 | ABCB3 |
tacaacacccgccatcaggaa |
tcataaaggaaagcaggctgc |
| 16 | ABCB4 |
gaaacaagagtgggagataag |
ctggacactgaccattgaaaa |
| 17 | ABCB5 |
tgcagcattgctgagaacatc |
actgactgtgcattcactaac |
| 18 | ABCB6 |
tttcactgtgatgcctggaca |
gatgcagccatccttgatgac |
| 19 | ABCB7 |
tatgatgaagctacttcatcg |
cgaacagtttccacagccttt |
| 20 | ABCB8 |
ctggaagcttccgatgaagag |
ttcaggagctcttcatgtgtc |
| 21 | ABCB9 |
attgatggcatcgtcatccag |
catgagaggctgaacatgaag |
| 22 | ABCB10 |
ctgcttctggaactattagtc |
actaacaccgttcttccatcc |
| 23 | ABCB11 |
gtgttgtttgcctgtagcata |
ccatgacagcaatgatatccg |
| 24 | ABCC1
(MRP1) |
gacacagtggactccatgatc |
ccaccaagccagcactgaggc |
| 25 | ABCC2
(MRP2) |
gcagcgatttctgaaacacaa |
tcaacagccacaatgttggtc |
| 26 | ABCC3 |
acctgcacacgtttgtgagct |
gaagatgcctctagctgcaat |
| 27 | ABCC4 |
caaatgtggatccaagaactg |
ggaagtgtttgtaaccatgtg |
| 28 | ABCC5 |
ctagagagactgtggcaagaa |
aaatgccatggttaggatggc |
| 29 | ABCC6 |
aagatccacgcaggagagaag |
cagacacaggagctgtttctg |
| 30 | ABCC7 |
cgaagatcttgctgcttgatg |
tcttgcacctcttcttctgtc |
| 31 | ABCC8 |
ctgagaggaagtgctcagata |
tgagcagcttctctggcttat |
| 32 | ABCC9 |
gcaggatccaatactattcag |
aagagacacggtgagctattg |
| 33 | ABCC10 |
tccctgttgttggtgctcttc |
tctgagttcaggatcgtgttg |
| 34 | ABCC11 |
tcccacatcctcaattctctg |
tgtgccttccatgtgtaaagg |
| 35 | ABCC12 |
gagagaacattcatgagagac |
cttctgctgctagtaacatcg |
| 36 | ABCD1 |
actcagtggaggacatgcaaa |
cgaactgtagcaagtgtgtgt |
| 37 | ABCD2 |
atgctgttatggactggaaag |
tccagctagctgagattctag |
| 38 | ABCD3 |
ggaagggaatttctgacctag |
catatgcaggtagtactcatg |
| 39 | ABCD4 |
cgatgatgagaggatcttgag |
ccacagagtttcagaaccaag |
| 40 | ABCE1 |
tcacccacaatttgtgaccga |
ggtttgaggactgtttgcaac |
| 41 | ABCF1 |
gcttcttcaaccagcagtatg |
agctggcaattggtttctgtg |
| 42 | ABCF2 |
agctggacttagatctctcac |
cacttggtgattgtctgcttc |
| 43 | ABCF3 |
ccttcatcaagagtaagcagg |
agactcgagatcagcagacac |
| 44 | ABCG1 |
ttcagatcatgttcccagtgg |
gaggacaaaataggcaatgag |
| 45 | ABCG2 |
tcaggaagacttatgttccac |
agctctgttctggattccagt |
| 46 | ABCG4 |
tatggctgagaagaagagcag |
aaggtgagcacagttggcatg |
| 47 | ABCG5 |
tagtcaacagtgtagtggctc |
ctaggatgacaagagctggaa |
| 48 | ABCG8 |
actgtgcctacatcatcatct |
ctgctgaactgaatcttcatc |
| 49 | GAPDH |
gtgtgaaccatgagaagtatg |
tttggcaggtttttctagacg |
Expression of ABCA1, ABCA3, ABCA7 and
ABCB10 mRNA in KCP-4R cells
We then investigated the mRNA expression of
ABCA1, ABCA3, ABCA7 and ABCB10 in
KCP-4R cells by RT-PCR. We found that mRNA expressions of these 4
genes were reduced in KCP-4R cells when compared with those in
KCP-4 cells (Fig. 3A). qRT-PCR
analysis revealed that the expression levels of ABCA1,
ABCA3, ABCA7 and ABCB10 mRNA in KCP-4 cells
were 77.2±14.5-, 7.5±1.9-, 11.5±1.7- and 9.9±0.8-fold higher,
respectively, than those in KB-3-1 cells. However, the levels in
KCP-4R cells were significantly reduced when compared with those in
KCP-4 cells (relative expression levels of KCP-4R vs. KCP-4 cells,
0.06±0.01-, 0.45±0.09-, 0.54±0.09- and 0.42±0.09-fold,
respectively; Fig. 3B).
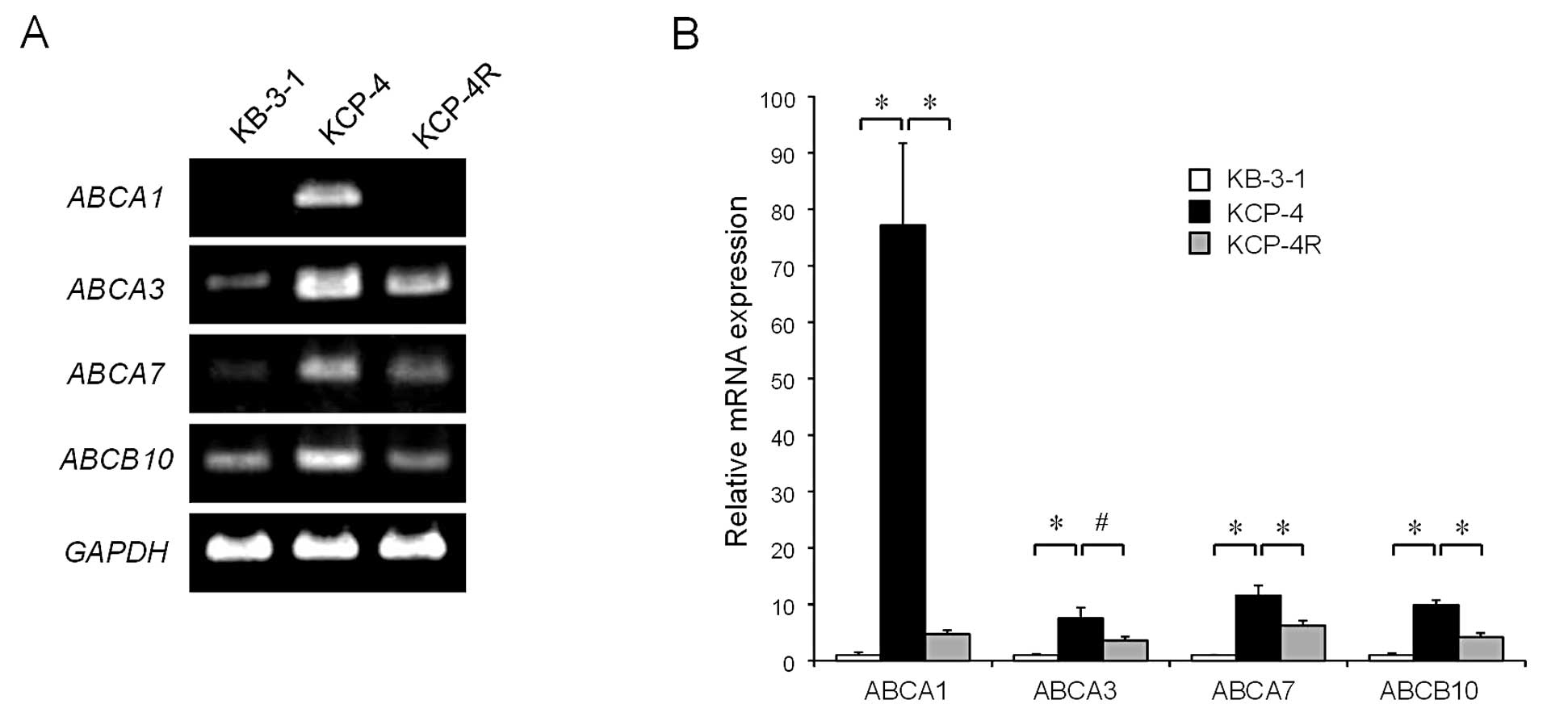 | Figure 3Comparison of mRNA expression levels
of ABCA1, ABCA3, ABCA7 and ABCB10 in
KB-3-1, KCP-4 and KCP-4R cells. (A) Messenger RNA expression of
ABCA1, ABCA3, ABCA7 and ABCB10 in
KB-3-1, KCP-4 and KCP-4R cells as detected by RT-PCR. (B) Relative
mRNA expression of ABCA1, ABCA3, ABCA7 and
ABCB10 in KB-3-1, KCP-4 and KCP-4R cells, as determined by
qRT-PCR. These data are expressed as the means ± SD of 3
independent experiments. *P<0.01;
#P<0.05. |
Expression of p53 in KB-3-1 and KCP-4
cells and sequence analysis
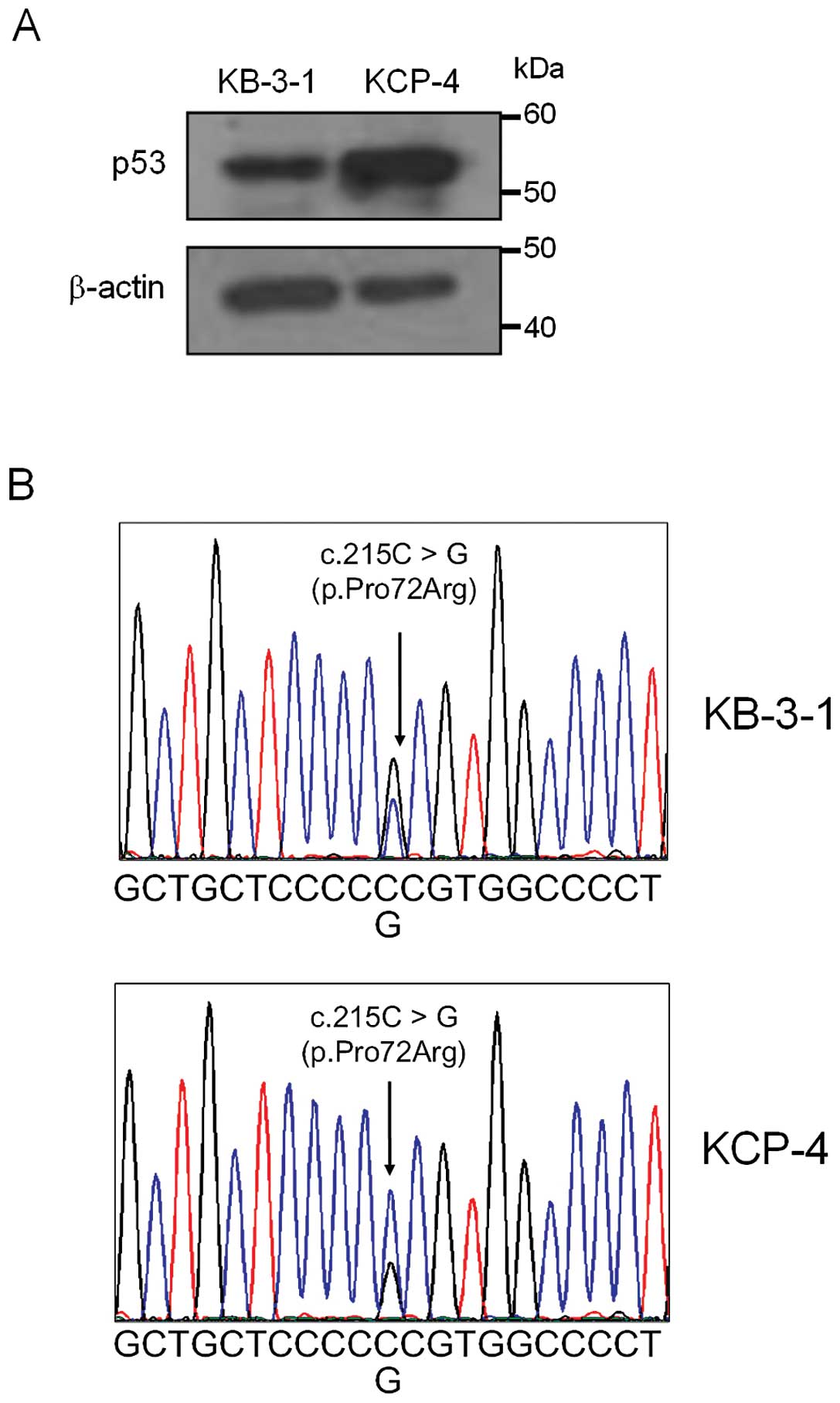
We also investigated whether p53 protein was
expressed in KB-3-1 and KCP-4 cells by immunoblot analysis. The
expression level of p53 in KCP-4 was higher than that in KB-3-1
(Fig. 4A). Sequence analyses of p53
genes prepared from KB-3-1 and KCP-4 cells revealed the existence
of a common heterozygous mutation (c.215C>G) in the p53-encoding
gene of both cells (Fig. 4B). This
missense mutation results in the substitution of proline at codon
72 for arginine (p.Pro72Arg).
Discussion
KCP-4 is a cisplatin-resistant cell line derived
from the human epidermoid carcinoma cell line KB-3-1. In the
present study, we showed that the resistance of KCP-4 cells to
cisplatin was approximately 1,000-fold greater than that of KB-3-1
cells and that the accumulation of cisplatin in KCP-4 cells was
markedly decreased when compared with that in KB-3-1 cells.
Furthermore, the cisplatin sensitivity of the KCP-4R cell line,
which represents revertant KCP-4 cells, recovered to nearly the
level of KB-3-1 cells and the accumulation of cisplatin in KCP-4R
cells was markedly higher than that in KCP-4 cells. These results
indicated that the cisplatin resistance of KCP-4 cells is
associated with the intracellular accumulation of cisplatin.
Previous studies showed that the accumulation of cisplatin in KCP-4
cells was markedly decreased; this appeared to be mediated by any
ATP-dependent efflux pump (21,22,24).
Although some studies have reported that overexpression of MDR1,
MRP1 and MRP2 is related to the mechanism of cisplatin resistance
(14,19,20),
other studies revealed that none of those were involved in the
cisplatin resistance of KCP-4 cells (24,25).
Our data showed that mRNA expression levels of MDR1,
MRP1 and MRP2 in KCP-4 cells were not higher than
those in KB-3-1 cells, which were consistent with the latter
proposal (24,25). In contrast, ABCA1,
ABCA3, ABCA7 and ABCB10 were highly expressed
in KCP-4 cells when compared with KB-3-1 cells. The expression
levels of ABCA1, ABCA3, ABCA7 and
ABCB10 in revertant KCP-4R cells were markedly reduced when
compared with those in KCP-4 cells, but were not equal to those of
KB-3-1 cells. Quantitative real-time RT-PCR analysis also showed
similar variations.
The sensitivity of KCP-4R cells to cisplatin as
determined by MTT assay and cisplatin accumulation levels in KCP-4R
cells was also markedly, but not completely recovered, to the
levels of KB-3-1 cells. The expression of ABCA1,
ABCA3, ABCA7 and ABCB10 in KB-3-1, KCP-4 and
KCP-4R cells varied in parallel with sensitivity to cisplatin and
the intracellular accumulation level of cisplatin. These results
suggested that ABCA1, ABCA3, ABCA7 and ABCB10 may contribute to the
cisplatin resistance of KCP-4 cells.
ABCA1 plays a role in phospholipid transport,
cholesterol homeostasis and high-density lipoprotein metabolism
(30,31). Previous studies showed that the
expression of ABCA1 may be associated with resistance to an
antitumor drug. The antitumor activity of nitidine, a
benzophenanthridine alkaloid that has antitumor effects via the
inhibition of topoisomerase I, was increased upon downregulation of
ABCA1 (32). In addition, it was
reported that ABCA1 may be related to the resistance to the
antitumor activity of curcumin (33). Moreover, it was also reported that
the grade of cancer is influenced by the cholesterol environment in
prostate cancer and that ABCA1 expression contributes to the
control of this environment (34,35).
However, ABCA1 has not been reported to be associated with
resistance to cisplatin.
ABCA3 is known to be expressed predominantly at the
limiting membrane of the lamellar bodies in lung alveolar type II
cells and is involved in surfactant secretion (36,37).
Regarding antitumor drug resistance, ABCA3 is reported to be
involved in multidrug resistance of some leukemia cells (38,39).
These studies showed that ABCA3 remains localized within the
limiting membranes of lysosomes and multivesicular bodies and
induces a phenotype of broad multidrug resistance, mediated by
subcellular drug sequestration to lysosomes. If ABCA3 is associated
with the cisplatin resistance of KCP-4 cells in the same manner,
the intracellular accumulation of cisplatin would be unchanged.
However, our data indicated that accumulation of cisplatin in KCP-4
cells is markedly decreased when compared with that in KB-3-1
cells. Therefore, ABCA3 would not be associated with the cisplatin
resistance of KCP-4 cells or would be involved via a different
mechanism from that in multidrug-resistant leukemia cells.
ABCA7 is reported to be associated with phospholipid
transport, similar to ABCA1 (40),
and is linked to Alzheimer's disease (41,42).
However, to date, no report has demonstrated the association of
ABCA7 with antitumor drug resistance.
ABCB10 has been identified as a mitochondrial
transporter induced by GATA-1 during erythroid differentiation
(43,44). It is known that ABCB10 is involved
in mitochondrial iron importation and heme biosynthesis by
interacting with an iron importer, mitoferrin-1, in the
mitochondrial membrane (45,46).
The association of ABCB10 with antitumor drug resistance has yet to
be clarified.
A previous study showed that the cisplatin efflux
pump in KCP-4 cells functions as a GS-X pump (25). Although MRP1 and MRP2, among ABC
proteins, are already known to function as GS-X pumps (47), it is unknown whether ABCA1, ABCA3,
ABCA7 and ABCB10 function as GS-X pumps. To clarify their potential
function as GS-X pumps and the association of each candidate ABC
protein to cisplatin resistance, further studies using a stable
cell line that expresses high levels of each candidate ABC protein
alone are required.
In the present study, we also investigated whether
there were any mutations in the p53-encoding genes of KB-3-1 and
KCP-4 cells. Sequence analyses demonstrated a common heterozygous
mutation, which results in the substitution of proline at codon 72
for arginine in p53 of both cells. Bergamaschi et al
(48) reported that head and neck
cancer expressing a p53 mutant involving arginine at codon 72 (72R)
had a lower response to chemotherapy than those expressing p53 with
proline at codon 72 (72P). The expression level of p53 protein in
KCP-4 cells was also higher than that in KB-3-1, so that 72R may
strongly influence the resistance of KCP-4 to cisplatin.
In the present study, we showed that enhanced
expression of ABCA1, ABCA3, ABCA7 and
ABCB10 and the mutation of p53 at codon 72, alone or in
combinations, may be candidate factors of the cisplatin resistance
mechanism of KCP-4 cells. We previously reported that one of the
mechanisms for cisplatin resistance in KCP-4 cells is the
activation of NF-κB (23). KCP-4
cells may be highly resistant to cisplatin due to multiple
mechanisms, such as increased cisplatin efflux, expression of
mutant p53 and activation of the NF-κB pathway.
Acknowledgements
The authors thank Dr Shin-ichi Akiyama for providing
KB-3-1, KCP-4 and KCP-4R cells.
References
|
1
|
Loehrer PJ and Einhorn LH: Drugs five
years later. Cisplatin Ann Intern Med. 100:704–713. 1984.PubMed/NCBI
|
|
2
|
Elit L, Oliver TK, Covens A, et al:
Intraperitoneal chemotherapy in the first-line treatment of women
with stage III epithelial ovarian cancer: a systematic review with
metaanalyses. Cancer. 109:692–702. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sculier JP and Moro-Sibilot D: First- and
second-line therapy for advanced nonsmall cell lung cancer. Eur
Respir J. 33:915–930. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vermorken JB and Specenier P: Optimal
treatment for recurrent/metastatic head and neck cancer. Ann Oncol.
21(Suppl 7): vii252–261. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ismaili N, Amzerin M and Flechon A:
Chemotherapy in advanced bladder cancer: current status and future.
J Hematol Oncol. 4:352011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Comess KM, Burstyn JN, Essigmann JM and
Lippard SJ: Replication inhibition and translesion synthesis on
templates containing site-specifically placed
cis-diamminedichloroplatinum(II) DNA adducts. Biochemistry.
31:3975–3990. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Suo Z, Lippard SJ and Johnson KA: Single
d(GpG)/cis-diammineplatinum(II) adduct-induced inhibition of DNA
polymerization. Biochemistry. 38:715–726. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang G, Reed E and Li QQ: Molecular basis
of cellular response to cisplatin chemotherapy in non-small cell
lung cancer (Review). Oncol Rep. 12:955–965. 2004.PubMed/NCBI
|
|
9
|
Siddik ZH: Cisplatin: mode of cytotoxic
action and molecular basis of resistance. Oncogene. 22:7265–7279.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kelland LR: New platinum antitumor
complexes. Crit Rev Oncol Hematol. 15:191–219. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Timmer-Bosscha H, Mulder NH and de Vries
EG: Modulation of cis-diamminedichloroplatinum(II) resistance: a
review. Br J Cancer. 66:227–238. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Borst P, Rottenberg S and Jonkers J: How
do real tumors become resistant to cisplatin? Cell Cycle.
7:1353–1359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Torigoe T, Izumi H, Ishiguchi H, et al:
Cisplatin resistance and transcription factors. Curr Med Chem
Anticancer Agents. 5:15–27. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stewart DJ: Mechanisms of resistance to
cisplatin and carboplatin. Crit Rev Oncol Hematol. 63:12–31. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Safaei R and Howell SB: Copper
transporters regulate the cellular pharmacology and sensitivity to
Pt drugs. Crit Rev Oncol Hematol. 53:13–23. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ishida S, Lee J, Thiele DJ and Herskowitz
I: Uptake of the anticancer drug cisplatin mediated by the copper
transporter Ctr1 in yeast and mammals. Proc Natl Acad Sci USA.
99:14298–14302. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miyashita H, Nitta Y, Mori S, et al:
Expression of copper-transporting P-type adenosine triphosphatase
(ATP7B) as a chemoresistance marker in human oral squamous cell
carcinoma treated with cisplatin. Oral Oncol. 39:157–162. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Samimi G, Safaei R, Katano K, et al:
Increased expression of the copper efflux transporter ATP7A
mediates resistance to cisplatin, carboplatin, and oxaliplatin in
ovarian cancer cells. Clin Cancer Res. 10:4661–4669. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Surowiak P, Materna V, Kaplenko I, et al:
ABCC2 (MRP2, cMOAT) can be localized in the nuclear membrane of
ovarian carcinomas and correlates with resistance to cisplatin and
clinical outcome. Clin Cancer Res. 12:7149–7158. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Taniguchi K, Wada M, Kohno K, et al: A
human canalicular multispecific organic anion transporter (cMOAT)
gene is overexpressed in cisplatin-resistant human cancer cell
lines with decreased drug accumulation. Cancer Res. 56:4124–4129.
1996.
|
|
21
|
Fujii R, Mutoh M, Sumizawa T, Chen ZS,
Yoshimura A and Akiyama S: Adenosine triphosphate-dependent
transport of leukotriene C4 by membrane vesicles
prepared from cisplatin-resistant human epidermoid carcinoma tumor
cells. J Natl Cancer Inst. 86:1781–1784. 1994.PubMed/NCBI
|
|
22
|
Fujii R, Mutoh M, Niwa K, Yamada K, Aikou
T, Nakagawa M, Kuwano M and Akiyama S: Active efflux system for
cisplatin in cisplatin-resistant human KB cells. Jpn J Cancer Res.
85:426–433. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Oiso S, Ikeda R, Nakamura K, Takeda Y,
Akiyama S and Kariyazono H: Involvement of NF-κB activation in the
cisplatin resistance of human epidermoid carcinoma KCP-4 cells.
Oncol Rep. 28:27–32. 2012.
|
|
24
|
Chen ZS, Mutoh M, Sumizawa T, et al: An
active efflux system for heavy metals in cisplatin-resistant human
KB carcinoma cells. Exp Cell Res. 240:312–320. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chuman Y, Chen ZS, Sumizawa T, et al:
Characterization of the ATP-dependent LTC4 transporter
in cisplatin-resistant human KB cells. Biochem Biophys Res Commun.
226:158–165. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Velculescu VE and El-Deiry WS: Biological
and clinical importance of the p53 tumor-suppressor gene.
Clin Chem. 42:858–868. 1996.PubMed/NCBI
|
|
27
|
El-Deiry WS: The role of p53 in
chemosensitivity and radiosensitivity. Oncogene. 22:7486–7495.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hussain SP and Harris CC: Molecular
epidemiology of human cancer: contribution of mutation spectra
studies of tumor suppressor genes. Cancer Res. 58:4023–4037.
1998.
|
|
29
|
Akiyama S, Fojo A, Hanover JA, Pastan I
and Gottesman MM: Isolation and genetic characterization of human
KB cell lines resistant to multiple drugs. Somat Cell Mol Genet.
11:117–126. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tanaka AR, Abe-Dohmae S, Ohnishi T, et al:
Effects of mutations of ABCA1 in the first extracellular domain on
subcellular trafficking and ATP binding/hydrolysis. J Biol Chem.
278:8815–8819. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nagao K, Takahashi K, Hanada K, Kioka N,
Matsuo M and Ueda K: Enhanced apoA–I-dependent cholesterol efflux
by ABCA1 from sphingomyelin-deficient Chinese hamster ovary cells.
J Biol Chem. 282:14868–14874. 2007.
|
|
32
|
Iwasaki H, Okabe T, Takara K, Yoshida Y,
Hanashiro K and Oku H: Down-regulation of lipids transporter ABCA1
increases the cytotoxicity of nitidine. Cancer Chemother Pharmacol.
66:953–959. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bachmeier BE, Iancu CM, Killian PH, et al:
Overexpression of the ATP binding cassette gene ABCA1 determines
resistance to Curcumin in M14 melanoma cells. Mol Cancer.
8:1292009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee BH, Taylor MG, Robinet P, et al:
Dysregulation of cholesterol homeostasis in human prostate cancer
through loss of ABCA1. Cancer Res. 73:1211–1218. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sekine Y, Demosky SJ, Stonik JA, et al:
High-density lipoprotein induces proliferation and migration of
human prostate androgen-independent cancer cells by an
ABCA1-dependent mechanism. Mol Cancer Res. 8:1284–1294. 2010.
View Article : Google Scholar
|
|
36
|
Matsumura Y, Sakai H, Sasaki M, Ban N and
Inagaki N: ABCA3-mediated choline-phospholipids uptake into
intracellular vesicles in A549 cells. FEBS Lett. 581:3139–3144.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Matsumura Y, Ban N, Ueda K and Inagaki N:
Characterization and classification of ATP-binding cassette
transporter ABCA3 mutants in fatal surfactant deficiency. J Biol
Chem. 281:34503–34514. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chapuy B, Koch R, Radunski U, et al:
Intracellular ABC transporter A3 confers multidrug resistance in
leukemia cells by lysosomal drug sequestration. Leukemia.
22:1576–1586. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chapuy B, Panse M, Radunski U, et al: ABC
transporter A3 facilitates lysosomal sequestration of imatinib and
modulates susceptibility of chronic myeloid leukemia cell lines to
this drug. Haematologica. 94:1528–1536. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Iwamoto N, Abe-Dohmae S, Sato R and
Yokoyama S: ABCA7 expression is regulated by cellular cholesterol
through the SREBP2 pathway and associated with phagocytosis. J
Lipid Res. 47:1915–1927. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Karch CM, Jeng AT, Nowotny P, Cady J,
Cruchaga C and Goate AM: Expression of novel Alzheimer's disease
risk genes in control and Alzheimer's disease brains. PLoS One.
7:e509762012.
|
|
42
|
Kim WS, Li H, Ruberu K, et al: Deletion of
Abca7 increases cerebral amyloid-β accumulation in the J20
mouse model of Alzheimer's disease. J Neurosci. 33:4387–4394.
2013.PubMed/NCBI
|
|
43
|
Graf SA, Haigh SE, Corson ED and Shirihai
OS: Targeting, import, and dimerization of a mammalian
mitochondrial ATP binding cassette (ABC) transporter, ABCB10
(ABC-me). J Biol Chem. 279:42954–42963. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shirihai OS, Gregory T, Yu C, Orkin SH and
Weiss MJ: ABC-me: a novel mitochondrial transporter induced by
GATA-1 during erythroid differentiation. EMBO J. 19:2492–2502.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen W, Dailey HA and Paw BH:
Ferrochelatase forms an oligomeric complex with mitoferrin-1 and
Abcb10 for erythroid heme biosynthesis. Blood. 116:628–630. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen W, Paradkar PN, Li L, et al: Abcb10
physically interacts with mitoferrin-1 (Slc25a37) to enhance its
stability and function in the erythroid mitochondria. Proc Natl
Acad Sci USA. 106:16263–16268. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ishikawa T, Li ZS, Lu YP and Rea PA: The
GS-X pump in plant, yeast, and animal cells: structure, function,
and gene expression. Biosci Rep. 17:189–207. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bergamaschi D, Gasco M, Hiller L, et al:
p53 polymorphism influences response in cancer chemotherapy via
modulation of p73-dependent apoptosis. Cancer Cell. 3:387–402.
2003. View Article : Google Scholar : PubMed/NCBI
|