Introduction
Gastric cancer is a common malignancy worldwide,
with eastern Asia considered to have the highest mortality rate.
The mainstay of treatment for gastric cancer is surgery (1). However, some patients experience a
recurrence after curative resection for advanced gastric cancer.
Various regimens with 5-fluorouracil (5-FU) have shown that
adjuvant chemotherapy is effective against gastric cancer (2–4). 5-FU
is the principal component of S-1 (TS-1; Taiho Pharmaceutical Co.,
Ltd., Tokyo, Japan). S-1 is an orally active combination of tegafur
(a prodrug that is converted by cells to fluorouracil), gimeracil
(an inhibitor of dihydropyrimidine dehydrogenase, which degrades
fluorouracil), and oteracil (which inhibits the phosphorylation of
fluorouracil in the gastrointestinal tract, thereby reducing the
gastrointestinal toxic effects of fluorouracil). The Adjuvant
Chemotherapy Trial of S-1 for Gastric Cancer (ACTS-GC) group
reported on the effects of postoperative S-1 chemotherapy compared
with surgery alone in pathological stage II or III gastric cancer
patients (5). Since then, S-1
chemotherapy has been recommended as adjuvant chemotherapy for
patients who have undergone curative surgery for advanced gastric
cancer (6).
Nevertheless, some patients suffer recurrence even
after S-1 adjuvant chemotherapy (5,7). There
was no clear predictive marker of its effects. If we identify
predictors of relapse after adjuvant chemotherapy, patients may be
able to receive more suitable regimen in their cancer
characteristics and avoid both side-effects and excessive
therapy.
microRNAs (miRNAs) regulate physiological processes,
cell and differentiation, and apoptosis through the suppression of
mRNA (8). As miRNAs are associated
with these tumor characteristics, specific miRNAs are thought to be
associated with sensitivity to chemotherapy (9,10).
We previously extracted high quality miRNAs from
formalin-fixed, paraffin-embedded (FFPE) samples obtained from
gastric cancer patients who had undergone operations, excluded
stage I and IV in non-radical surgery (11) and we detected specific miRNAs
associated with the prognosis of these patients using
high-sensitivity DNA chips. Therefore, we hypothesized that
patients who may require further chemotherapy can be identified by
miRNA expression profile analysis in patients who showed relapse
after adjuvant chemotherapy for gastric cancer.
Materials and methods
Patients and tissue specimens
FFPE specimens of gastric cancer and associated
patient information were collected. In total, 458 patients with
p-stage II or III gastric cancer underwent curative surgery between
2000 and 2012, including 161 patients from Toyama University
Hospital, 114 from Itoigawa General Hospital, 126 from Saiseikai
Toyama Hospital and 57 from Murakami Memorial Hospital Asahi
University; the latter 57 patients underwent surgery between 2006
and 2012. Of these, 398 were ineligible as they did not undergo
adjuvant chemotherapy (173 patients), were treated with a
chemotherapy other than S-1 (105 patients), underwent S-1
chemotherapy for <3 months (22 patients), died from a cause
other than gastric cancer (1 case), underwent neoadjuvant
chemotherapy (20 cases), were followed up for <5 years (52
cases), or relapsed in <6 months (10 cases). S-1 treatment was
continued for at least 3 months in 452 patients (87.4%) in the
ACTS-GC study (5). Therefore, we
excluded patients treated with S-1 for <3 months and T1 cases
were excluded as the tumors were too small for extraction of miRNA
(15 patients). Thus, a total of 60 specimens were selected from 458
gastric cancer patients. We obtained approval from the Ethics
Committee of the University of Toyama (Approval number, 20–63),
Itoigawa General Hospital, Saiseikai Toyama Hospital, and Murakami
Memorial Hospital Asahi University.
RNA extraction from FFPE specimens
RNA was extracted from FFPE specimens of tumor and
normal tissue from each patient, as previously reported (12). Extracted RNA was processed using a
silica-based spin column (Toray Industries, Tokyo, Japan) to obtain
purified total RNA. The degree of RNA cross-linking and RNA
degradation was analyzed using electrophoresis with an Agilent 2100
Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The RNA
quality criteria were as previously reported (12). In addition, we analyzed samples with
the same degree of total RNA decay in this experiment so as to
examine the relationship between tumor and normal tissues. Thus, we
determined whether RNAs could be used for microarray analysis.
miRNA assays
RNAs were labeled with Hy5 (miRCURY LNA microRNA Hy5
Power labeling kit; Exiqon A/S Corp., Vedbaek, Denmark). miRNAs
were examined with Toray 3D-Gene® miRNA oligo chips
(ver. 17; Toray Industries). A solution adjusted for the DNA chips
was applied, (including 500 ng of total RNA) and hybridization was
performed. Fluorescent signals were scanned using a 3D-Gene Scanner
(Toray Industries) and analyzed with 3D-Gene Extraction software
(Toray Industries). Quality evaluations were performed for the
averages of the tumor/normal (T/N ratio) as previously reported
(12).
Statistical analysis
P-values for gender, histological type, depth of
invasion, lymph node metastasis, p-stage, pre-operative serum
carcinoembryonic antigen (CEA) levels and pre-operative serum
carbohydrate antigen 19-9 (CA 19-9) levels were calculated using
the Chi-square test. P-values for age, maximum tumor diameter and
the total S-1 dose were calculated using the Student’s t-test. The
T/N ratio between the groups that did and did not relapse were
calculated and compared with the Student’s t-test. We constructed
receiver operating characteristic (ROC) curves and calculated the
area under the curve (AUC) to evaluate the specificity and
sensitivity of predicting cases. We divided gastric cancer patients
into two groups: the miRNA high-expression group and the
low-expression group, according to ROC curves. Positive predictive
value and negative predictive value were calculated as previously
reported (13). The disease-free
survival curve and overall survival curve were plotted according to
the Kaplan-Meier method. The differences in survival rates among
the groups with high and low miRNA expression were analyzed using
the log-rank test and the generalized Wilcoxon test. Disease-free
survival time was calculated from the date of surgery until the
patient relapsed or until the last follow-up. Overall survival time
was calculated from the date of surgery until the patient succumbed
to the disease or until the last follow-up. All statistical
analyses were performed using Microsoft Office Excel 2007 and JMP
10. P<0.05 was considered to indicate a statistically
significant result.
Results
RNA was extracted from tumor and normal tissue
specimens obtained from 60 patients. Twenty-five cases were
excluded due to poor RNA quality. Of the remaining 35 cases, 15
relapsed within 5 years. Characteristics of the patients are shown
in Table I. There were no
significant differences in age, gender, histological type, maximum
tumor diameter, depth of invasion, lymph node metastasis, p-stage,
pre-operative serum CEA levels, pre-operative serum CA 19-9 levels,
or S-1 total dose between patients who relapsed and those who did
not.
 | Table ICharacteristics of the evaluable
gastric cancer patients. |
Table I
Characteristics of the evaluable
gastric cancer patients.
| Characteristics | Relapse (n=15) | Non-relapse
(n=20) | P-value |
|---|
| Age (years, mean ±
SD) | 71.0±2.88 | 63.4±2.49 | 0.0543 |
| Gender |
| Male | 13 | 12 | 0.0746 |
| Female | 2 | 8 | |
| Histological
type |
|
Differentiateda | 6 | 8 | 1.0000 |
|
Undifferentiatedb | 9 | 12 | |
| Maximum tumor
diameter (mm, mean) | 62 | 48 | 0.0639 |
| Depth of
invasion |
| T2 | 1 | 6 | 0.0716 |
| ≥T3 | 14 | 14 | |
| Lymph node
metastasis |
| <3 | 6 | 10 | 0.5560 |
| ≥3 | 9 | 10 | |
| p-stage |
| II | 4 | 9 | 0.2623 |
| III | 11 | 11 | |
| Carcinoembryonic
antigen (ng/ml) |
| <5 | 12 | 13 | 0.2502 |
| ≥5 | 2 | 7 | |
| Carbohydrate antigen
19-9 (U/ml) |
| <37 | 14 | 17 | 0.2513 |
| ≥37 | 0 | 3 | |
| S-1 total dose (mg,
mean) | 15970 | 22263 | 0.1892 |
A total of 218 miRNAs were evaluated for T/N ratio
in each of the 15 relapse and 20 non-relapse cases. For these 218
miRNAs, microarray analysis revealed 5 miRNAs (miR-92b, 422a,
4732-5p, 4758-3p and 221) that were highly expressed according to
the T/N ratio in the patients who relapsed but not in those who did
not relapse (P-value <0.05), as shown in Table II (from a group of 9 miRNAs). The
results of ROC curves are shown in Table III. We divided 35 gastric cancer
patients into two groups, the miRNA high-expression group and the
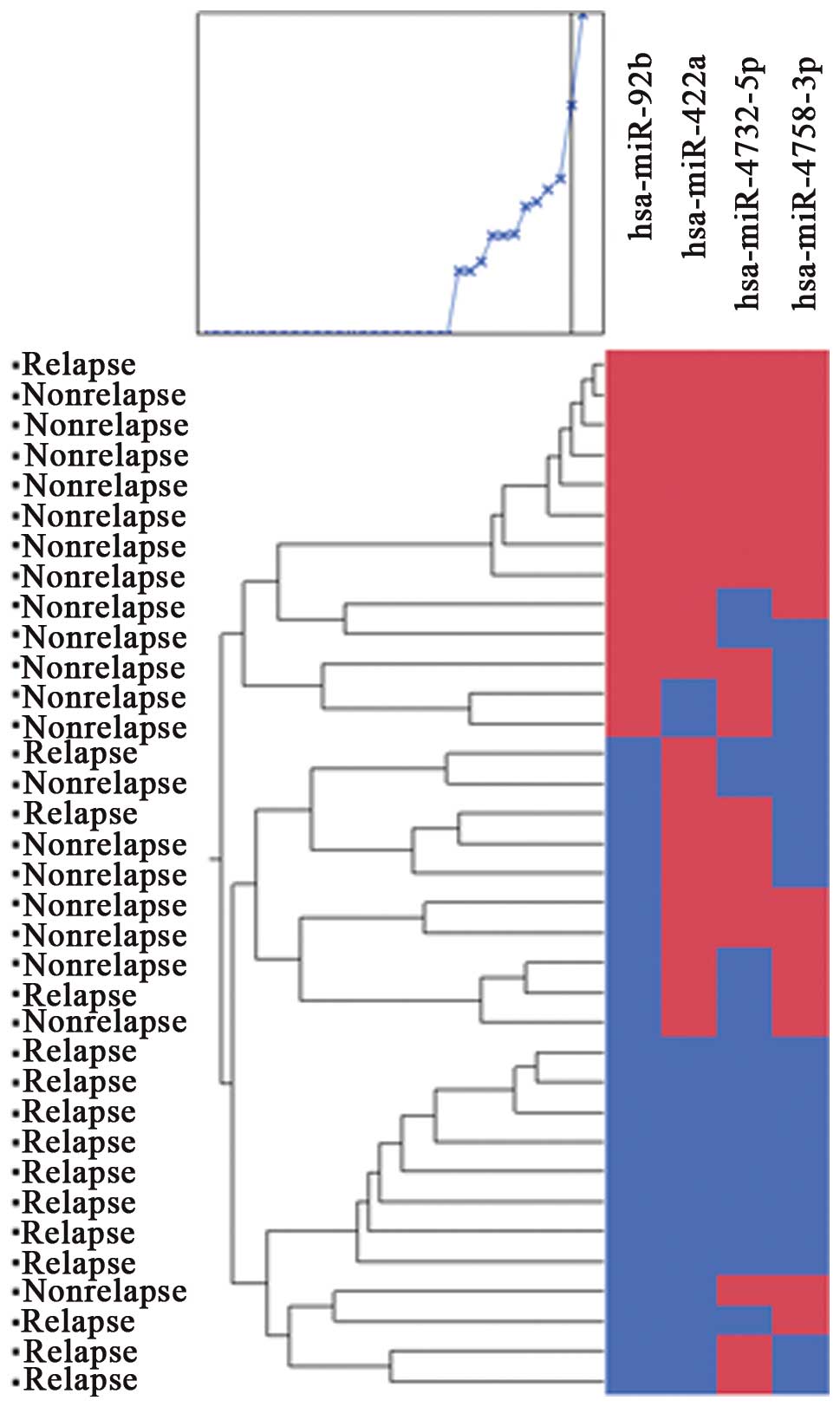
low-expression group, according to ROC curves. Cluster analysis of
5 miRNAs (miR-92b, 422a, 4732-5p, 4758-3p and 221) classified two
groups. Their positive predictive value was 85.1% and the negative
predictive value was 82.4%. Cluster analysis of 4 miRNAs (miR-92b,
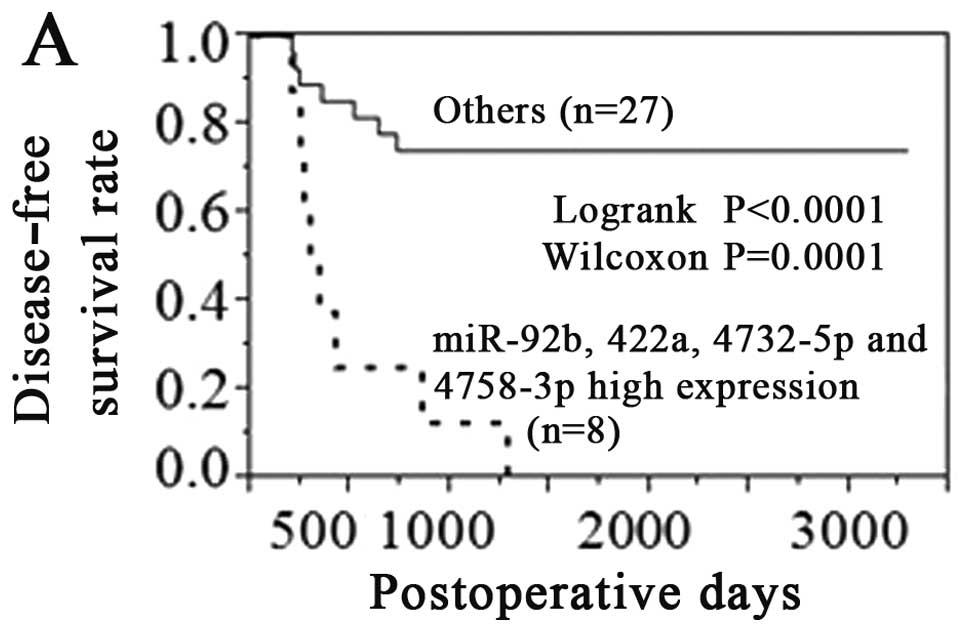
422a, 4732-5p and 4758-3p) classified two groups (Fig. 1). Their positive predictive value
was 93.8% and negative predictive value was 92.3%. If tumors showed
high expression of 4 miRNAs (miR-92b, 422a, 4732-5p and 4758-3p),
their disease-free survival rate and overall survival rate were
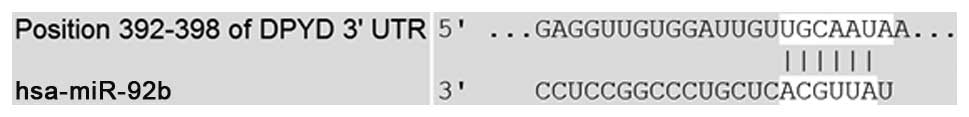
very poor (Fig. 2). We searched for
potential target of mRNA using bioinformatic logarithms available
on-line (TargetScan). According to TargetScan, dihydropyrimidine
dehydrogenase gene (DPYD) may be one of the predictable target
genes of miR-92b (Fig. 3).
 | Table IImiRNA expression of 35 patients
analyzed using miRNA oligo chips. |
Table II
miRNA expression of 35 patients
analyzed using miRNA oligo chips.
| Name | Relapse signal | Relapse T/N
ratio | Non-relapse
signal | Non-relapse T/N
ratio | P-value |
|---|
| hsa-miR-92b | 15 | 1.27 | 20 | 0.95 | 0.0143 |
| hsa-miR-422a | 15 | 0.89 | 20 | 0.61 | 0.0174 |
| hsa-miR-4732-5p | 15 | 2.00 | 20 | 1.09 | 0.0203 |
| hsa-miR-4758-3p | 15 | 1.36 | 20 | 0.98 | 0.0218 |
| hsa-miR-221 | 15 | 1.61 | 20 | 1.03 | 0.0384 |
| hsa-miR-1246 | 15 | 1.98 | 20 | 1.06 | 0.0703 |
|
hsa-miR-3663-3p | 15 | 1.03 | 20 | 0.86 | 0.0764 |
| hsa-miR-3135b | 15 | 1.03 | 20 | 0.80 | 0.0845 |
| hsa-miR-30d | 15 | 1.06 | 20 | 0.68 | 0.0993 |
 | Table IIICut-off value for predicting relapse
analyzed using receiver operating characteristic (ROC) curves. |
Table III
Cut-off value for predicting relapse
analyzed using receiver operating characteristic (ROC) curves.
| Name | Cut-off | AUC | Sensitivity | Specificity | True positive | True negative | False positive | False negative |
|---|
| hsa-miR-92b | 1.046232 | 0.75 | 0.93 | 0.6 | 14 | 12 | 8 | 1 |
| hsa-miR-422a | 0.815585 | 0.75 | 0.73 | 0.85 | 11 | 17 | 3 | 4 |
|
hsa-miR-4732-5p | 1.146845 | 0.77 | 0.73 | 0.75 | 11 | 15 | 5 | 4 |
|
hsa-miR-4758-3p | 1.028134 | 0.75 | 0.80 | 0.65 | 12 | 13 | 7 | 3 |
| hsa-miR-221 | 1.000008 | 0.70 | 0.80 | 0.65 | 12 | 13 | 7 | 3 |
| hsa-miR-1246 | 1.223516 | 0.67 | 0.73 | 0.7 | 11 | 14 | 6 | 4 |
|
hsa-miR-3663-3p | 0.778234 | 0.70 | 0.93 | 0.45 | 14 | 9 | 11 | 1 |
| hsa-miR-3135b | 1.170270 | 0.65 | 0.40 | 0.95 | 6 | 19 | 1 | 9 |
| hsa-miR-30d | 1.068176 | 0.63 | 0.53 | 0.85 | 8 | 17 | 3 | 7 |
Discussion
In the present study, we identified 5 specific
miRNAs related to gastric cancer recurrence after adjuvant S-1
chemotherapy. If tumors showed high expression of 4 miRNAs,
miR-92b, 422a, 4732-5p and 4758-3p, their disease-free survival
rate and overall survival rate were very poor. Combination of
miRNAs may be possible to predict recurrence after S-1
chemotherapy.
No study has reported the relationship between
gastric cancer, S-1 resistance, and miRNAs. In the study of the
relationship between S-1 resistivity and miRNAs in colon cancer,
Nakajima et al (14)
reported that let-7g and miR-181b are strongly associated with the
response to S-1 chemotherapy. In their report, 69.6% (32/46)
patients underwent S-1 chemotherapy and 30.4% (14/46) patients
underwent S-1 plus cisplatin chemotherapy. Thus, the results of
their study may be affected by cisplatin combination therapy.
There are no previous reports on the relationship
between miR-92b expression and gastric cancer recurrence after
adjuvant S-1 chemotherapy. In addition, no study has reported the
relationship between S-1 and miR-92b expression or between gastric
cancer and miR-92b expression. Haug et al (15) reported that MYCN-regulated miRNA-92
inhibits the secretion of the tumor suppressor Dickkopf-3 (DKK3) in
neuroblastoma. Also, Wang et al (16) reported that miR-92b binds to 3′UTR
of Nemo-like kinase that acts as a negative regulator of Wnt
signaling and activates Wnt/β-catenin signaling. As a result,
miR-92b activates glioma proliferation and invasion. Since miR-92b
was highly expressed according to the T/N ratio in the patients who
relapsed, it may be considered to promote tumor cell growth.
Dihydropyrimidine dehydrogenase gene (DPYD) is key
in the antitumor effect of 5-fluorouracil and S-1.
Dihydropyrimidine dehydrogenase (DPD) is the rate-limiting enzyme
in the catabolism of pyrimidine. However, Ichikawa et al
(17) and Jeung et al
(18) reported that S-1 responders
had higher DPD expression than S-1 non-responders in gastric cancer
patients. If miR-92b downregulates this gene, these reports support
our results that miR-92b was highly expressed according to the T/N
ratio average in the gastric cancer patients who relapsed but not
in those who did not relapse after S-1 chemotherapy. Kai et
al (19) reported that 5-FU
responders showed lower DPD expression than 5-FU non-responders in
gastric cancer patients. Regarding DPD expression, there might be a
difference between S-1 therapy and 5-FU therapy.
In the study of the relationship between miR-422a
and cancer, Gougelet et al (20) reported that miR-422a was
overexpressed in good responders for ifosfamide therapy in
osteosarcoma. Faltejskova et al (21) reported that miR-422a was
significantly decreased in colorectal cancer (CRC) tissues compared
to normal tissues. Thus, these results differed from our
observation that miR-422a was overexpressed in the relapse group.
These findings together indicate a dual role of miR-422a as either
a tumor-promoting or a tumor-suppresive miRNA in certain types of
cancer.
In a study of the relationship between miR-221 and
gastric cancer, Kim et al (22) reported that miR-221 downregulates
both p27 and p57 that delay cell cycle. Chun-Zhi et al
(23) reported that miR-221
upregulates gastric carcinoma cell proliferation and
radioresistance by targeting the tumor suppressor gene PTEN. These
findings support our results that miR-221 was overexpressed in the
relapsed group.
There are no previous reports on miR-4732-5p and
miR-4758-3p expression. We found that both were highly expressed in
gastric cancer patients who relapsed after adjuvant S-1
chemotherapy. We, therefore, consider both miR-4732-5p and
miR-4758-3p expression to be related to recurrence after adjuvant
S-1 chemotherapy. Further larger and more detailed studies are
required to confirm these findings.
Combination of miRNAs is a very useful technique to
improve the prediction of recurrence (24). From a group of 9 miRNAs (as shown in
Table II), various permutations
and combinations demonstrated that the combination of 4 miRNAs
showed better positive and negative predictive values and predicted
relapse more accurately after S-1 chemotherapy compared with a
combination of other miRNAs. Although miR-221 was highly expressed
in gastric cancer patients who relapsed after adjuvant S-1
chemotherapy, it was not useful in the prediction of relapse
compared with the other 4 specific miRNAs.
In conclusion, this is the first report on miRNAs
that are relapse-associated in gastric cancer patients after S-1
adjuvant chemotherapy. In the present study, we identified 5
specific miRNAs (miR-92b, 422a, 4732-5p, 4758-3p and 221) related
to gastric cancer recurrence after adjuvant S-1 chemotherapy. The
combination of 4 miRNAs, miR-92b, 422a, 4732-5p and 4758-3p, may
possibly help to predict recurrence after S-1 chemotherapy.
Acknowledgements
We received research grants from the Japanese
Ministry of Education, Culture, Sports, Science and Technology
(MEXT/JSPS KAKENHI grant no. B: 23390320). We also received a
research grant from the Japan Society for the Promotion of Science
(JSPS) Funding Program for World-Leading Innovative R&D on
Science and Technology (FIRST Program).
References
|
1
|
Terashima M, Kitada K, Ochiai A, Ichikawa
W, Kurahashi I, Sakuramoto S, Katai H, Sano T, Imamura H and Sasako
M: Impact of expression of human epidermal growth factor receptors
EGFR and ERBB2 on survival in stage II/III gastric cancer. Clin
Cancer Res. 18:5992–6000. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fujimoto S, Akao T, Itoh B, Koshizuka I,
Koyano K, Kitsukawa Y, Takahashi M, Minami T, Ishigami H, Miyazaki
M, Amamiya K, Ohyama Y, Ono K, Kure M, Kenjiro Itoh and Hikosaka T:
Protracted oral chemotherapy with fluorinated pyrimidines as an
adjuvant to surgical treatment for stomach cancer. Ann Surg.
185:462–466. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Earle CC and Maroun JA: Adjuvant
chemotherapy after curative resection for gastric cancer in
non-Asian patients: revisiting a meta-analysis of randomized
trials. Eur J Cancer. 35:1059–1064. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mari E, Floriani I, Tinazzi A, Buda A,
Belfiglio M, Valentini M, Cascinu S, Barni S, Labianca R and Torri
V: Efficacy of adjuvant chemotherapy after curative resection for
gastric cancer: a metaanalysis of published randomized trials. Ann
Oncol. 11:837–843. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sakuramoto S, Sasako M, Yamaguchi T,
Kinoshita T, Fujii M, Nashimoto A, Furukawa H, Nakajima T, Ohashi
Y, Imamura H, Higashino M, Yamamura Y, Kurita A and Arai K:
Adjuvant chemotherapy for gastric cancer with S-1, an oral
fluoropyrimidine. N Engl J Med. 357:1810–1820. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Japanese Gastric Cancer Association.
Japanese gastric cancer treatment guidelines 2010 (ver. 3). Gastric
Cancer. 14:113–123. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sasako M, Sakuramoto S, Katai H, Kinoshita
T, Furukawa H, Yamaguchi T, Nashimoto A, Fujii M, Nakajima T and
Ohashi Y: Five-year outcomes of a randomized phase III trial
comparing adjuvant chemotherapy with S-1 versus surgery alone in
stage II or III gastric cancer. J Clin Oncol. 29:4387–4393.
2011.PubMed/NCBI
|
|
8
|
Calin G and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
Downing JR, Jacks T, Horvitz HR and Golub TR: MicroRNA expression
profiles classify human cancers. Nature. 435:834–838. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hummel R, Hussey DJ and Haier J:
MicroRNAs: predictors and modifiers of chemo- and radiotherapy in
different tumour types. Eur J Cancer. 46:298–311. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Koizumi W, Narahara H, Hara T, Takagane A,
Akiya T, Takagi M, Miyashita K, Nishizaki T, Kobayashi O, Takiyama
W, Toh Y, Nagaie T, Takagi S, Yamamura Y, Yanaoka K, Orita H and
Takeuchi M: S-1 plus cisplatin versus S-1 alone for first-line
treatment of advanced gastric cancer (SPIRITS trial): a phase III
trial. Lancet Oncol. 9:215–221. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Osawa S, Shimada Y, Sekine S, Okumura T,
Nagata T, Fukuoka J and Tsukada K: MicroRNA profiling of gastric
cancer patients from formalin-fixed paraffin-embedded samples.
Oncol Lett. 2:613–619. 2011.PubMed/NCBI
|
|
13
|
Xing Y, Bronstein Y, Ross MI, Askew RL,
Lee JE, Gershenwald JE, Royal R and Cormier JN: Contemporary
diagnostic imaging modalities for the staging and surveillance of
melanoma patients: a meta-analysis. J Natl Cancer Inst.
103:129–142. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nakajima G, Hayashi K, Xi Y, Kudo K,
Uchida K, Takasaki K, Yamamoto M and Ju J: Non-coding MicroRNAs
hsa-let-7g and hsa-miR-181b are associated with chemoresponse to
S-1 in colon cancer. Cancer Genomics Proteomics. 3:317–324.
2006.PubMed/NCBI
|
|
15
|
Haug BH, Henriksen JR, Buechner J, Geerts
D, Tømte E, Kogner P, Martinsson T, Flægstad T, Sveinbjørnsson B
and Einvik C: MYCN-regulated miRNA-92 inhibits secretion of the
tumor suppressor DICKKOPF-3 (DKK3) in neuroblastoma.
Carcinogenesis. 32:1005–1012. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang K, Wang X, Zou J, Zhang A, Wan Y, Pu
P, Song Z, Qian C, Chen Y, Yang S and Wang Y: miR-92b controls
glioma proliferation and invasion through regulating
Wnt/beta-catenin signaling via Nemo-like kinase. Neuro Oncol.
15:578–588. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ichikawa W, Takahashi T, Suto K, Shirota
Y, Nihei Z, Shimizu M, Sasaki Y and Hirayama R: Simple combinations
of 5-FU pathway genes predict the outcome of metastatic gastric
cancer patients treated by S-1. Int J Cancer. 119:1927–1933. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jeung HC, Rha SY, Shin SJ, Lim SJ, Roh JK,
Noh SH and Chung HC: Predictive values of 5-fluorouracil pathway
genes for S-1 treatment in patients with advanced gastric cancer.
Anticancer Drugs. 22:801–810. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kai K, Kitajima Y, Hiraki M, Satoh S,
Tanaka M, Nakafusa Y, Tokunaga O and Miyazaki K: Quantitative
double-fluorescence immunohistochemistry (qDFIHC), a novel
technology to assess protein expression: a pilot study analyzing
5-FU sensitive markers thymidylate synthase, dihydropyrimidine
dehydrogenase and orotate phosphoribosyl transferases in gastric
cancer tissue specimens. Cancer Lett. 258:45–54. 2007.
|
|
20
|
Gougelet A, Pissaloux D, Besse A, Perez J,
Duc A, Dutour A, Blay JY and Alberti L: Micro-RNA profiles in
osteosarcoma as a predictive tool for ifosfamide response. Int J
Cancer. 129:680–690. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Faltejskova P, Svoboda M, Srutova K,
Mlcochova J, Besse A, Nekvindova J, Radova L, Fabian P, Slaba K,
Kiss I, Vyzula R and Slaby O: Identification and functional
screening of microRNAs highly deregulated in colorectal cancer. J
Cell Mol Med. 16:2655–2666. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim YK, Yu J, Han TS, Park SY, Namkoong B,
Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK and Kim VN: Functional links
between clustered microRNAs: suppression of cell-cycle inhibitors
by microRNA clusters in gastric cancer. Nucleic Acids Res.
37:1672–1681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang CZ, Han L, Zhang AL, Fu YC, Yue X,
Wang GX, Jia ZF, Pu PY, Zhang QY and Kang CS: MicroRNA-221 and
microRNA-222 regulate gastric carcinoma cell proliferation and
radioresistance by targeting PTEN. BMC Cancer. 10:367–376. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang X, Yan Z, Zhang J, Gong L, Li W, Cui
J and Liu Y: Combination of hsa-miR-375 and hsa-miR-142-5p as a
predictor for recurrence risk in gastric cancer patients following
surgical resection. Ann Oncol. 22:2257–2266. 2011. View Article : Google Scholar : PubMed/NCBI
|