Introduction
Head and neck cancer, which is the sixth most common
cancer in the world among human malignant disorders, is an
aggressive and life-threatening disease with poor prognosis,
morbidity and high mortality in advanced disease. Survival rates
have not improved significantly for patients with head and neck
squamous cell carcinoma (HNSCC) in the past 30 years despite active
clinical and basic research addressing this issue. More than 40,000
new cases of HNSCC are diagnosed in the United States each year,
with a mortality rate of 12,000 in the USA annually (1). Treatment for HNSCC includes surgical
resection, chemotherapy and radiation therapy; however,
approximately 50% of all patients have advanced disease at the time
of diagnosis often requiring use of all three treatment modalities.
Cancer-specific molecular biomarkers, which have the ability to
warn the clinicians in the earlier stage before the disease
advances, or to provide insight regarding the prognosis of the
disease or outcome of the patients, are required. In addition, it
is important to develop new methods that provide sensitive and
reliable biomarkers of HNSCC for detection, treatment response and
prognosis.
Epigenetic alterations are a recent attractive
phenomenon of human cancer, with the activation of proto-oncogenes
and inactivation of tumor suppressor genes, either through
hypomethylation or hypermethylation in the promoter regions of the
genes, respectively (2).
Transcriptional silencing of tumor suppressor genes by means of
promoter hypermethylation plays an important role in head and neck
carcinogenesis (3). Methylation of
the CpG islands in the promoter regions of tumor suppressor genes
is frequently observed with reduced gene expression (4,5).
From a previous study using the gene expression
profiling via oligonucleotide microarray-based approach to discover
the new cancer-specific methylated genes (6), in the present study, we evaluated the
hypermethylation of 10 genes [nucleolar protein 4 (NOL4),
iroquois homeobox 1 (IRX1), sodium-coupled monocarboxylate
transporter 1 (SLC5A8), leucine rich repeat containing 3B
(LRRC3B), functional smad-suppressing element on chromosome
18 (FUSSEL18), early B-cell factor 3 (EBF3),
gastrulation brain homeobox 2 (GBX2), H6 family homeobox 2
(HMX2), septin 9 (SEPT9), ALX homeobox 3
(ALX3)] identified by Restriction Landmark Genomic Scanning
(RLGS) in previous studies (7,8), and
by personal communication with Bennett et al (7,8), and
two other genes [suppressor of cytokine signaling 3 (SOCS3)
and LIM homeobox 6 (LHX6)] selected from the literature via
candidate gene approach (9–14). IRX1, SLC5A8,
FUSSEL18, EBF3, GBX2, HMX2,
SEPT9 and ALX3 genes showed tumor suppressor activity
in previous cancer studies (15–19)
and were involved in transforming growth factor (TGF) signaling
pathway which has a high frequency of alteration in HNSCC (15,20–23).
To measure methylation levels, real-time quantitative
methylation-specific PCR (QMSP) was performed to provide an
objective, robust and rapid assessment of promoter methylation
status (24–27).
Materials and methods
Tissue samples
Following institutional review board approval and
after obtaining appropriate informed consent, the HNSCC patients
and control population from healthy subjects enrolled in a
community screening study were recruited from the Johns Hopkins
School of Medicine, Department of Otolaryngology-Head and Neck
Surgery. Mucosal samples and salivary rinses from healthy
population and HNSCC tissue samples were collected. In the present
study, salivary rinses were obtained by brushing oral cavity and
oropharyngeal surfaces with an exfoliating brush followed by rinse
and gargle with 20 ml normal saline solution. The brush was gently
agitated to release the obtained material into saline. After
centrifugation, the supernatant was discarded and DNA was isolated
from the pellet. Tumors were snap frozen and microdissected on a
cryostat to ≥75% purity. DNA from 16 salivary rinse samples from
non-cancer individuals were analyzed as a control, to investigate
the normal promoter methylation status of two newly identified
candidate genes, IRX1 and NOL4. The methylation
status of these genes was analyzed in 33 fresh tumor samples from
patients with head and neck cancer and 14 normal mucosa samples
from healthy individuals.
DNA extraction and bisulfite
treatment
DNA was isolated as previously described (28). In brief, DNA was obtained by
phenol/chloroform extraction after overnight incubation with
proteinase K (Boehringer-Mannheim, Germany) at 48°C. DNA from tumor
and control samples was subjected to bisulfite treatment using
EpiTect Bisulfite modification kit (Qiagen, Valencia, CA, USA) as
per the manufacturer’s protocol.
Bisulfite sequencing
The bisulfite sequence analysis was performed to
determine the methylation status in the promoter regions of 12
genes. Bisulfite-treated DNA was amplified for the 5′ region that
included at least a portion of the CpG island within 1–2 kb of the
first exon of the genes. The promoter regions of the genes were
found from the database of the University of California, Santa Cruz
(UCSC) (http://genome.ucsc.edu/). Primer
sequences were determined by MethPrimer program (29) showing the CpG islands in the
promoter regions of 12 genes for bisulfite sequencing (Table I). A thousand base pair region of
the genes’ promoters and some part of the first exon were sequenced
by the specific primers producing 400–500 bp PCR fragments. The
primers for bisulfite sequencing were designed to hybridize to
regions in the promoter without CpG dinucleotides. PCR products
were gel-purified using the QIAquick Gel Extraction kit (Qiagen)
according to the manufacturer’s instructions. Each amplified DNA
sample was sequenced by the Applied Biosystems 3700 DNA Analyzer
using nested, forward or reverse primers and BD terminator dye
(Applied Biosystems, Foster City, CA, USA).
 | Table IPrimer sequences of 12 selected genes
for bisulfite sequencing. |
Table I
Primer sequences of 12 selected genes
for bisulfite sequencing.
| Gene name | 5′ Primer sequence
3′ | Primer
sequence |
|---|
| LRRC3B |
TAAAGAGAGGGGAAAGATTTTTGTT |
AATCAATTTCCCCTACAATTCTAAAA |
|
GGAAAATTGAATTTTATTTTTTTT |
AAAATATTAACTCCCTCTACTACTCTC |
|
AATAGGAGAAAGAATGGGGTTATAGTT |
TAACCTTACAAAAAAAACAAACAAAA |
| NOL4 |
GGAAGTTTTGAATGGAGTAATTGTT |
CAAATACATTTTAAATAAATTCCAACC |
|
GTTTGGGGTATTATAATTTATTTTGTAGAA |
CTCTCCTTCCTCCTAAATCCTACTT |
|
TAGGATTTAGGAGGAAGGAGAGATT |
ACACCATTCTAACCCAAAAAAACTA |
|
GTTGGGATGGTTTTGGTTATAAA |
CACCTATTCACCCTAAACTCATAAAA |
|
FUSSEL18 |
TTATTTAATTATTTGAGATTAGAATTA |
AATAAATTCCTAAAAAACCTAAAACCTTAT |
|
GGTTTTAGGTTTTTTAGGAATTTAT |
TCACCTAACCCACCTAATTAAATTTAA |
| EBF3 |
TTTTAGGATAAGTTGTAGTTTTTTGTATTT |
ATTTAACCCCTTAATACCTCCCTAC |
|
GTAGGGAGGTATTAAGGGGTTAAAT |
CACAAAACTCAACCCTCTCTCCC |
|
GGGAGAGAGGGTTGAGTTTTGTG |
CCCAAACATAAAAACTACTAAC |
| GBX2 |
GAGGGGTAGGATTTTGTTTTTAATT |
AACCTTAAAACCCTACAACCTTATC |
|
GAGGTTAGTTTGGGTGGAAAG |
ATAAAACATAAACATAAAATAACC |
|
GGTAGGTAAAATGTGAATGAGAAAGAGGAG |
ATAAAACATAAACATAAAATAACC |
| IRX1 |
TGGGTGAAGAGAAAGTTTTTTTT |
AAACATCTTTAACAAAAATACACCC |
|
TTTTGTTAAAGATGTTTTTTGGAGG |
TACTTTAATTAACATCCCCTTAAAC |
| HMX2 |
GTTTTGTTATTAGTTTTTTATTTTTTTT |
AACCTCATCCCTATCACAAATTCTA |
|
GGAATTTGAATTTAGATTTTTTG |
TTAAACCCCTTAAACCCTTCTCT |
|
GAGAGAAGGGTTTAAGGGGTTTA |
TACAACAAACAAACAATAAAAAAAA |
|
GGAGGATGGTGGAGTAGTTTGTATA |
TCTAACCAAAAACAACCAAAACTAAA |
| SLC5A8 |
GTGGATTGTTTATTTAGGATTAGATGG |
AACCTTCATATAACACATACATACTTAACA |
|
GTGTTAAGTATGTATGTGTTATATGAAGGT |
AAAAACTAAAACCTCCAACTACTTCC |
|
GATTGTTGAATTGGAAAGTTAAAATTTA |
CCTCAAACCCAAATATAAAACCTC |
| SEPT9 |
GGAAGATGTTTTTTTTGTTAAGGAG |
TCAATCTATACTACTCCCCAAAACC |
|
TTGGGGAGTAGTATAGATTGAAAAGT |
TTAAACTTCACCTCAAAAATTCATT |
|
GGATGAATAGTGGGGAATAGTATTG |
CCAAAAAAAACCCTAAAAAATCAC |
| ALX3 |
GTGGGTTTTTAGATATTTGGGTTATT |
CAAACAAACAAACCTTAAACTACAATTT |
|
ATTTTTAATAGTTTTTTTTATTGTG |
CTAACTTAATACTAAAACCATCCAC |
|
TTTGAGTTGTTTGGGATTGG |
CTCTAAAAAATAAAACTCCAAAAACC |
| SOCS3 |
GAGAGTATTTGGTTTAATTTATA |
CTTCCCCTTCCCCTTTTCCC |
|
TGTAGTTTTGGGTTTTTTTTT |
CAACTTCTCATTCACATTTCC |
|
GATTTGGATTTTTTGTTT |
CTCCTCCTTCCTACCTAATC |
| LHX6 |
GTAGATGGTATGGTTATGGGT |
ACCTCCCTAACTACTACC |
|
AGGAGGATAAGGAGGAGGGAG |
CTCATACTTCCAATACATAAACC |
|
GTTTTTGTAGTAGTTTTTGT |
CCAACATTTACATAATATATTCC |
|
GGTTTATGTATTGGAAGTATGAG |
AAAAAAAAACACCCTCCAACC |
|
GGTTGGAGGGTGTTTTTTTTT |
AATTTTTTCTCTCTCCACC |
Quantitative methylation-specific
PCR
Primer and probe sequences were determined by
MethPrimer program showing the CpG islands in the promoter regions
of two genes selected after bisulfite sequencing (Table II). To determine if the methylated
genes in tumor samples were cancer-specific, we investigated
promoter methylation in 16 normal saliva, 14 age-matched normal
mucosa from healthy individuals that were analyzed as a control, to
investigate the normal promoter methylation status of two newly
identified candidate genes (NOL4, IRX1) and in 33
HNSCC tumor samples by QMSP. Lymphocytes obtained from a healthy
individual were in vitro methylated using excess SssI
methyltransferase (New England Biolabs Inc., Beverly, MA, USA) to
generate completely methylated DNA that was used as a positive
control standard. To quantitate the relative percent of
methylation, we computed the ratio between the QMSP values of the
gene of interest relative to an internal control, ACTB (gene
of interest/reference gene ×100) (30). Fluorogenic PCR was carried out in a
reaction volume of 20 μl consisting of 600 nM of each primer; 200
nM of probe; 0.6 U of platinum Taq polymerase (Invitrogen,
Carlsbad, CA, USA); 200 μM of each dATP, dCTP, dGTP and dTTP; 1X
ROX Dye reference and 1X buffer [16.6 mM of ammonium sulfate; 67 mM
of Trizma (Sigma, St. Louis, MO, USA); 6.7 mM of magnesium
chloride; 10 mM of mercaptoethanol and 0.1% dimethylsulfoxide].
Thirty nanograms of bisulfite treated DNA were used in each
real-time QMSP reaction. Amplifications were carried out in
384-well plates in a 7900 Sequence Detector system (Perkin-Elmer
Applied Biosystems, Norwalk, CT, USA) and were analyzed by SDS 2.3
(sequence detector system) (Applied Biosystems). Each reaction was
performed in triplicate.
 | Table IIPrimers and probe sequences of 2
selected genes for validation by QMSP. |
Table II
Primers and probe sequences of 2
selected genes for validation by QMSP.
| Gene name | Probe sequence | 5′ Primer
sequence | 3′ Primer
sequence | Temp (°C) | bp |
|---|
| NOL4 |
GGGGAGGCGGCGTTGCGTTTTAT |
TTTTCGGGGTTTAAAGGCGTTG |
AAATAATCCCTAAACGCCTCGC | 60 | 178 |
| IRX1 |
AGTAGTTGGTCGGGTCGGTACGG |
GGGGATATATTTCGGTCGCGA |
TCCCGCGAACACGTAATACC | 60 | 198 |
Results
Clinicopathological characteristics of
control subjects and patients with HNSCC
Table III
describes the demographic parameters of the sample populations used
in the present study. The mean age of normal mucosal samples was
43.4 years (range, 24–65). Tobacco users were observed as 42%. Both
normal mucosal and tumor samples had a similar male and Caucasian
predominance. Smoking rate was 78% and alcohol consumption was 69%.
Tumor samples (n=33) were obtained from patients with stage I
(7.4%), stage II (22%), stage III (26%) and stage IV (44%) lesions.
These were from primary tumors of the oral cavity (n=9), oropharynx
(n=7), hypopharynx (n=2), larynx (n=8), maxillary sinus (n=2),
nasal floor (n=1), salivary gland (n=1) and unknown primary/neck
(n=3). The male and Caucasian prevalence was smaller in the normal
salivary rinse samples and tobacco users were found as 31.25%. The
ages of individuals from which the normal salivary rinse was
obtained were slightly lower than the population of head and neck
cancer patients, mean ages 53.06 years (range, 33–83) and 61.4
years (range, 36–88), respectively. Due to our small cohort, we did
not perform any statistics between the clinical parameters and QMSP
results of tumor and normal populations.
 | Table IIIQMSP results and demographics of the
patients with HNSCC. |
Table III
QMSP results and demographics of the
patients with HNSCC.
| Tumor samples | IRX1 | NOL4 | Age (years) | Gender | Race | Smoking | Alcohol | Tumor anatomic
site | Overall stage |
|---|
| 1 | N | N | 67 | M | C | Yes | Yes | Nasal floor | 2 |
| 2 | Y | Y | 57 | M | C | Yes | Yes | Larynx | 2 |
| 3 | N | N | 61 | M | C | No | No | Neck | 3 |
| 4 | Y | Y | 60 | F | C | Yes | Yes | Larynx | 4 |
| 5 | Y | Y | 55 | M | A | Yes | Yes | Larynx | 2 |
| 6 | Y | Y | 54 | M | C | No | Yes | Oropharynx | 4 |
| 7 | Y | N | 64 | M | A | Yes | Yes | Hypopharynx | 3 |
| 8 | Y | Y | 55 | F | A | Yes | No | Oral cavity | 1 |
| 9 | N | Y | 80 | M | C | Yes | No | Oral cavity | NA |
| 10 | Y | Y | 54 | F | C | No | Oral | cavity | 4 |
| 11 | Y | Y | 62 | M | C | Yes | Yes | Oropharynx | 4 |
| 12 | Y | Y | 72 | M | C | Yes | Yes | Hypopharynx | 3 |
| 13 | Y | Y | 42 | M | C | Yes | No | Larynx | 2 |
| 14 | Y | Y | 66 | M | C | Yes | No | Oropharynx | 4 |
| 15 | Y | Y | 74 | M | C | Yes | Larynx | 2 | |
| 16 | Y | Y | 58 | M | A | Yes | Yes | Oropharynx | 4 |
| 17 | Y | Y | 56 | F | C | Yes | Yes | Oral cavity | 2 |
| 18 | Y | Y | 43 | M | C | Yes | Yes | Oropharynx | 4 |
| 19 | Y | Y | 68 | M | C | Yes | Yes | Oropharynx | 4 |
| 20 | Y | Y | 63 | M | A | Yes | | Oral cavity | NA |
| 21 | Y | Y | 64 | F | C | No | Yes | Oral cavity | 3 |
| 22 | N | Y | 88 | M | C | Yes | Yes | Oral cavity | NA |
| 23 | Y | Y | 42 | M | C | Yes | No | Oral cavity | 3 |
| 24 | Y | Y | 51 | M | C | Yes | Yes | Larynx | 4 |
| 25 | Y | Y | 80 | M | C | No | Yes | Neck | NA |
| 26 | Y | Y | 58 | M | C | Yes | Yes | Larynx | 3 |
| 27 | Y | Y | 71 | M | C | Yes | Yes | Neck | NA |
| 28 | Y | Y | 48 | M | C | Yes | Yes | Oropharynx | 4 |
| 29 | Y | Y | 61 | M | C | Yes | No | Maxillary
sinus | 1 |
| 30 | Y | Y | 77 | M | C | No | No | Salivary gland | NA |
| 31 | Y | Y | 67 | M | C | Larynx | 3 | | |
| 32 | Y | Y | 36 | M | C | Yes | No | Oral cavity | 4 |
| 33 | Y | Y | 74 | F | C | No | Yes | Maxillary
sinus | 4 |
Genes specifically methylated in HNSCC
tumors
In the present study, we investigated methylation of
the 12 gene promoters by bisulfite modification and QMSP. In the
initial screening of the methylated CpG islands, bisulfite
sequencing was performed by using four HNSCC cell lines (JHU-06,
JHU-022, JHU-022B and JHU-028), eight normal salivary rinses and
eight HNSCC samples. Only two genes, NOL4 (NM_003787) and
IRX1 (NM_024337) (http://www.genenames.org), had methylated CpG
dinucleotides on their promoter regions in tumor samples but
absence of methylated CpGs was found in normal salivary rinse
samples (Table IV). We
investigated the methylation frequency in a larger cohort of normal
salivary rinses, mucosal and HNSCC specimens, in order to find a
biomarker candidate.
 | Table IVInformation regarding candidate tumor
suppressor genes and the bisulfite sequencing results. |
Table IV
Information regarding candidate tumor
suppressor genes and the bisulfite sequencing results.
| Gene ref. ID | Chromosomal
location | Candidate TSGs | Normal salivary
rinses n (%) | HNSCC tissue n
(%) | HNSCC cell lines n
(%) |
|---|
| NM_052953 |
chr3:26,664,300–26,752,265 | LRRC3B | 1/3 (33) | 3/4 (75) | 4/4 (100) |
|
NM_003787 |
chr18:31,431,070–31,803,446 | NOL
4 | 0/6 (0) | 6/6
(100) | 4/4
(100) |
| NM_001037802 |
chr18:44,757,495–44,775,554 |
FUSSEL18 | 5/8 (62.5) | 4/7 (57) | 3/4 (75) |
| NM_001005463 |
chr10:131,633,547–131,762,091 | EBF3 | 2/4 (50) | 1/3 (33) | 3/4 (75) |
| NM_001485 |
chr2:237,074,307–237,076,652 | GBX2 | 3/5 (60) | 4/6 (67) | 4/4 (100) |
|
NM_024337 |
chr5:3,596,168–3,601,517 |
IRX1 | 0/6 (0) | 6/6
(100) | 4/4
(100) |
| NM_005519 |
chr10:124,907,638–124,910,188 | HMX2 | 2/4 (50) | 1/3 (33) | 4/4 (100) |
| NM_145913 |
chr12:101,549,994–101,604,016 | SLC5A8 | 3/5 (60) | 2/3 (67) | 2/2 (100) |
| NM_006640 |
chr17:75,315,597–75,496,678 | SEPT9 | 4/7 (57) | 2/3 (67) | 2/4 (50) |
| NM_006492 |
chr1:110,602,997–110,613,322 | ALX3 | 1/3 (33) | 0/3 (0) | 3/4 (75) |
| NM_003955 |
chr17:76,352,859–76,356,158 | SOCS3 | 0/8 (0) | 0/8 (0) | 1/2 (50) |
| NM_014368 |
chr9:124,964,858–124,991,019 | LHX6 | 0/8 (0) | 0/8 (0) | 1/1 (100) |
In the second stage of the present study, we
performed QMSP on 16 normal salivary rinse and 14 normal mucosal
samples from healthy individuals and 33 HNSCC tumor samples for two
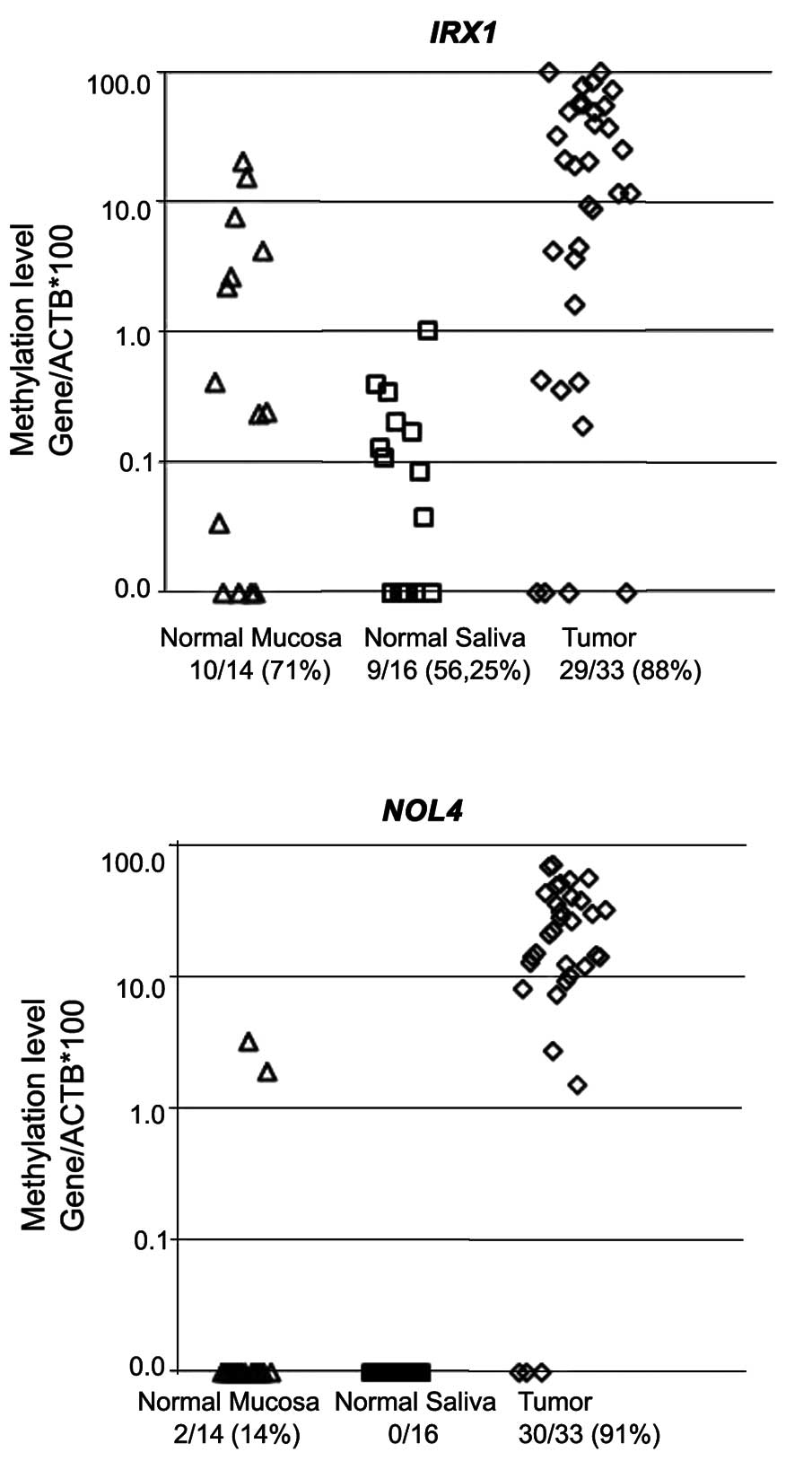
selected genes. The NOL4 gene showed no methylation (0/16)
in normal salivary rinses and two out of 14 (14%) mucosal samples
were minimally methylated between 1 and 3% methylation values,
whereas the methylation rate was 91% (30/33) on the promoter region
of the NOL4 gene in HNSCC tumor samples showing high
methylation values are 79% (26/33) of the patients between 10 and
100%. The IRX1 gene [10/14 (71%), 9/16 (56.25%) and 29/33
(88%)] demonstrated varying degrees of methylation on their
promoter regions in normal mucosa, normal salivary rinses and HNSCC
tumor samples, respectively (Fig.
1). Although normal salivary methylation rate was observed as
high, IRX1 methylation values were <1% in normal salivary
rinses and were mostly (8/14, 57%) between 0.1 and 10% in normal
mucosal samples indicating a very strong marker which may define
HNSCC tumor tissues as a potential biomarker (Fig. 1).
Discussion
In the present study, we investigated methylation
status of the promoter regions of 12 genes by bisulfite
modification, bisulfite sequencing and QMSP techniques. Only two of
them (IRX1 and NOL4) showed methylation in their CpG
islands located on promoter region of these genes, indicating a
characteristic of a biomarker molecule. NOL4 gene encoding a
nucleolar protein which is expressed predominantly in brain and
testis, was identified by Ueki et al (31) and there is only one study showing
high methylation status of 20 patients with cervical cancer by
MethyLight assays (32) in
concordance with our results.
The IRX1 gene is a member of the iroquois
homeobox gene family and plays a role during pattern formation of
vertebrate embryos. In published literature, there are only four
studies investigating the effects of epigenetically silencing
IRX1 gene, in the patients with gastric cancer (33) and the studies of Bennett et
al with HNSCC (7,8,34).
Bennett et al (7) reported
an association between the HNSCC and hypermethylation of
IRX1 in accordance with our results, and four other genes
(FUSSEL18, EBF3, SLC5A8 and SEPT9), but
not SLC5A8; we did not find any methylated CpG on the
promoter regions of the last five genes. In two other studies,
Bennett et al also showed the association between
IRX1 methylation and recurrence and between four other genes
and clinicopathological parameters such as HPV status, alcohol and
tobacco usage (8) and they
investigated the interactions with some molecules of the TGF-β
pathway, and their transcriptional inactivation by methylation in
HNSCC caused the decrease in apoptosis and differentiation, and
increased proliferation (34). In
the present study, we only observed the methylation of the
IRX1 gene which was the one of the five frequently
methylated genes identified by Restriction Landmark Genomic
Scanning (RLGS) method in metastatic HNSCC samples compared to
primary tumors. Mass array methylation (7) and COBRA (8) analysis were used in
fresh/paraffin-embeded HNSCC tumors and matched normal mucosa
samples in the previous studies of Bennett et al (7,8). We
screened the methylated CpG islands on the promoter regions of
these genes by bisulfite sequencing using frozen HNSCC tumors,
HNSCC cell lines and normal salivary rinses in order to select the
genes that have the methylated CpG islands on their promoter
regions in tumor samples but no methylation in normal mucosal and
normal salivary rinse samples. Then, the methylation levels were
measured by QMSP technique in 33 HNSCC tumors, 16 salivary rinses
and 14 normal mucosa as reported in our previous study (35). Our aim was to find a cancer-specific
biomarker and to detect the tumor tissues in salivary rinses of the
patients earlier and to screen the normal population. In addition,
in a previous study (34),
tonsillar carcinoma samples were used, whereas we quantified the
methylation in the primary tumors of the oral cavity, oropharynx,
hypopharynx, larynx, maxillary sinus, nasal floor and unknown
primary/neck. Therefore, discrepant observations between these
studies by Bennett et al (7,8,34) and
our data may be due to confounding factors including anatomic site,
methodology and sampling method. Therefore, the four other genes
described by Bennett et al (7,8), may
not be good biomarker candidates in salivary rinse samples despite
being differentially methylated in primary tumor samples. The
present study is the first to investigate the methylation levels of
NOL4 gene and to show high methylation in the patients with
HNSCC by QMSP technique.
In addition, it would be helpful to increase sample
size to facilitate a more precise determination of accuracy of
these biomarkers in detection of the disease and to evaluate the
association of the results with the clinical parameters of the
disease. These newly identified silenced genes here remain to be
tested in saliva, serum or plasma samples from HNSCC patients in a
larger sample size.
Acknowledgements
This study was supported by the National Cancer
Institute SPORE (5P50CA096784-05) and the Scientific Research
Projects Coordination Unit of Istanbul University (UDP-30980). This
study/analysis is based on a web database application provided by
Research Information Technology Systems (RITS)-https://www.rits.onc.jhmi.edu/.
References
|
1
|
Jemal A, Siegel R, Ward E, Murray T, Xu J,
Smigal C and Thun MJ: Cancer statistics, 2006. CA Cancer J Clin.
56:106–130. 2006. View Article : Google Scholar
|
|
2
|
Baylin SB, Herman JG, Graff JR, Vertino PM
and Issa JP: Alterations in DNA methylation: a fundamental aspect
of neoplasia. Adv Cancer Res. 72:141–196. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dulaimi E, Hillinck J, Ibanez de Caceres
I, Al-Saleem T and Cairns P: Tumor suppressor gene promoter
hypermethylation in serum of breast cancer patients. Clin Cancer
Res. 10:6189–6193. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leonhardt H and Cardoso MC: DNA
methylation, nuclear structure, gene expression and cancer. J Cell
Biochem Suppl. 35:78–83. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yamashita K, Upadhyay S, Osada M, et al:
Pharmacologic unmasking of epigenetically silenced tumor suppressor
genes in esophageal squamous cell carcinoma. Cancer Cell.
2:485–495. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bennett KL, Karpenko M, Lin MT, et al:
Frequently methylated tumor suppressor genes in head and neck
squamous cell carcinoma. Cancer Res. 68:4494–4499. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bennett KL, Lee W, Lamarre E, et al: HPV
status-independent association of alcohol and tobacco exposure or
prior radiation therapy with promoter methylation of
FUSSEL18, EBF3, IRX1, and SEPT9, but
not SLC5A8, in head and neck squamous cell carcinomas. Genes
Chromosomes Cancer. 49:319–326. 2010.PubMed/NCBI
|
|
9
|
Estécio MR, Youssef EM, Rahal P, et al:
LHX6 is a sensitive methylation marker in head and neck carcinomas.
Oncogene. 25:5018–5026. 2006.PubMed/NCBI
|
|
10
|
Pierconti F, Martini M, Pinto F, et al:
Epigenetic silencing of SOCS3 identifies a subset of
prostate cancer with an aggressive behavior. Prostate. 71:318–325.
2011.
|
|
11
|
Isomoto H: Epigenetic alterations in
cholangiocarcinoma-sustained IL-6/STAT3 signaling in cholangio-
carcinoma due to SOCS3 epigenetic silencing. Digestion. 79(Suppl
1): 2–8. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fourouclas N, Li J, Gilby DC, et al:
Methylation of the suppressor of cytokine signaling 3 gene
(SOCS3) in myeloproliferative disorders. Haematologica.
93:1635–1644. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Martini M, Pallini R, Luongo G, Cenci T,
Lucantoni C and Larocca LM: Prognostic relevance of SOCS3
hypermethylation in patients with glioblastoma multiforme. Int J
Cancer. 123:2955–2960. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weber A, Hengge UR, Bardenheuer W, et al:
SOCS-3 is frequently methylated in head and neck squamous cell
carcinoma and its precursor lesions and causes growth inhibition.
Oncogene. 24:6699–6708. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thangaraju M, Gopal E, Martin PM, Ananth
S, Smith SB, Prasad PD, Sterneck E and Ganapathy V: SLC5A8 triggers
tumor cell apoptosis through pyruvate-dependent inhibition of
histone deacetylases. Cancer Res. 66:11560–11564. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao LY, Niu Y, Santiago A, Liu J, Albert
SH, Robertson KD and Liao D: An EBF3-mediated transcriptional
program that induces cell cycle arrest and apoptosis. Cancer Res.
66:9445–9452. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu YY, Ji J, Lu Y, Bu L, Liu BY, Zhu ZG
and Lin YZ: High-resolution analysis of chromosome 5 and
identification of candidate genes in gastric cancer. Zhonghua Zhong
Liu Za Zhi. 28:84–87. 2006.(In Chinese).
|
|
18
|
Jönsson G, Staaf J, Olsson E, et al:
High-resolution genomic profiles of breast cancer cell lines
assessed by tiling BAC array comparative genomic hybridization.
Genes Chromosomes Cancer. 46:543–558. 2007.PubMed/NCBI
|
|
19
|
Burrows JF, Chanduloy S, McIlhatton MA, et
al: Altered expression of the septin gene, SEPT9, in ovarian
neoplasia. J Pathol. 201:581–588. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Amir S, Wang R, Matzkin H, Simons JW and
Mabjeesh NJ: MSF-A interacts with hypoxia-inducible factor-1α and
augments hypoxia-inducible factor transcriptional activation to
affect tumorigenicity and angiogenesis. Cancer Res. 66:856–866.
2006.PubMed/NCBI
|
|
21
|
Endo S, Zeng Q, Burke NA, et al: TGF-α
antisense gene therapy inhibits head and neck squamous cell
carcinoma growth in vivo. Gene Ther. 7:1906–1914. 2000.
|
|
22
|
Le QT, Kong C, Lavori PW, et al:
Expression and prognostic significance of a panel of tissue hypoxia
markers in head-and-neck squamous cell carcinomas. Int J Radiat
Oncol Biol Phys. 69:167–175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sánchez-Elsner T, Botella LM, Velasco B,
Corbí A, Attisano L and Bernabéu C: Synergistic cooperation between
hypoxia and transforming growth factor-β pathways on human vascular
endothelial growth factor gene expression. J Biol Chem.
276:38527–38535. 2001.PubMed/NCBI
|
|
24
|
Bernard PS and Wittwer CT: Real-time PCR
technology for cancer diagnostics. Clin Chem. 48:1178–1185.
2002.PubMed/NCBI
|
|
25
|
Eads CA, Danenberg KD, Kawakami K, et al:
MethyLight: a high-throughput assay to measure DNA methylation.
Nucleic Acids Res. 28:E322000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cottrell SE and Laird PW: Sensitive
detection of DNA methylation. Ann NY Acad Sci. 983:120–130. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jerónimo C, Usadel H, Henrique R, Oliveira
J, Lopes C, Nelson WG and Sidransky D: Quantitation of GSTP1
methylation in non-neoplastic prostatic tissue and organ-confined
prostate adenocarcinoma. J Natl Cancer Inst. 93:1747–1752.
2001.PubMed/NCBI
|
|
28
|
Tokumaru Y, Yamashita K, Osada M, et al:
Inverse correlation between cyclin A1 hypermethylation and p53
mutation in head and neck cancer identified by reversal of
epigenetic silencing. Cancer Res. 64:5982–5987. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li LC and Dahiya R: MethPrimer: designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Park HL, Kim MS, Yamashita K, et al: DCC
promoter hypermethylation in esophageal squamous cell carcinoma.
Int J Cancer. 122:2498–2502. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ueki N, Kondo M, Seki N, Yano K, Oda T,
Masuho Y and Muramatsu M: NOLP: identification of a novel human
nucleolar protein and determination of sequence requirements for
its nucleolar localization. Biochem Biophys Res Commun. 252:97–102.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang SS, Smiraglia DJ, Wu YZ, et al:
Identification of novel methylation markers in cervical cancer
using restriction landmark genomic scanning. Cancer Res.
68:2489–2497. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo X, Liu W, Pan Y, et al: Homeobox gene
IRX1 is a tumor suppressor gene in gastric carcinoma. Oncogene.
29:3908–3920. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bennett KL, Romigh T and Eng C: Disruption
of transforming growth factor-β signaling by five frequently
methylated genes leads to head and neck squamous cell carcinoma
pathogenesis. Cancer Res. 69:9301–9305. 2009.
|
|
35
|
Demokan S, Chuang AY, Chang X, et al:
Identification of guanine nucleotide-binding protein γ-7 as an
epigenetically silenced gene in head and neck cancer by gene
expression profiling. Int J Oncol. 42:1427–1436. 2013.
|