Introduction
Preoperative chemoradiotherapy is widely used as an
effective method in treatment of locally advanced rectal cancer
(1,2). However, treatment responses among
patients differ, even in those with similar tumor histopathological
types and clinical stages (3). In
addition, radiation exposure of the rectum, a critical organ, can
induce radiation injury of varying degrees. These complications of
radiation lead to poorer patient quality of life and may even be
life-threatening (4). Conventional
preoperative evaluation approaches, such as computed tomography
(CT) and magnetic resonance imaging (MRI), cannot predict the
effects of radiotherapy. Therefore, a better method of assessing
potential tumor response should be established, to improve
individualized therapy (5).
Peripheral blood detection is a comparatively
non-invasive method, therefore circulating cell-free DNA detection
in evaluating prognosis is a promising concept (6–10).
Previous studies demonstrated the concentration of plasma
circulating cell-free DNA in cancer patients was higher than that
in healthy controls (11). Some
studies have also shown that cell-free DNA was closely related to
tumor status (12). In general,
cell-free DNA fragments in healthy individuals are small and
uniform, and mainly originate from normal cell apoptosis. However,
in cancer patients, cell-free DNA fragments are usually incomplete
and random, with different fragment lengths, and are considered to
originate from necrosis of tumor cells (13). A high concentration of
tumor-associated cell-free DNA and high mutant KRAS levels
are related to poor prognosis, outcome and recurrence risk in
patients with colorectal cancer (14). In addition, a previous study
demonstrated that plasma cell-free DNA concentration showed dynamic
changes during radiotherapy (15).
Another study revealed that baseline DNA integrity and variations
induced by treatment were closely related to pathologic tumor
response (16). These data suggest
that plasma cell-free DNA detection may have potential clinical
utility in cancer treatment. However, these previous studies were
limited as they did not take into account the variability of TNM
status, chemotherapy drugs or radiotherapy doses. Therefore, the
relationship between cell-free DNA and the response to preoperative
chemoradiotherapy in rectal cancer remains unclear.
In the present study, we investigated whether the
detection and analysis of plasma cell-free DNA, including
concentration and tumor-associated DNA, may be used to predict the
response to preoperative chemoradiotherapy in patients with rectal
cancer.
Materials and methods
Patient and control sample
collection
Thirty-four patients with locally advanced rectal
cancer without distant metastases (Fudan University Shanghai Cancer
Center, Shanghai, China) were enrolled in the present study.
Following approval by our Institutional Research Ethics Committee,
informed consent was obtained from all patients. The patient
characteristics are shown in Table
I. All patients received preoperative chemoradiotherapy (pelvic
radiation therapy, total dose of 50 Gy, 2 Gy/fraction/day, 5
days/week, 25 fractions by using a high-energy linear accelerator;
concurrent chemotherapy, capecitabine 625 mg/m2, bid,
day 1–5/week and oxaliplatin 85 mg/m2, qw) followed by
radical surgery after 6–8 weeks. Blood samples were collected 7
days prior to and following chemoradiotherapy. Control samples were
obtained from 10 healthy volunteers (5 men and 5 women with mean
age, 37.2±15.2 years).
 | Table IRectal cancer patient characteristics
and radiotherapy treatment information. |
Table I
Rectal cancer patient characteristics
and radiotherapy treatment information.
| Patient no. | Cancer
location | Gender | Age (years) | Tumor stage | Pathology | Rt dose (Gy) | Fractions |
|---|
| 1 | Rectum | F | 52 | T3N2M0 | Adenocarcinoma | 50 | 25 |
| 2 | Rectum | M | 66 | T3N2M0 | Adenocarcinoma | 50 | 25 |
| 3 | Rectum | F | 54 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 4 | Rectum | M | 66 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 5 | Rectum | F | 65 | T3N2M0 | Adenocarcinoma | 50 | 25 |
| 6 | Rectum | M | 29 | T4N0M0 | Adenocarcinoma | 50 | 25 |
| 7 | Rectum | M | 47 | T2N1M0 | Adenocarcinoma | 50 | 25 |
| 8 | Rectum | M | 54 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 9 | Rectum | F | 35 | T3N2M0 | Adenocarcinoma | 50 | 25 |
| 10 | Rectum | F | 66 | T3N2M0 | Adenocarcinoma | 50 | 25 |
| 11 | Rectum | F | 64 | T4N2M0 | Adenocarcinoma | 50 | 25 |
| 12 | Rectum | F | 64 | T4N1M0 | Adenocarcinoma | 50 | 25 |
| 13 | Rectum | F | 68 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 14 | Rectum | F | 69 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 15 | Rectum | M | 45 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 16 | Rectum | M | 60 | T4N1M0 | Adenocarcinoma | 50 | 25 |
| 17 | Rectum | M | 71 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 18 | Rectum | M | 55 | T3N2M0 | Adenocarcinoma | 50 | 25 |
| 19 | Rectum | F | 46 | T4N1M0 | Adenocarcinoma | 50 | 25 |
| 20 | Rectum | M | 62 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 21 | Rectum | M | 56 | T3N2M0 | Adenocarcinoma | 50 | 25 |
| 22 | Rectum | F | 44 | T4N0M0 | Adenocarcinoma | 50 | 25 |
| 23 | Rectum | F | 48 | T3N2M0 | Adenocarcinoma | 50 | 25 |
| 24 | Rectum | F | 49 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 25 | Rectum | M | 52 | T4N2M0 | Adenocarcinoma | 50 | 25 |
| 26 | Rectum | M | 58 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 27 | Rectum | M | 73 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 28 | Rectum | M | 60 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 29 | Rectum | M | 61 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 30 | Rectum | F | 60 | T2N1M0 | Adenocarcinoma | 50 | 25 |
| 31 | Rectum | M | 42 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 32 | Rectum | M | 39 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 33 | Rectum | F | 58 | T3N1M0 | Adenocarcinoma | 50 | 25 |
| 34 | Rectum | M | 42 | T3N2M0 | Adenocarcinoma | 50 | 25 |
DNA extraction from plasma samples
Free DNA in plasma was extracted from samples by
using the QIAamp® DNA Blood Mini kit (Qiagen, Hilden,
Germany) according to the manufacturer’s instructions. Each 200 μl
of free DNA was extracted from 200 μl plasma.
Detection of plasma cell-free DNA
concentration
Quantitative real-time polymerase chain reaction
(QRT-PCR) analyses were performed to quantify the concentration of
circulating DNA. Human genome DNA was extracted and diluted to
2,000, 500, 100, 20 and 5 ng/ml by using a spectrophotometer
(BioTek, Winooski, VT, USA) in order to establish a standard curve.
Each PCR reaction was performed with 10 μl SYBR® Premix
Ex Taq™ (Takara, Otsu, Shiga, Japan) and 5 μl circulating DNA.
Human β-actin genomic DNA fragments were quantified by using PCR
with the following primers: actin-100 primers (forward,
5′-GCACCACACCTTCTACAATGA-3′ and reverse,
5′-GTCATCTTCTCGCGGTTGGC-3′), and actin-400 primers (forward,
5′-GCACCACACCTTCTACAATGA-3′ and reverse, 5′-TGTCACGCACGATTTCCC-3′).
The PCR conditions were: 40 cycles at 95°C for 10 sec and 60°C for
30 sec. All PCR amplification was performed by using the
LightCycler® 480 Real-Time PCR System (Roche, Basel,
Switzerland).
Detection of mutations of the KRAS
gene
The detection of KRAS mutations at codon 12 was
performed by using a 2-step PCR-restriction fragment length
polymorphism (PCR-RFLP) method with circulating DNA. In the first
step, 10 μM of each primer (17),
P1 (5′-TCAAAGAATGGTCCTGGACC-3′) and P2
(5′-ACTGAATATAAACTTGTGGTAGTTGGACCT-3′) and 20 ng of circulating DNA
combined was used in a 25 μl reaction mixture by using Premix Taq™
Hot Start Version (Takara). The PCR conditions were 95°C for 6 min,
35 cycles at 95°C for 30 sec, 58°C for 30 sec, and 72°C for 30 sec,
and 72°C for 5 min. The first step PCR products were digested with
the enzyme Mva I (Thermo Fisher Scientific, West Palm Beach, FL,
USA) at 37°C for 90 min according to the manufacturer’s
instructions. Enzyme-digested products were diluted 20-fold and 1.5
μl were used as a template in the second PCR step with the
following primers: P2 and P3
(5′-TAATATGTCGACTAAAACAAGATTTACCTC-3′); the PCR reaction system was
the same as step 1. The PCR conditions were: 95°C for 5 min, 35
cycles at 95°C for 30 sec, 64°C for 30 sec, and 72°C for 30 sec,
and at 72°C for 5 min. The second step PCR products were digested
with the enzyme Mva I under the same conditions as the first step
PCR products. The PCR products of the KRAS analysis were
separated by using electrophoresis on a 3% agarose gel and stained
with ethidium bromide.
DNA bisulfite modification and
methylation-specific PCR (MSP)
Methylation status in the promoter region of
O6-methylguanine-DNA methyltransferase (MGMT) gene was detected by
MSP using bisulfite-treated DNA. DNA (500 ng) was treated with
bisulfite by using CpGenome™ Universal DNA Modification kit
according to the manufacturer’s instructions (Millipore, Billerica,
MA, USA). In order to increase the sensitivity of detecting
hypermethylation, we applied a 2-step PCR approach. In the first
PCR step, the promoter regions of the tumor suppressor gene were
amplified with the following primers (17): MGMT-F, 5′-TGGTAATTA AGGTATAGAG-3′
and MGMT-R, 5′-CCAATCCACAATCA CTCA-3′. In this step, 10 μM of each
primer, 50 ng of bisulfite-modified DNA and 12.5 ml Premix Taq™ Hot
Start Version (Takara) were added to 25 μl PCR reaction. The PCR
conditions were: 95°C for 6 min, 35 cycles at 95°C for 30 sec, 53°C
for 45 sec, 72°C for 30 sec, and 72°C for 10 min. All PCR
amplification was performed in a MasterCycler® Pro
(Eppendorf, Hamburg, Germany). The first step PCR products were
diluted 20-fold and then 2 μl were used as a template in the second
PCR step with the following MSP primers (18): MGMT-M-F,
5′-TTTCGACGTTCGTAGGTTTTCGC-3′ and MGMT-M-R,
5′-GCACTCTTCCGAAAACGAAACG-3′; MGMT-U-F,
5′-TTTGTGTTTTGATGTTTGTAGGTTTTT GT-3′ and MGMT-U-R,
5′-AACTCCACACTCTTCCAAAA ACAAAACA-3′. The PCR conditions were: 95°C
for 10 min, 35 cycles at 95°C for 30 sec, 62°C for 45 sec, and 72°C
for 30 sec, followed by 72°C for 10 min. The 81-base pair (bp) and
93-bp PCR products of the MGMT analysis were separated by using
electrophoresis on a 3% agarose gel and stained with ethidium
bromide.
Pathological assessment
An experienced pathologist who was blinded to the
clinical results and cell-free DNA information comprehensively
evaluated each specimen following surgery. Tumor regression was
graded by performing histological evaluation of the surgical
specimens according to the criteria described by Dworak et
al (19). The grade of tumor
regression grading (TRG) was defined as follows: grade 0, no
regression; grade 1, dominant tumor mass with obvious fibrosis
and/or vasculopathy; grade 2, dominantly fibrotic changes with few
tumor cells or groups (easy to find); grade 3, very few tumor cells
(difficult to find microscopically) in fibrotic tissue with or
without mucous substance; grade 4, no tumor cells, only fibrotic
mass (total regression or response).
Statistical analysis
The R Project Software (version 2.14.1) was used for
statistical analyses. Significant differences in levels of plasma
cell-free DNA were determined by using the Wilcoxon rank sum test.
Significant differences in KRAS mutation and MGMT
methylation changes were determined by using Fisher’s exact test. A
value of p<0.05 was considered to indicate a statistically
significant difference.
Results
Detection of plasma cell-free DNA
concentration in rectal cancer patients and healthy controls
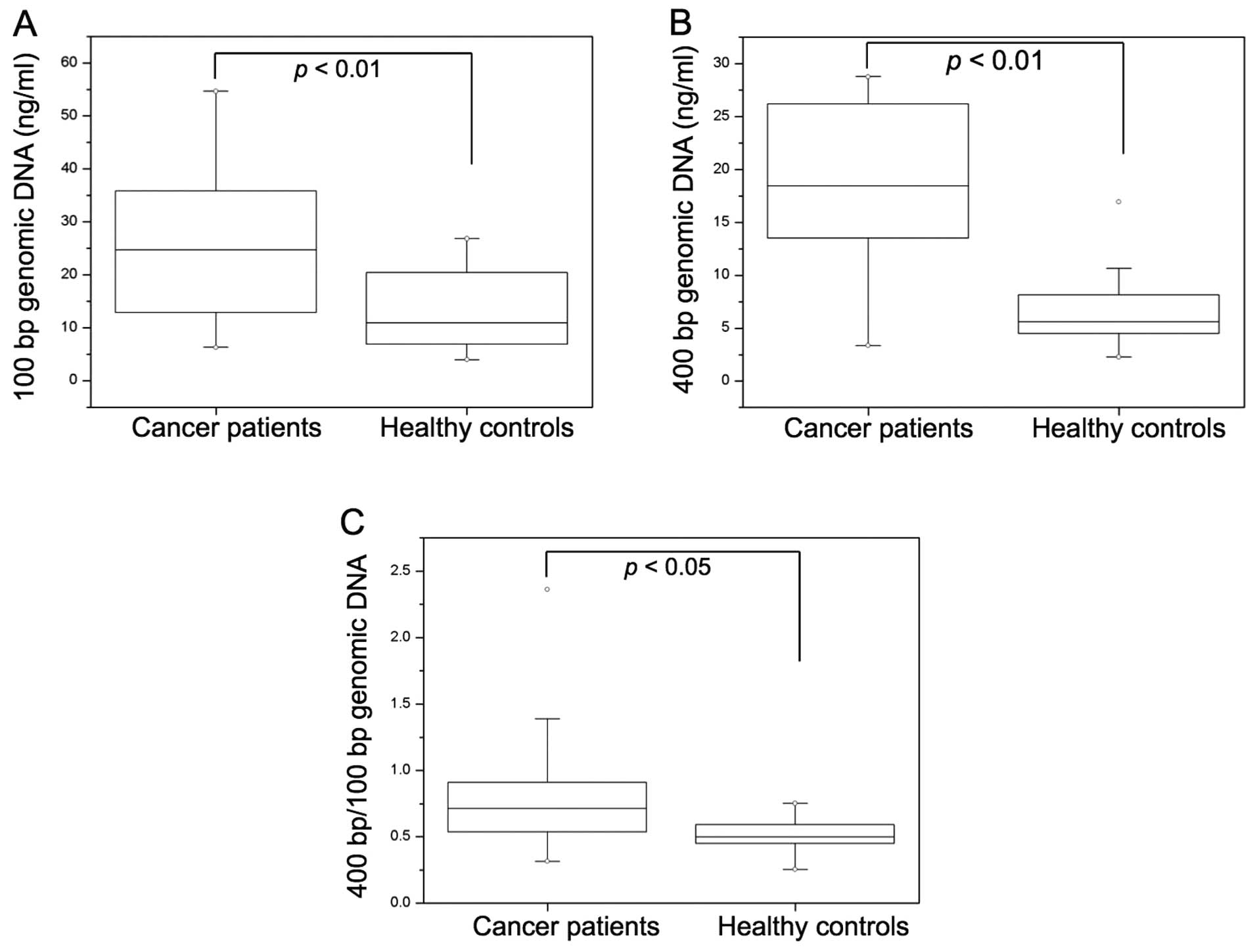
The concentrations of 100- or 400-bp genomic
fragments from 34 patients with rectal cancer and 10 healthy
controls were detected and compared. The concentrations of both
100-bp (p<0.01) and 400-bp (p<0.01) fragment DNA in cancer
patients were significantly higher than those in healthy controls
(Fig. 1A and B). The ratio of
400-/100-bp DNA concentrations, which showed cell-free plasma DNA
integrity, was also significantly higher in cancer patients than in
healthy controls (p<0.05) (Fig.
1C).
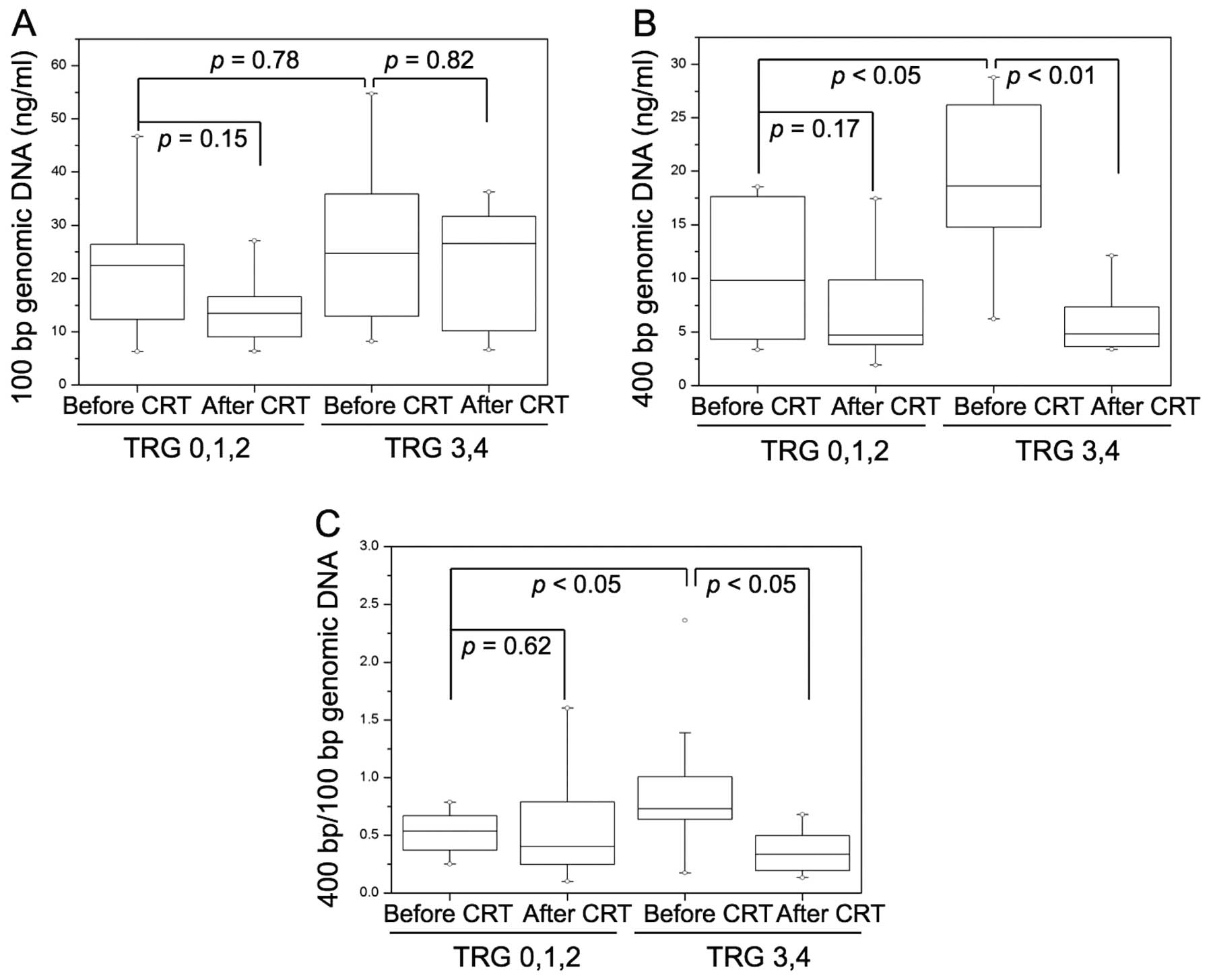
Correlation between plasma cell-free DNA
concentration and TRG score
The concentration of genomic DNA from 34 patients
with rectal cancer before and after chemoradiotherapy was screened.
No significant difference was found in DNA concentration of 100-bp
fragments before vs. after chemoradiotherapy (TRG 0,1,2 group,
p=0.15; TRG 3,4 group, p=0.82) (Fig.
2A). The 400-bp fragment DNA concentration was significantly
lower after chemoradiotherapy (TRG 0,1,2 group, p=0.17; TRG 3,4
group, p<0.01) in the good response group (Fig. 2B). The ratio of 400-/100-bp DNA
concentrations, which showed cell-free plasma DNA integrity, was
also significantly lower after chemoradiotherapy compared to before
chemoradiotherapy (TRG 0,1,2 group, p=0.62; TRG 3,4 group,
p<0.05) in the good response group (Fig. 2C).
The correlation between tumor response to
chemoradiotherapy and baseline level of plasma cell-free DNA was
studied. The 100-bp fragment DNA concentration at baseline was not
significantly different between the poor response group (TRG 0,1,2)
and the good response group (TRG 3,4) (p=0.78) (Fig. 2A). The good response group had
significantly higher 400-bp plasma cell-free DNA levels (p<0.05)
(Fig. 2B) and ratio of
400-bp/100-bp DNA concentrations (p<0.05) compared with those
from the poor response group (Fig.
2C).
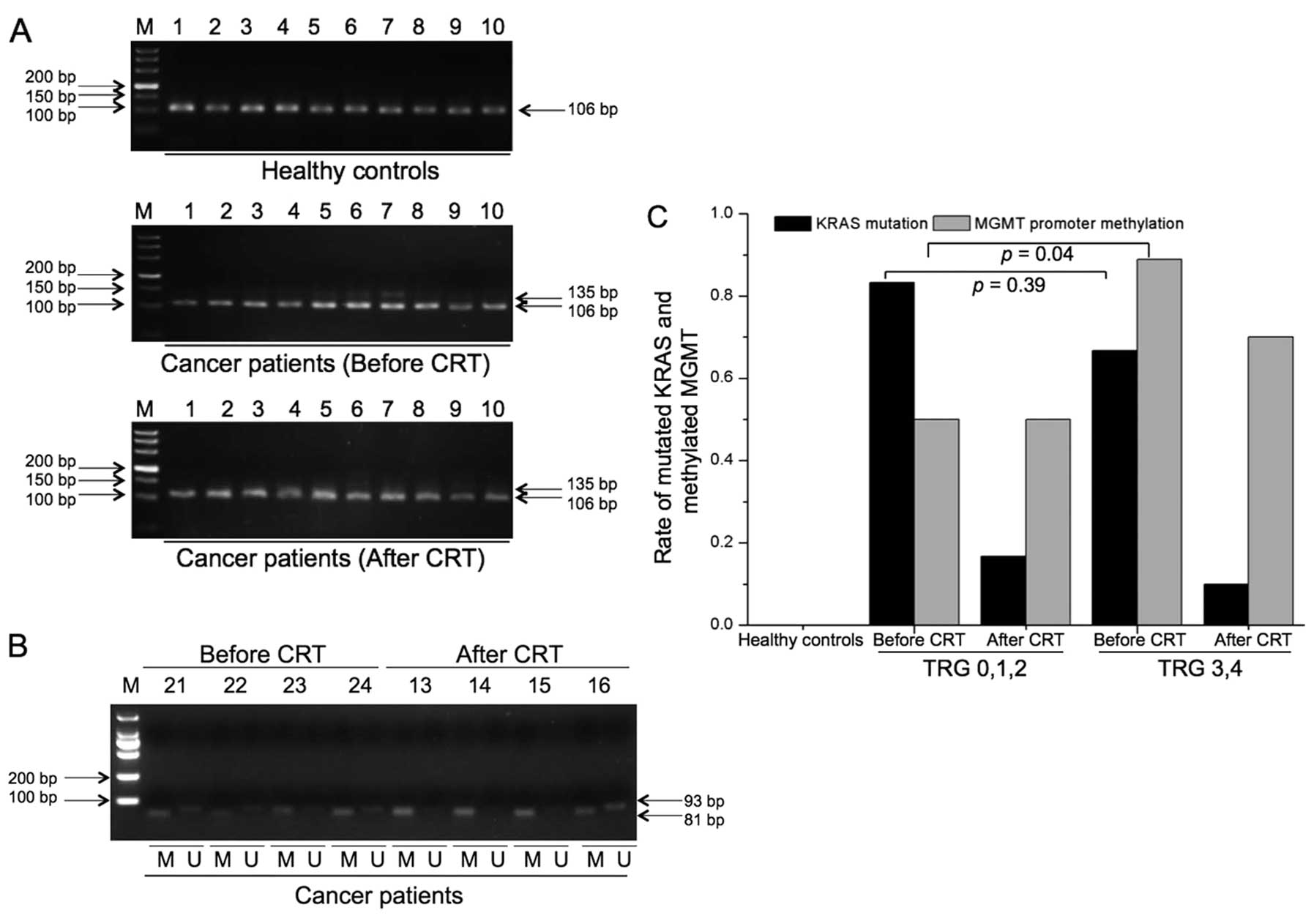
Correlation of KRAS mutation, MGMT
promoter methylation, and response to preoperative
chemoradiotherapy in rectal cancer patients
The KRAS mutation and MGMT promoter
methylation level in plasma cell-free DNA from 34 patients with
rectal cancer and 10 healthy controls were analyzed. Mutated
KRAS and methylated MGMT were not found in healthy controls
(Fig. 3A and B). The rate of
KRAS mutation decreased significantly after
chemoradiotherapy (TRG 0,1,2 group, 83.3 vs. 16.7%, p=0.01; TRG 3,4
group, 66.7 vs. 10.0%, p<0.001) (Fig. 3C). There was no significant
difference in methylation status of MGMT before and after
chemoradiotherapy (TRG 0,1,2 group, 50.0 vs. 50.0%, p=1.0; TRG 3,4
group, 88.9 vs. 70.0%, p=0.09) (Fig.
3C). The rate of KRAS mutation was not significantly
different between the poor response group (TRG 0,1,2) and the good
response group (TRG 3,4) (83.3 vs. 66.7%, p=0.39). The rate of
methylation status of MGMT was significantly higher in the good
response group than that in the poor response group (88.9 vs. 50%,
p=0.04) (Fig. 3C).
Discussion
Rectal cancer is one of the most common malignancies
and the majority of cases are locally advanced (20,21).
Treatment of locally advanced rectal cancer consists of surgery,
radiotherapy and chemotherapy. Preoperative chemoradiotherapy is
now considered as standard treatment for patients with locally
advanced rectal cancer (22).
Preoperative radiochemotherapy was shown to improve local disease
control and even long-term survival (23). It was also beneficial in reducing
tumor spread, increasing pathological complete response rates and
improving sphincter preservation after surgery (24). The rectum is a critical organ and is
radiosensitive. Radiation induces rectal damage, which can impair
tissue function and lead to poorer patient quality of life.
Therefore, prediction of treatment response to preoperative
chemoradiotherapy is necessary for patients with rectal cancer.
The potential value of cell-free nucleic acids in
plasma and serum for disease diagnosis has been demonstrated
(25). Higher levels of cell-free
DNA have been found in cancer patients and in tumor-bearing animals
compared with healthy controls (26,27). A
large quantity of circulating cell-free DNA in cancer patients was
shown to be associated with the presence of tumor (28,29).
The cell-free DNA in plasma from cancer patients was considered to
originate from necrotic tumor cells, active tumor liberation,
micrometastases, tumor cell apoptosis, and circulating tumor cells,
while cell-free DNA in the plasma of healthy individuals was
considered to originate mainly from apoptotic normal cells
(30). A previous study showed the
circulating cell-free DNA derived from tumors varied in size,
whereas that from non-tumoral apoptotic cells was uniformly
truncated into fragments and shorter than 200 bp. Circulating
cell-free DNA fragments from tumor necrosis were variable in size
and generally larger than 200 bp. In addition, the ratio between
longer fragments and shorter fragments, which was termed the
integrity index, was more reliable in reflecting tumor status
(16). In the present study, we
also demonstrated that plasma cell-free DNA levels were higher in
patients with rectal cancer than in healthy individuals, which was
consistent with results of previous reports (31). A previous study demonstrated that
the concentration of plasma cell-free DNA showed dynamic changes
during radiotherapy (15). In
addition, the plasma cell-free DNA level decreased after treatment
but increased in patients with recurrent cancer (32). These data suggested the cell-free
DNA level may be associated with clinical status, effect of
treatment and tumor prognosis. However, plasma cell-free DNA
consists of various DNA fragments of different lengths. Compared
with the total cell-free DNA concentration, the proportion of
longer DNA fragments (300–400 bp) was more closely associated with
the clinical status of cancer patients (33). Here, we also found that not only
longer lengths of cell-free DNA but also the ratio of larger
fragments decreased after chemoradiotherapy in the good response
group. This suggested that the decrease in cell-free DNA levels may
have resulted from tumor regression induced by chemoradiotherapy.
In addition, larger lengths of cell-free DNA and the ratio of
larger fragments both increased in the higher TRG scores group,
which suggested that these parameters were associated with a better
response to preoperative chemoradiotherapy. However, this is not
consistent with a previous study, in which lower levels of
circulating DNA and lower integrity were found in the responsive
group, although this was not statistically significant (16). This difference suggested circulating
plasma DNA may originate mainly from tumor necrosis but not active
liberation of the tumor itself in rectal cancer.
Further studies demonstrated that tumor-associated
plasma cell-free DNA was of value in clinical research, not only
through detection of its concentration (34). KRAS is a part of the
epidermal growth factor (EGF) system, as a downstream mediator
following EGF binding to its receptor EGFR (35). Thus, EGF is considered an important
target of chemoradiotherapy. KRAS mutation can decrease the
response to EGFR tyrosine kinase inhibitors (TKIs) and enhance
repair of radiation-induced double-strand breaks by the
EGFR-phosphatidylinositol-3-kinase-AKT pathway, thereby inducing
drug and radiation-resistance (36). The codon 12 mutation of KRAS
is the most common mutation pattern, and was used to detect
KRAS mutation in tumor tissue and plasma DNA (37,38).
Previous studies demonstrated that plasma KRAS detection was
not only convenient but also highly sensitive in detection of
KRAS mutation as compared with tumor tissue assessment
(39,40). In the present study, we found that
the incidence of KRAS mutation was significantly higher in
cancer patients than in healthy controls. We also found that the
ratio of KRAS mutation decreased significantly after
treatment in both the good and poor response groups. A higher
incidence of plasma KRAS mutation was associated with a
lower survival rate in colorectal cancer and non-small cell lung
cancer (41). Higher levels of
plasma KRAS mutation were also correlated with poor tumor
control with treatment of cetuximab and irinotecan (42). However, the relationship between
tumor regression response induced by preoperative chemoradiotherapy
and the incidence of KRAS mutation in baseline plasma DNA
was not found.
MGMT is an important protein, which acts by removing
alkyl products from the O6 position on guanine. It is widely found
in gliomas, colon and head and neck cancer (18,43,44).
The methylation of CpG islands in the MGMT gene promoter can induce
loss of MGMT gene transcription and protein expression. Loss of
MGMT expression leads to decrease DNA repair. Here, we found the
ratio of MGMT promoter methylation in cancer patients was
significantly higher than in healthy controls. After treatment, the
MGMT promoter methylation ratio did not change significantly
compared with the methylation ratio before treatment in both the
good and the poor response group. However, we found higher MGMT
promoter methylation was associated with better tumor regression
response induced by preoperative chemoradiotherapy. A previous
study also demonstrated that MGMT promoter methylation was
associated with higher radiosensitivity (45). These data suggested that plasma MGMT
promoter methylation levels may serve as a potential biomarker of
radiation response.
Preoperative chemoradiotherapy is an indispensable
treatment for rectal cancer. However, few data focus on
personalized responses to preoperative chemoradiotherapy in rectal
cancer. Plasma detection is a less invasive and convenient method
and may be used in the assessment of prognostic and treatment
impact. In the present study, we found the level, integrity and
variations of larger cell-free DNA fragments in plasma were closely
associated with treatment response to preoperative
chemoradiotherapy. Further investigation showed that patient groups
with higher MGMT promoter methylation status had a better tumor
regression response. This suggested plasma cell-free DNA detection
also has potential in the prediction of response to preoperative
chemoradiotherapy. However, several problems, including the origins
of plasma cell-free DNA and its dynamic changes, remain unclear.
Further study based on larger numbers of patients is necessary. A
better understanding of plasma cell-free DNA is required to confirm
its clinical value in treatment response prediction and prognostic
evaluation of cancer patients.
Acknowledgements
This research was sponsored by the Young Scientist
Foundation from Shanghai Municipal Health Bureau (no. 157, 2011),
the Scientific Research Foundation for the Returned Overseas
Chinese Scholars (no. N130204) from China State Education Ministry,
the National Natural Science Foundation of China (no. 81202148),
the Foundation of Fudan University 985 Project (985IIIYPT06), the
Young Scientist Foundation of Fudan University Shanghai Cancer
Center (Y.L., 2011), the Shanghai Pujiang Program (no.
13PJ1401600), and the Foundation of Shanghai Committee of Science
and Technology of China (no. 12DZ2260100).
References
|
1
|
Tural D, Ozturk M, Selcukbiricik F, et al:
Preoperative chemoradiotherapy improves local recurrence free
survival in locally advanced rectal cancer. J BUON. 18:385–390.
2013.PubMed/NCBI
|
|
2
|
Valentini V, Coco C, Picciocchi A, et al:
Does downstaging predict improved outcome after preoperative
chemoradiation for extraperitoneal locally advanced rectal cancer?
A long-term analysis of 165 patients. Int J Radiat Oncol Biol Phys.
53:664–674. 2002. View Article : Google Scholar
|
|
3
|
Ho-Pun-Cheung A, Assenat E,
Bascoul-Mollevi C, et al: A large-scale candidate gene approach
identifies SNPs in SOD2 and IL13 as predictive markers of response
to preoperative chemoradiation in rectal cancer. Pharmacogenomics
J. 11:437–443. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wong MT, Lim JF, Ho KS, et al: Radiation
proctitis: a decade’s experience. Singapore Med J. 51:315–319.
2010.
|
|
5
|
Torres-Roca JF and Stevens CW: Predicting
response to clinical radiotherapy: past, present, and future
directions. Cancer Control. 15:151–156. 2008.PubMed/NCBI
|
|
6
|
Benn P, Cuckle H and Pergament E:
Non-invasive prenatal testing for aneuploidy: current status and
future prospects. Ultrasound Obstet Gynecol. 42:15–33. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liao GJ, Chiu RW and Lo YM: Prenatal
assessment of fetal chromosomal and genetic disorders through
maternal plasma DNA analysis. Pathology. 44:69–72. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hahn S, Rusterholz C, Hösli I and Lapaire
O: Cell-free nucleic acids as potential markers for preeclampsia.
Placenta. 32(Suppl 1): S17–S20. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
De Mattos-Arruda L, Cortes J, Santarpia L,
et al: Circulating tumour cells and cell-free DNA as tools for
managing breast cancer. Nat Rev Clin Oncol. 10:377–389.
2013.PubMed/NCBI
|
|
10
|
Ghorbian S and Ardekani AM: Non-invasive
detection of esophageal cancer using genetic changes in circulating
cell-free DNA. Avicenna J Med Biotechnol. 4:3–13. 2012.PubMed/NCBI
|
|
11
|
Kamat AA, Baldwin M, Urbauer D, et al:
Plasma cell-free DNA in ovarian cancer: an independent prognostic
biomarker. Cancer. 116:1918–1925. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ellinger J, Müller SC, Stadler TC, et al:
The role of cell-free circulating DNA in the diagnosis and
prognosis of prostate cancer. Urol Oncol. 29:124–129. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
van der Vaart M and Pretorius PJ: The
origin of circulating free DNA. Clin Chem. 53:22152007.PubMed/NCBI
|
|
14
|
Lecomte T, Berger A, Zinzindohoué F, et
al: Detection of free-circulating tumor-associated DNA in plasma of
colorectal cancer patients and its association with prognosis. Int
J Cancer. 100:542–548. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheng C, Omura-Minamisawa M, Kang Y, et
al: Quantification of circulating cell-free DNA in the plasma of
cancer patients during radiation therapy. Cancer Sci. 100:303–309.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Agostini M, Pucciarelli S, Enzo MV, et al:
Circulating cell-free DNA: a promising marker of pathologic tumor
response in rectal cancer patients receiving preoperative
chemoradiotherapy. Ann Surg Oncol. 18:2461–2468. 2011. View Article : Google Scholar
|
|
17
|
Mulcahy HE, Lyautey J, Lederrey C, et al:
A prospective study of K-ras mutations in the plasma of pancreatic
cancer patients. Clin Cancer Res. 4:271–275. 1998.PubMed/NCBI
|
|
18
|
Koutsimpelas D, Pongsapich W, Heinrich U,
et al: Promoter methylation of MGMT, MLH1 and
RASSF1A tumor suppressor genes in head and neck squamous
cell carcinoma: Pharmacological genome demethylation reduces
proliferation of head and neck squamous carcinoma cells. Oncol Rep.
27:1135–1141. 2012.
|
|
19
|
Dworak O, Keilholz L and Hoffmann A:
Pathological features of rectal cancer after preoperative
radiochemotherapy. Int J Colorectal Dis. 12:19–23. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sameer AS: Colorectal cancer: molecular
mutations and polymorphisms. Front Oncol. 3:1142013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gagliardi G, Biricotti M, Failli A, et al:
Colorectal carcinoma and folate. Ann Ital Chir. 84:123–131.
2013.PubMed/NCBI
|
|
22
|
Wong RK, Tandan V, De Silva S and
Figueredo A: Pre-operative radiotherapy and curative surgery for
the management of localized rectal carcinoma. Cochrane Database
Syst Rev. 2:CD0021022007.PubMed/NCBI
|
|
23
|
Kilic D, Yalman D, Aksu G, et al: Impact
of adjuvant chemoradiotherapy for rectal cancer on the long-term
quality of life and late side effects: a multicentric clinical
evaluation by the Turkish Oncology Group. Asian Pac J Cancer Prev.
13:5741–5746. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
De Caluwé L, Van Nieuwenhove Y and Ceelen
WP: Preoperative chemoradiation versus radiation alone for stage II
and III resectable rectal cancer. Cochrane Database Syst Rev.
2:CD0060412013.
|
|
25
|
González-Masiá JA, García-Olmo D and
García-Olmo DC: Circulating nucleic acids in plasma and serum
(CNAPS): applications in oncology. Onco Targets Ther. 6:819–832.
2013.PubMed/NCBI
|
|
26
|
Gorges TM, Schiller J, Schmitz A, et al:
Cancer therapy monitoring in xenografts by quantitative analysis of
circulating tumor DNA. Biomarkers. 17:498–506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mouliere F, Robert B, Arnau Peyrotte E, et
al: High fragmentation characterizes tumour-derived circulating
DNA. PLoS One. 6:e234182011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thierry AR, Mouliere F, Gongora C, et al:
Origin and quantification of circulating DNA in mice with human
colorectal cancer xenografts. Nucleic Acids Res. 38:6159–6175.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mouliere F and Thierry AR: The importance
of examining the proportion of circulating DNA originating from
tumor, microenvironment and normal cells in colorectal cancer
patients. Expert Opin Biol Ther. 12(Suppl 1): S209–S215. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zitt M, Müller HM, Rochel M, et al:
Circulating cell-free DNA in plasma of locally advanced rectal
cancer patients undergoing preoperative chemoradiation: a potential
diagnostic tool for therapy monitoring. Dis Markers. 25:159–165.
2008. View Article : Google Scholar
|
|
31
|
Zitt M, Zitt M, Müller HM, et al:
Disseminated tumor cells in peripheral blood: a novel marker for
therapy response in locally advanced rectal cancer patients
undergoing preoperative chemoradiation. Dis Colon Rectum.
49:1484–1491. 2006. View Article : Google Scholar
|
|
32
|
Liggett TE, Melnikov AA, Marks JR and
Levenson VV: Methylation patterns in cell-free plasma DNA reflect
removal of the primary tumor and drug treatment of breast cancer
patients. Int J Cancer. 128:492–499. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jing R, Cui M, Wang H and Ju S: Cell-free
DNA: characteristics, detection and its applications in myocardial
infarction. Curr Pharm Des. 19:5135–5145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lilleberg SL, Durocher J, Sanders C, et
al: High sensitivity scanning of colorectal tumors and matched
plasma DNA for mutations in APC, TP53, K-RAS, and BRAF genes with a
novel DHPLC fluorescence detection platform. Ann NY Acad Sci.
1022:250–256. 2004. View Article : Google Scholar
|
|
35
|
Wang JY, Hsieh JS, Chang MY, et al:
Molecular detection of APC, K-ras, and p53
mutations in the serum of colorectal cancer patients as circulating
biomarkers. World J Surg. 28:721–726. 2004.
|
|
36
|
Misale S, Yaeger R, Hobor S, et al:
Emergence of KRAS mutations and acquired resistance to
anti-EGFR therapy in colorectal cancer. Nature. 486:532–536.
2012.
|
|
37
|
Dobrzycka B, Terlikowski SJ, Mazurek A, et
al: Mutations of the KRAS oncogene in endometrial hyperplasia and
carcinoma. Folia Histochem Cytobiol. 47:65–68. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dobrzycka B, Terlikowski SJ, Mazurek A, et
al: Circulating free DNA, p53 antibody and mutations of KRAS
gene in endometrial cancer. Int J Cancer. 127:612–621. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Roberts PJ and Stinchcombe TE: KRAS
mutation: should we test for it, and does it matter? J Clin Oncol.
31:1112–1121. 2013. View Article : Google Scholar
|
|
40
|
Morgan SR, Whiteley J, Donald E, et al:
Comparison of KRAS mutation assessment in tumor DNA and
circulating free DNA in plasma and serum samples. Clin Med Insights
Pathol. 5:15–22. 2012.
|
|
41
|
Nygaard AD, Garm Spindler KL, Pallisgaard
N, et al: The prognostic value of KRAS mutated plasma DNA in
advanced non-small cell lung cancer. Lung Cancer. 79:312–317. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Spindler KL, Pallisgaard N, Vogelius I and
Jakobsen A: Quantitative cell-free DNA, KRAS, and
BRAF mutations in plasma from patients with metastatic
colorectal cancer during treatment with cetuximab and irinotecan.
Clin Cancer Res. 18:1177–1185. 2012.
|
|
43
|
Fukushima T, Takeshima H and Kataoka H:
Anti-glioma therapy with temozolomide and status of the DNA-repair
gene MGMT. Anticancer Res. 29:4845–4854. 2009.PubMed/NCBI
|
|
44
|
Psofaki V, Kalogera C, Tzambouras N, et
al: Promoter methylation status of hMLH1, MGMT, and
CDKN2A/p16 in colorectal adenomas. World J Gastroenterol.
16:3553–3560. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rivera AL, Pelloski CE, Gilbert MR, et al:
MGMT promoter methylation is predictive of response to radiotherapy
and prognostic in the absence of adjuvant alkylating chemotherapy
for glioblastoma. Neuro Oncol. 12:116–121. 2010. View Article : Google Scholar : PubMed/NCBI
|