Introduction
Gastric cancer (GC) is the 4th most common cancer
and the 2nd leading cause of cancer-related mortality worldwide
(1). Although treatment measures
are being developed, the prognosis of GC remains poor and is
correlated with tumor invasion (2).
Development and oncogenesis of GC remains controversial and
involves diverse factors, due to complex genetic backgrounds.
Semaphorins are a large family of membrane-bound and cytoplasmic
proteins, with up to 20 members (3). Semaphorins were originally identified
to play a role in axonal guidance (4–7).
However, further evidence showed that members of the semaphorins
family also regulate cell migration, cell differentiation, growth
and angiogenesis in a variety of tissues (8).
Semaphorin 3B (SEMA3B) is a member of the semaphorin
family, located on 3p21.3. SEMA3B was identified as a tumor
suppressor gene that is inactivated in lung cancer (9,10), HEY
ovarian carcinoma (11),
hepatocellular carcinomas and cholangiocarcinomas (12). Multiple mechanisms are involved in
the tumor suppressive role of SEMA3B that result in a two-hit
theory, i.e., silencing of SEMA3B through epigenetic changes and
allelic loss is a common and important event in its tumorigenesis
procedure. Furthermore, various regulatory factors participate in
the expression pattern of SEMA3B. Research shows that the Sema3b
protein may play a role in regulating cell growth and SEMA3B has
been identified as a mediator of p53 tumor-suppressor (13). SEMA3B is a potential tumor
suppressor that induces apoptosis in SEMA3B-inactivated tumor cells
through the Np-1 Receptor through inactivating the Akt signaling
pathway (14). As a carcinogenic
factor, vascular endothelial growth factor (such as VEGF165) acts
as an autocrine survival factor that could block SEMA3B-mediated
tumor-suppressing effects in tumor cells (15). Koyama et al (16) demonstrated that insulin-like growth
factor-binding protein-6 (IGFBP-6) is the effector of tumor
suppressor activity of SEMA3B. However, upregulation of furin-like
pro-protein convertases in malignant cells may enable tumors to
evade the antiangiogenic effects of Sema3b protein (17). SEMA3B also exerts unexpected
functions in cancer progression by fostering a prometastatic
environment through elevated IL-8 secretion and recruitment of
macrophages coupled to suppress tumor growth (18).
In the present study, we characterized the gene
expression and clinical meaning of SEMA3B in GC. Differential GC
cell lines and tissues from gastric tumor were used for the
exploration of the relationship between methylation status and
expression of the SEMA3B gene. Our findings indicated that the
SEMA3B gene was expressed low in GC and contributed to the growth
of tumor. The demethylation effect of 5-Aza-2′-deoxycytidine
restored the expression of SEMA3B. We suggest that the antitumor
effect of SEMA3B could be used as a new approach in the diagnosis
and treatment of GC.
Materials and methods
Patients and tissues samples
Thirty patients with primary GC (22 males, 8
females; median age, 64.5 years; range, 42–80 years) undergoing
gastrectomy (Billroth II or Radical Gastrectomy) between 2011 and
2013 at the First Affiliated Hospital of Wenzhou Medical University
were included in the present retrospective study. Tumor samples
were compared with normal gastric mucosa specimens (obtained from
negative resection margin) of the same individuals. No patient
received preoperative or adjuvant radio- or chemotherapy. TNM
staging of tumor was performed according to the UICC/AJCC (2010)
criteria. Specimens were snap frozen in liquid nitrogen within 30
min after the resection for storage.
All patients were informed of the special
examination of tumor samples in accordance with the ethical
standards of the Committee on Human Experimentation of the First
Affiliated Hospital of Wenzhou Medical University.
Cell culture and 5-Aza-2′-deoxycytidine
treatment
Human GC cell lines BGC-823, MGC-803 and SGC-7901
were grown in RPMI-1640 plus 10% fetal bovine serum (FBS;
Invitrogen, Cergy Pontoise, France) at 37°C and 5% CO2.
The cell line AGS was grown in F12K plus 10% FBS. Cells were seeded
at a density of 5×105 cells/10-cm plate on the first
day. 5-Aza-2′-deoxycytidine (Sigma, St. Louis, MO, USA) was added
at the final concentration of 0, 2 and 10 μM after 10-h starvation.
Cells were harvested on days 3 and 5 for analysis of SEMA3B mRNA,
protein expression and methylation status.
RNA extraction and quantitative real-time
reverse transcription PCR (qRT-PCR) analysis
Total RNA was isolated from cultured cells with
TRIzol reagent (Invitrogen Life Technologies, Grand Island, NY,
USA) following the manufacturer’s protocol and mRNA levels of
SEMA3B, p53 and GAPDH were evaluated with the real-time PCR system
by Bio-Rad (Bio-Rad CFX96; Bio-Rad Laboratories Hercules, CA, USA)
using cDNA reverse transcription kit (High Capacity type; Applied
Biosystems) and RNA-direct™ SYBR® Green Real-time PCR
Master Mix (Toyobo). Melting curves were generated for each
real-time PCR to verify the specificity of each PCR reaction. An
independent experiment was performed 3 times of real-time PCR for
accuracy. The GAPDH gene was used as a reference gene. The
2−ΔΔCT method was used to calculate the relative cDNA
amounts between different cell lines and this application was based
on assumptions stated by Livak and Schmittgen (19). All primer sequences and PCR
conditions used for mRNA expression and methylation analysis are
shown in Table I.
 | Table IPrimers used for SEMA3B mRNA and
methylation analysis. |
Table I
Primers used for SEMA3B mRNA and
methylation analysis.
| Gene (method) | Primer sequence
(5′-3′) | Annealing temperature
(°C) | Product size
(bp) |
|---|
| SEMA3B (qRT) | F:
CCAGTGCCAAGAGGCGGTTC
R: AGCACCTGGGTGTGGGCTGT | 62 | 214 |
| GAPDH (qRT) | F:
GACTCATGACCACAGTCCATGC
R: AGAGGCAGGGATGATGTTCTG | 62 | 113 |
| p53 (qRT) | F:
GGAGCCGCAGTCAGATCCTAG
R: CAAGGGGGACAGAACGTTG | 60 | 217 |
| SEMA3B (BSP) | F:
GGATTTTTTGTAGTTGGTGGAGT
R: GAGAGAGGAGTTAGATAGATTTTGA | 60 | 249 |
Western blot analysis
Total protein was obtained from cell culture by cell
lysis buffer (Beyotime Institute of Biotechnology, Shanghai,
China). Then, a small volume (50 μl) of lysate was removed to
perform a protein assay. To the remaining volume of cell lysate, we
added an equal volume of 2X Laemmli sample buffer. Each cell lysate
was boiled in sample buffer at 100°C for 5 min and aliquoted.
Lysates were stored at −20°C and equal amounts of protein were
loaded into the wells of the SDS-PAGE gel, along with molecular
weight markers. After transferring the protein from the gel to the
PVDF membrane, the membrane was blocked overnight at 4°C using 5%
blocking solution. We respectively incubated the membranes with
SEMA3B antibody (50 kDa, 1 μg/ml; Abcam, Cambridge, MA, USA) and
β-actin antibody (42 kDa; Beyotime Institute of Biotechnology) in
2% blocking solution overnight at 4°C. After washing the membrane
with TBST, horseradish-peroxidase (HRP) was added for signal
development. Images were acquired using darkroom development
techniques for chemiluminescence by ECL Plus (Amersham
Biosciences).
Genomic DNA isolation, sodium bisulfite
modification and bisulfite sequencing PCR
Genomic DNA was obtained from GC cell lines and
paired specimens of normal/tumor gastric tissues. Bisulfite
modification was performed by EZ DNA Methylation-Gold kit (Zymo
Research, Irvine, CA, USA) based on the manufacturer’s protocol.
The treated DNA was amplified by PCR using the following primer
with product size of 249: F, 5′-GGATTTTTTGTAGTTGGTGGAGT-3′ and R,
5′-GAGAGAGGAGTTAGATAGATTTTGA-3′ (Table
I). Amplified PCR products were analyzed by electrophoresis on
a 2% agarose gel and were purified with a TIANgel Midi Purification
kit (Tiangen Biotech, Co., Ltd., Beijing, China). The PCR fragments
were ligated to pMD 19-T Vector (Takara Biotechnology, Dalian,
China) and transformed into E. coli DH5α Electro-Cells
(Takara Biotechnology). Ten subcloned colonies were chosen at
random from each gastric cell line and 15 paired gastric tumor and
normal tissues. PCR products were sequenced by the 3730×l DNA
Analyzer (Applied Biosystems, Foster City, CA, USA).
Statistical and bioinformatic
analysis
Statistical significance of two groups was
determined by Student’s t-test using Stata/SE (Version 11.0;
StataCorp, College Station, TX, USA). The difference of DNA
methylation in paired specimens was analyzed by the χ2
test. The correlation between the DNA methylation level and mRNA
expression fold was determined by Spearman correlation analysis.
The 2−ΔΔCT method was adopted to quantify the mRNA
expression level between the tumor group and the normal tissue
group, where ΔΔCT was defined by the following equation:
ΔΔCT= [(CT of SEMA3B - CT of
GAPDH)Treatment - (CT of SEMA3B -
CT of GAPDH)Control]. Network relationships
between SEMA3B and its correlated regulators were analyzed by
Agilent Literature Search Software (20), which is a meta-search tool for
automatically querying multiple text-based search engines (both
public and proprietary) in order to extract associations among
genes/proteins of interest. All P-values were based on two-sided
tests, and the differences were considered statistically
significant when the P-value was ≤0.05.
Results
Prediction of CpG islands based on SEMA3B
gene DNA sequence
The SEMA3B gene is located in 3p21.3 of human
chromosome with exons length of ~3.4 kb, coding 749 amino acids.
Using MethPrimer online software (21), 4 CpG island regions were identified
based on SEMA3B gene sequence (NCBI Reference Sequence: NM_004636).
Conditions on CpG island identification had been set as default
terms that island size >100, GC Percent >50.0 and Obs/Exp
>0.6. Bisulfite PCR primer in island 3 was adopted in our
research, including 24 CpG dinucleotides in the 5′ region from exon
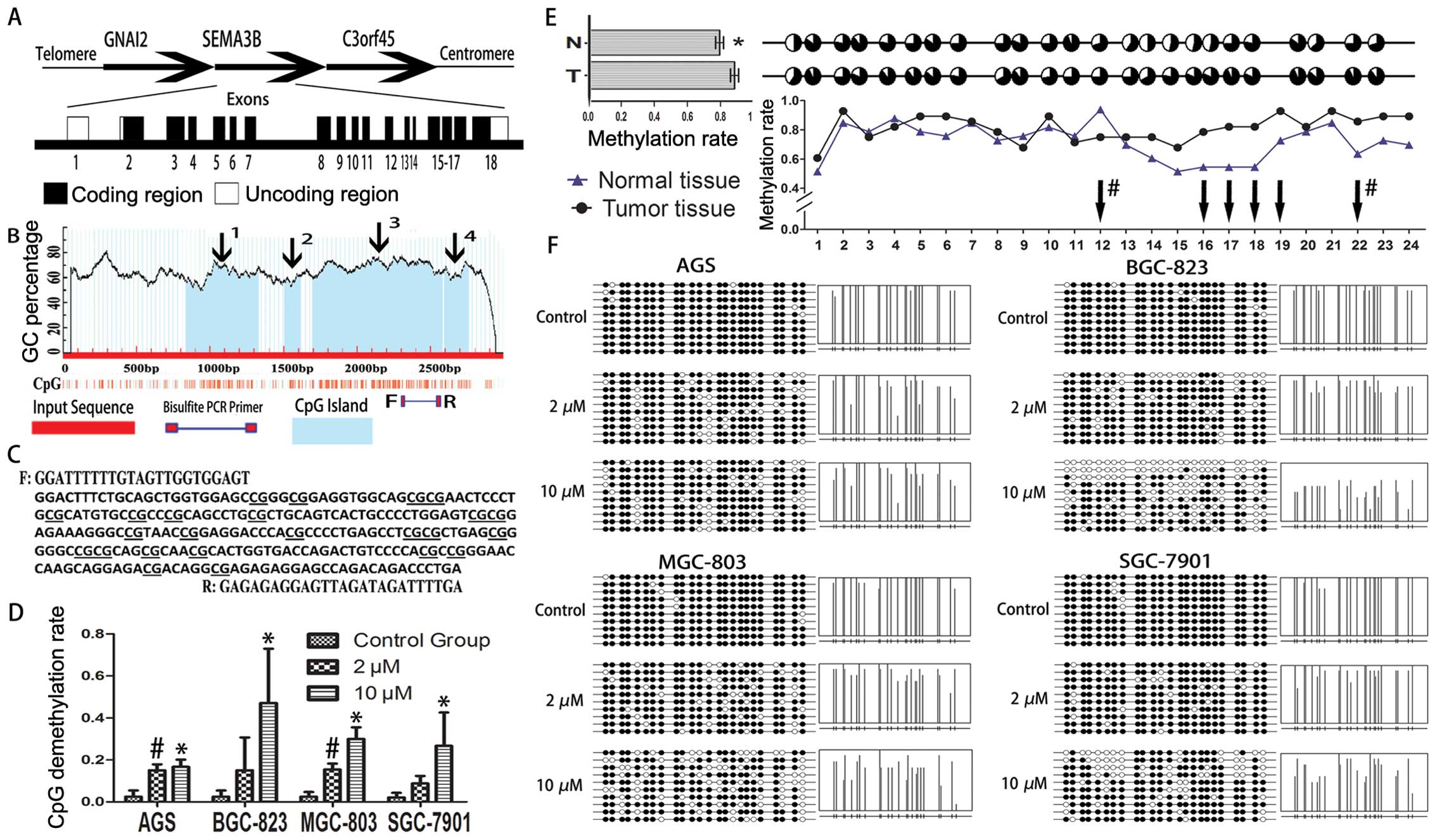
18 of SEMA3B gene (Fig. 1A–C).
SEMA3B expression is downregulated in GC
tissues
Results of qRT-PCR showed 63% of SEMA3B mRNA level
was downregulated in primary GC samples, compared to the matched
normal samples (19/30, P=0.034; Fig.
2C). Correlation analysis identified that SEMA3B mRNA
expression is negatively correlated with tumor size and tumor
N-stage (Table II). We noted that
advanced N stage showed less SEMA3B mRNA expression pattern
(P<0.01).
 | Table IIClinicopathological correlation of
SEMA3B mRNA expression status. |
Table II
Clinicopathological correlation of
SEMA3B mRNA expression status.
| SEMA3B mRNA
expression status (N) | Correlation
analysis |
|---|
|
|
|
|---|
| Clinical
characteristics | Upregulation | Downregulation | P-valuea | r, P-value |
|---|
| Gender | | | | −0.017, 0.927 |
| Male | 8 | 14 | 0.383b | |
| Female | 3 | 5 | | |
| Age (mean ± SD,
years) | 66.73±11.4
(11) | 63.47±7.6 (19) | 0.355 | 0.076, 0.689 |
| Tumor size (mean ±
SD, cm) | 3.73±1.3 (11) | 4.95±2.7 (19) | 0.063 | −0.386,
0.035c |
| Infiltration
depth | | | | 0.043, 0.821 |
| Submucosa | 1 | 4 | 0.529b | |
| Lamina
propria | 0 | 1 | | |
| Muscular
layer | 0 | 1 | | |
| Serosa | 10 | 13 | | |
| Lymphatic
metastasis | | | | −0.063, 0.741 |
| Yes | 8 | 13 | 0.804b | |
| No | 3 | 6 | | |
| Borrmann type | | | | 0.017, 0.928 |
| I + II | 10 | 17 | 0.321b | |
| III + IV | 1 | 2 | | |
|
Differentiation | | | | −0.172, 0.365 |
| Low | 9 | 11 | 0.246b | |
| Medium-high | 2 | 8 | | |
| T stage | | | | 0.023, 0.905 |
| T1–T2 | 1 | 3 | 0.570b | |
| T3 | 5 | 11 | | |
| T4 | 5 | 5 | | |
| N stage | | | | −0.491,
0.006d |
| N0–N1 | 9 | 6 |
0.003b | |
| N2–N3 | 2 | 13 | | |
| Nerve tract
involved | | | | 0.14, 0.461 |
| Yes | 5 | 6 | 0.447 | |
| No | 6 | 13 | | |
| Vessel
involved | | | | −0.023, 0.905 |
| Yes | 2 | 2 | 0.611b | |
| No | 9 | 17 | | |
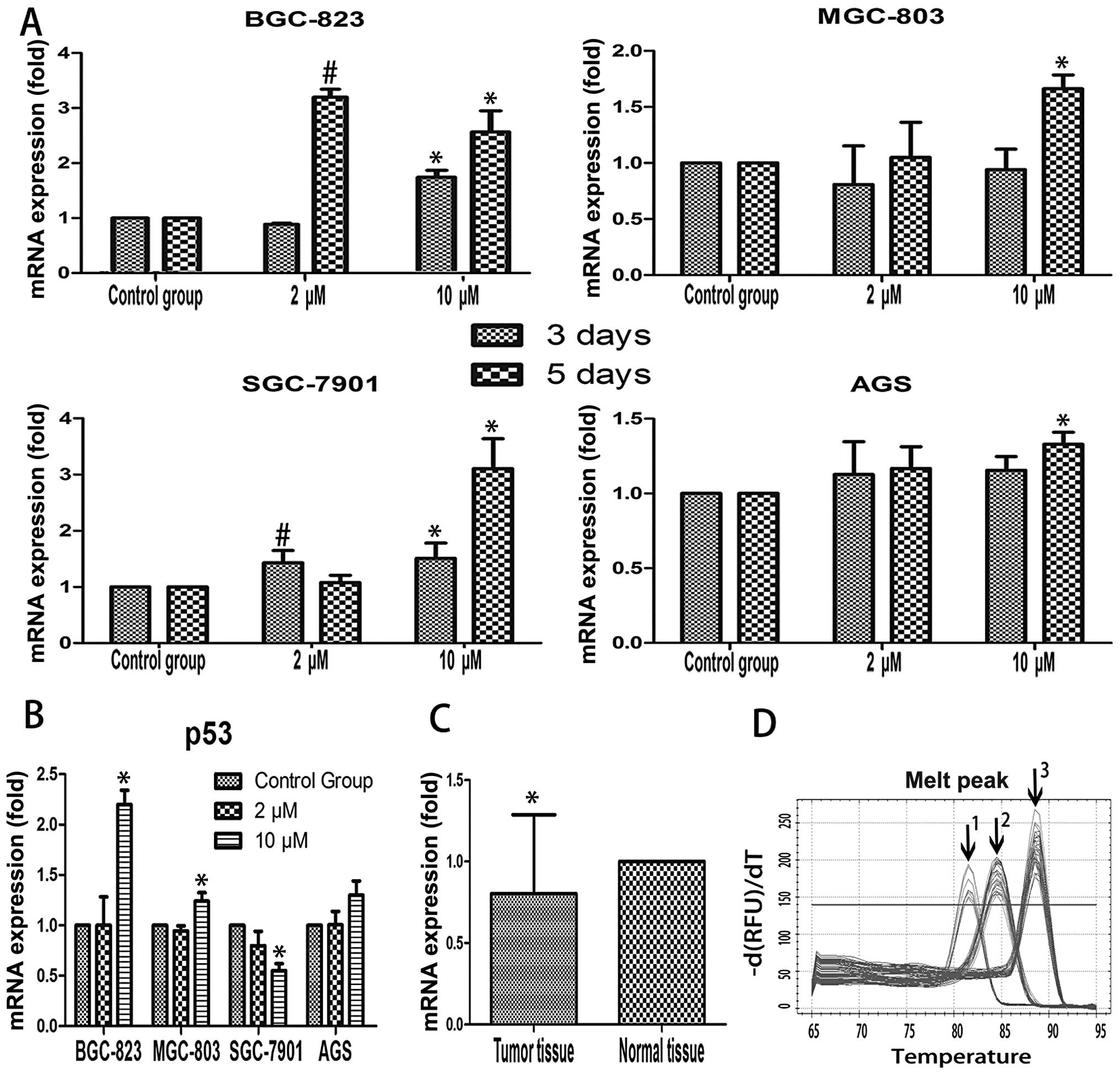
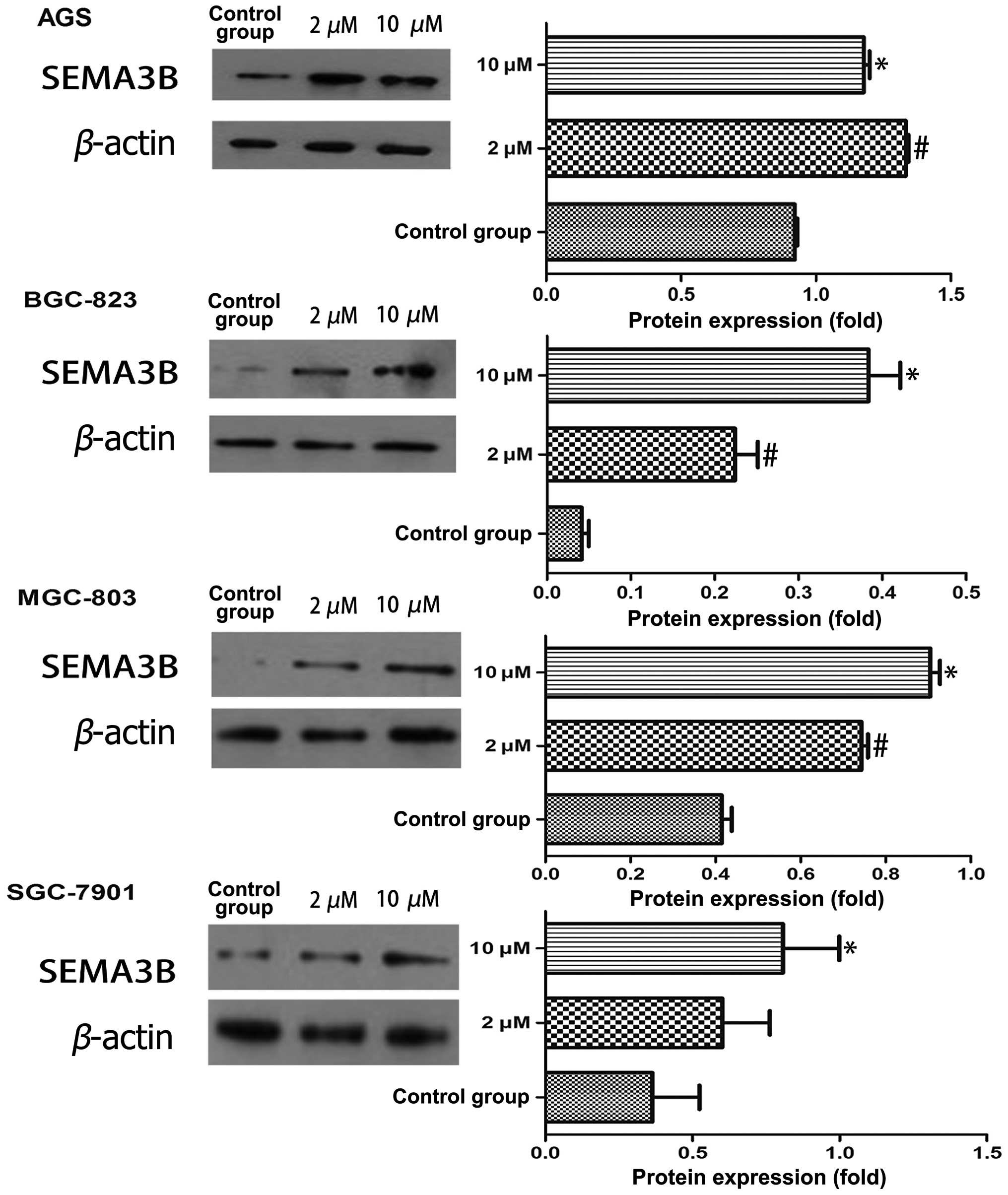
5-Aza-2′-deoxycytidine restores SEMA3B
gene expression based on different time and concentration
patterns
Expression of SEMA3B gene was evaluated by qRT-PCR
and western blotting, both of which were detected in 4 GC cell
lines (BGC-823, MGC-803, SGC-7901 and AGS). In 3-day treatment,
compared to the control group, only BGC-823 and SGC-7901 showed
increased mRNA expression following the dose cumulation of
5-Aza-2′-deoxycytidine (P<0.05). In 5-day treatment, all the
gastric cell lines showed the tendency of increased mRNA expression
at a concentration of 10 μM of 5-Aza-2′-deoxycytidine except for
BGC-823 (P<0.05; Fig. 2A). We
further explored the Sema3b protein expression in 5-day treatment.
BGC-823 and MGC-803 showed a significant increased Sema3b protein
expression when dosage of 5-Aza-2′-deoxycytidine was elevated
(P<0.05). This effect was only identified in SGC-7901 when it
was compared with the control group at the 10 μM concentration. AGS
showed the opposite expression pattern when the concentration of
5-Aza-2′-deoxycytidine was up to 10 μM, the Sema3b protein
expression was <2 μM (Fig.
3).
Elevated p53 expression is associated
with SEMA3B re-expression
In order to explore the role of p53 on SEMA3B mRNA
expression, we examined the p53 mRNA level by qRT-PCR, also with
the concentration of 5-Aza-2′-deoxycytidine at 0, 2 and 10 μM
(Fig. 2B). Results showed that p53
mRNA expression level was elevated in BGC-823, MGC-803 and AGS,
compared to the control group. However, AGS failed to reach
statistical significance (P>0.05). SGC-7901 showed decreased p53
mRNA expression level in 10 μM of 5-Aza-2′-deoxycytidine compared
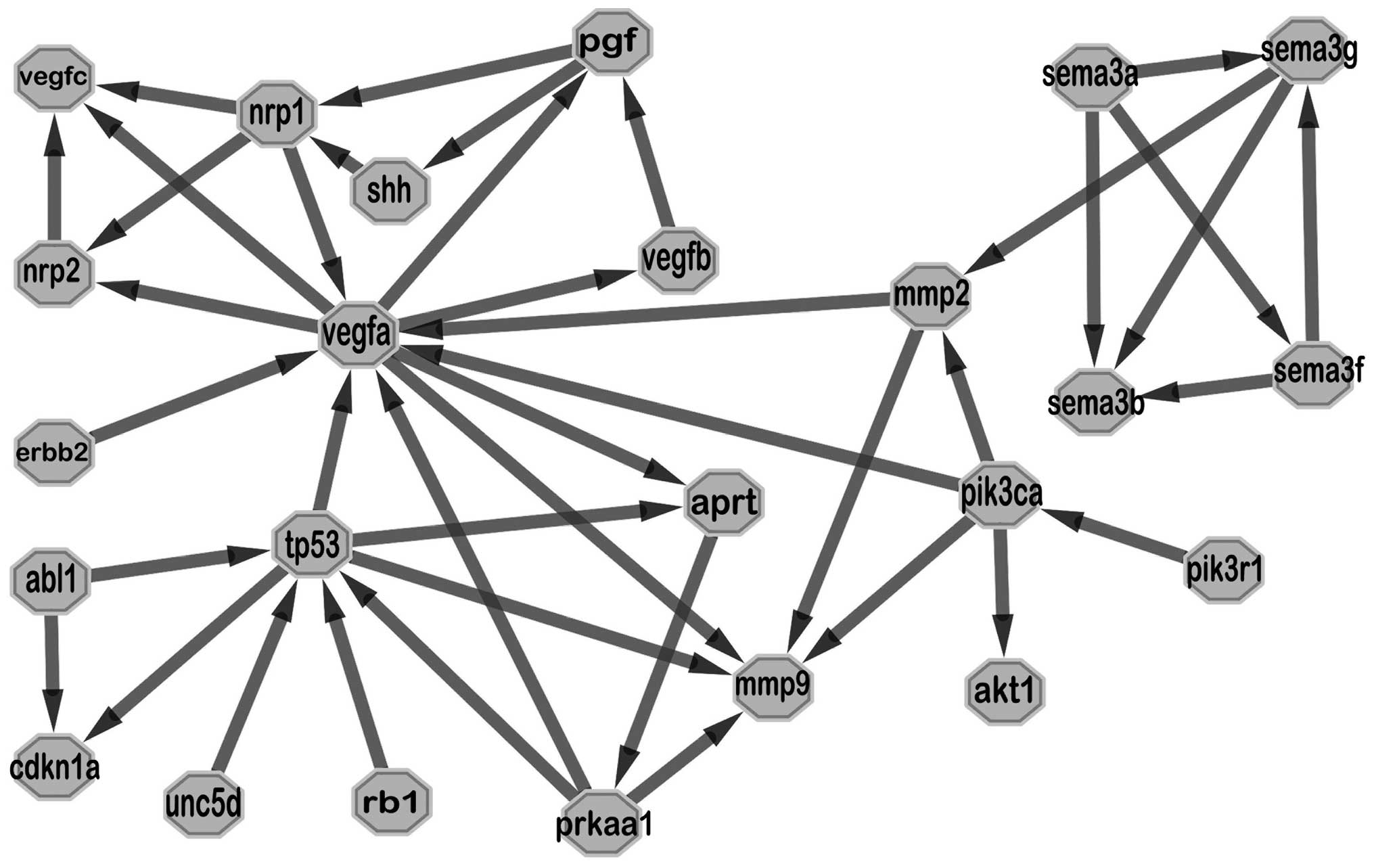
to the control group (P<0.05). By searching available databases,
Agilent Literature Search tool finds publications related to search
terms and creates interaction networks based on the search result.
We used the key words ‘Sema3b, p53, neuropilins, vascular
endothelial growth factor, cancer’ and 20 articles were found. A
clear relationship was shown in the generated network, in which
vegfa and tp53 exchanged information with outside more frequently
and mmp2 was an important intermediary agent connecting semaphoring
with vegfa (Fig. 4).
Methylation status of SEMA3B gene exon 18
affects transcript activities of SEMA3B
Bisulfite sequencing is regarded as the gold
standard to analyze the methylation status of one target sequence.
We performed bisulfite sequencing on gastric cell lines and tissues
of tumor and normal gastric mucosa. Nearly 100% of CpGs in exon 18
of SEMA3B showed methylated status in all 4 gastric cell lines.
After treatment of 5-Aza-2′-deoxycytidine for 5 days with the
concentration at 2 and 10 μM, CpG demethylation rate improved
significantly in all 4 gastric cell lines, compared to the control
group (P<0.05; Fig. 1D and F).
Furthermore, we detected the methylation levels of the exon 18 of
SEMA3B, compared among the available 15 paired tissues of tumor and
normal gastric mucosa (Fig. 1E). It
was observed that the methylation level was slightly but
significantly higher in tumor tissue compared to normal gastric
mucosa (P<0.05). Regarding the methylation level of each paired
CpG island which had different effect on methylation status in
cancer, CpG island -16, -17, -18, -19, -22 in the tumor group
showed hypermethylated status (P<0.05), while CpG island -12 in
paired normal tissue group showed hypermethylated status
(P<0.05).
Discussion
The present study was conducted to determine the
role of SEMA3B in GC with regard to the relationship between
methylation status and gene expression pattern.
Quantitative RT-PCR was used to assess mRNA level of
SEMA3B gene in primary gastric tumor and cell lines (BGC-823,
SGC-7901, MGC-803 and AGS). Data showed that the SEMA3B gene was
downregulated in gastric tissue compared to normal gastric mucosa
tissue. This suggested a decrease in SEMA3B expression may be
involved in the development of GC. Correlation analysis between
mRNA expression of SEMA3B and clinicopathological parameters
revealed that SEMA3B was involved in tumor growth and also affected
regional lymph node infiltration. As a type of secreted
semaphorins, Sema3s bind to a holoreceptor complex that consists of
Nrps as ligand binding. With the exception of binding to Sema3s,
Nrps could serve as VEGF family protein co-receptors (22). It was suggested that Sema3s compete
for the binding to Nrp1 (neuropilins) with VEGF to inhibit
VEGF-induced angiogenesis (23).
Lymph node infiltration was related to lymphatic vasculature
building, which was attributed to signaling by VEGF-C and regulated
by Nrp2 (24). This suggested that
Sema3s could also possibly function as anti-lymphangiogenic
signaling molecules through the binding affinity to Nrp2. Previous
studies showed that Sema3s are also involved in the lymph node
metastasis of prostate cancer, and were likely to modulate the
behavior of prostate cancer with an antitumor effect (25). In the present study, with the
deficiency of SEMA3B mRNA expression, its anti-angiogenic and
anti-lymphangiogenic effect was too low to induce tumor growth, and
lymph node infiltration was more common in GC. According to earlier
studies, SEMA3B mRNA level was also observed to be downregulated in
different types of tumors. Pronina et al (26) reported that SEMA3B expression levels
were frequently decreased in primary tumor extracts of kidney,
lung, breast, ovarian and colorectal cancer. Furthermore, this
phenomenon was confirmed in cell lines of breast cancer, epithelial
ovarian carcinoma and renal cell carcinoma.
We further explored the potential influencing
factors that affect the expression level of SEMA3B, in which
methylation status of SEMA3B in gastric tissue was detected.
Analysis of exon 18 of SEMA3B indicated that tissues from gastric
tumor showed higher methylated status than normal tissues
(P<0.05), in which methylation in CpG site -16, -17, -18, -19,
-22 weighted heavily. Epigenetic alteration in SEMA3B DNA sequence
is regarded as an important gene expression silencing pathway
demonstrated in several types of tumor, such as that in gallbladder
carcinoma (27). Kuroki et
al (28) reported that SEMA3B
hypermethylation is responsible for silencing SEMA3B expression in
NSCLC cell line by methylation-specific PCR. However, this result
was not in accordance with that in gliomas which showed SEMA3B was
not frequently methylated (29).
Different methylation rate has been reported in GC which varied
from 20% (30) to 60% (31). In our cases, the methylation rate of
SEMA3B in exon 18 from gastric tumor tissue was 88%, significantly
higher than the paired normal tissue (79%; P<0.05).
5-Aza-2′-deoxycytidine has been used as a
demethylation agent in recent years. After treatment with
5-Aza-2′-deoxycytidine, a demethylation state was detected in the 4
GC cell lines (BGC-823, MGC-803, SGC-7901 and AGS). Sema3b protein
expression was significantly higher in the
5-Aza-2′-deoxycytidine-treated group compared with that in the
control group. qRT-PCR further confirmed that in 5-day treatment,
5-Aza-2′-deoxycytidine could come to the largest effect confronted
with the hypermethylation status of SEMA3B in gastric cell lines.
Meanwhile, p53 expression was detected following the impact of
5-Aza-2′-deoxycytidine. Except for SGC-7901, all remaining gastric
cell lines showed higher p53 mRNA expression after the treatment of
5-Aza-2′-deoxycytidine at the concentration of 10 μM. Ochi et
al (13) demonstrated that
introduction of exogenous p53 into a glioblastoma cell line which
is lacking wild-type p53 markedly induced expression of SEMA3B
mRNA. This result suggested that SEMA3B plays some role as a direct
target of p53 and it was consistently comparable to our data.
Notably, SGC-7901 showed the opposite outcome that remains to be
demonstrated in future studies, which indicated a more complex
mechanism that had an effect on the relationship between p53 and
SEMA3B in different genetic backgrounds. Network analysis related
to sema3b, VEGF, neuropilins and p53 showed complex pathways
between them, in which mmp2 seemed to be an important channel
joining sema3b with vegfa. As an index of tumor invasion ability,
mmp2 has been shown to be inhibited by SEMA3G (32).
In summary, we confirmed that the methylation status
of the SEMA3B gene in gastric tumor was significantly higher than
that in paired normal gastric mucosa. We showed the epigenetic
inactivation of SEMA3B in several types of GC cell lines and
5-Aza-2′-deoxycytidine could reverse the hypermethylation status of
SEMA3B, which may benefit future studies exploring the application
of demethylating agents in clinical usage for GC.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Rivera F, Vega-Villegas ME and Lopez-Brea
MF: Chemotherapy of advanced gastric cancer. Cancer Treat Rev.
33:315–324. 2007. View Article : Google Scholar
|
|
3
|
Yazdani U and Terman JR: The semaphorins.
Genome Biol. 7:2112006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chilton JK and Guthrie S: Cranial
expression of class 3 secreted semaphorins and their neuropilin
receptors. Dev Dyn. 228:726–733. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Giger RJ, Cloutier JF, Sahay A, et al:
Neuropilin-2 is required in vivo for selective axon guidance
responses to secreted semaphorins. Neuron. 25:29–41. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Falk J, Bechara A, Fiore R, et al: Dual
functional activity of semaphorin 3B is required for positioning
the anterior commissure. Neuron. 48:63–75. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nawabi H, Briancon-Marjollet A, Clark C,
et al: A midline switch of receptor processing regulates
commissural axon guidance in vertebrates. Genes Dev. 24:396–410.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tamagnone L and Comoglio PM: To move or
not to move? Semaphorin signalling in cell migration. EMBO Rep.
5:356–361. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sekido Y, Bader S, Latif F, et al: Human
semaphorins A(V) and IV reside in the 3p21.3 small cell lung cancer
deletion region and demonstrate distinct expression patterns. Proc
Natl Acad Sci USA. 93:4120–4125. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tomizawa Y, Sekido Y, Kondo M, et al:
Inhibition of lung cancer cell growth and induction of apoptosis
after reexpression of 3p21.3 candidate tumor suppressor gene
SEMA3B. Proc Natl Acad Sci USA. 98:13954–13959. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tse C, Xiang RH, Bracht T and Naylor SL:
Human Semaphorin 3B (SEMA3B) located at chromosome 3p21.3
suppresses tumor formation in an adenocarcinoma cell line. Cancer
Res. 62:542–546. 2002.PubMed/NCBI
|
|
12
|
Tischoff I, Markwarth A, Witzigmann H, et
al: Allele loss and epigenetic inactivation of 3p21.3 in malignant
liver tumors. Int J Cancer. 115:684–689. 2005. View Article : Google Scholar
|
|
13
|
Ochi K, Mori T, Toyama Y, Nakamura Y and
Arakawa H: Identification of semaphorin3B as a direct target of
p53. Neoplasia. 4:82–87. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Castro-Rivera E, Ran S, Brekken RA and
Minna JD: Semaphorin 3B inhibits the phosphatidylinositol
3-kinase/Akt pathway through neuropilin-1 in lung and breast cancer
cells. Cancer Res. 68:8295–8303. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Castro-Rivera E, Ran S, Thorpe P and Minna
JD: Semaphorin 3B (SEMA3B) induces apoptosis in lung and breast
cancer, whereas VEGF165 antagonizes this effect. Proc Natl Acad Sci
USA. 101:11432–11437. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Koyama N, Zhang J, Huqun, et al:
Identification of IGFBP-6 as an effector of the tumor suppressor
activity of SEMA3B. Oncogene. 27:6581–6589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Varshavsky A, Kessler O, Abramovitch S, et
al: Semaphorin-3B is an angiogenesis inhibitor that is inactivated
by furin-like pro-protein convertases. Cancer Res. 68:6922–6931.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rolny C, Capparuccia L, Casazza A, et al:
The tumor suppressor semaphorin 3B triggers a prometastatic program
mediated by interleukin 8 and the tumor microenvironment. J Exp
Med. 205:1155–1171. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
|
|
20
|
Vailaya A, Bluvas P, Kincaid R, Kuchinsky
A, Creech M and Adler A: An architecture for biological information
extraction and representation. Bioinformatics. 21:430–438. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li LC and Dahiya R: MethPrimer: designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gluzman-Poltorak Z, Cohen T, Herzog Y and
Neufeld G: Neuropilin-2 is a receptor for the vascular endothelial
growth factor (VEGF) forms VEGF-145 and VEGF-165. J Biol Chem.
275:299222000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Soker S, Miao HQ, Nomi M, Takashima S and
Klagsbrun M: VEGF165 mediates formation of complexes containing
VEGFR-2 and neuropilin-1 that enhance VEGF165-receptor binding. J
Cell Biochem. 85:357–368. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Haiko P, Makinen T, Keskitalo S, et al:
Deletion of vascular endothelial growth factor C (VEGF-C) and
VEGF-D is not equivalent to VEGF receptor 3 deletion in mouse
embryos. Mol Cell Biol. 28:4843–4850. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li K, Chen MK, Li LY, et al: The
predictive value of semaphorins 3 expression in biopsies for
biochemical recurrence of patients with low- and intermediate-risk
prostate cancer. Neoplasma. 60:683–689. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pronina IV, Loginov VI, Prasolov VS, et
al: Alteration of SEMA3B gene expression levels in epithelial
tumors. Mol Biol (Mosk). 43:439–445. 2009.(In Russian).
|
|
27
|
Riquelme E, Tang M, Baez S, et al:
Frequent epigenetic inactivation of chromosome 3p candidate tumor
suppressor genes in gallbladder carcinoma. Cancer Lett.
250:100–106. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kuroki T, Trapasso F, Yendamuri S, et al:
Allelic loss on chromosome 3p21.3 and promoter hypermethylation of
semaphorin 3B in non-small cell lung cancer. Cancer Res.
63:3352–3355. 2003.PubMed/NCBI
|
|
29
|
Hesson L, Bieche I, Krex D, et al:
Frequent epigenetic inactivation of RASSF1A and BLU
genes located within the critical 3p21.3 region in gliomas.
Oncogene. 23:2408–2419. 2004.PubMed/NCBI
|
|
30
|
Bernal C, Vargas M, Ossandon F, et al: DNA
methylation profile in diffuse type gastric cancer: evidence for
hypermethylation of the BRCA1 promoter region in early-onset
gastric carcinogenesis. Biol Res. 41:303–315. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bernal C, Aguayo F, Villarroel C, et al:
Reprimo as a potential biomarker for early detection in gastric
cancer. Clin Cancer Res. 14:6264–6269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou X, Ma L, Li J, Gu J, Shi Q and Yu R:
Effects of SEMA3G on migration and invasion of glioma cells. Oncol
Rep. 28:269–275. 2012.PubMed/NCBI
|