Introduction
The study of genes that are differentially expressed
during tumor progression may identify key players in this complex
and multistep biological process. Polyomavirus large-T antigen
(PyLT-Ag) transgenic mice have been used as a model system for
studying differentially expressed genes during tumorigenesis
(1). With the pre- and
post-adenomatous testis cell lines derived from the PyLT-Ag
transgenic mice, as well as the differential display technique, a
testicular tumor differentially expressed gene (TDE1) was
identified to be upregulated during transformation (2–15 times
compared to the control cell line), indicating a potential
oncogenic property possessed by it (2–4). This
gene was also independently cloned by other researchers, and was
designated TMS-1, AIGP1 and/or Serinc3
(5–7). Subsequently, increasingly more
homologous and orthologous genes of TDE1 were cloned and
identified in species such as yeast, worm, fruit fly, zebrafish,
mouse, rat and human, giving rise to a large TDE1/TMS protein
family (also known as the Serinc family for serine incorporator)
(5,7–9). This
novel TDE1/TMS1/Serinc family appears in eukaryotic organisms and
contains no amino acid sequence homology with other known protein
families. At the same time, the TDE/Serinc proteins themselves are
highly conservative, sharing between 30 and 80% homology (8). There is a total five TDE/Serinc
proteins (Serinc1–5) identified in mammals while baking yeast
harbors only one ortholog (TMS1). The TDE/Serinc proteins contain
multiple transmembrane domains (up to 11) and often an N-terminal
signal peptide.
The characteristic multiple transmembrane domains of
TDE/Serinc proteins by peptide analysis suggested that they were
most closely related to proteins involved in ion-channel formation
or amino acid transportation. However, all attempts to detect amino
acid transport with mouse Serinc1 and Serinc3 failed (5,7). In
2005, Inuzuka et al (7)
studied the Serinc function by searching the yeast proteome
database for its interacting partners. Two proteins involved in
serine synthesis, SER3 and YGP1, were identified to interact with
TMS1 (the only Serinc protein in yeast). Yeast dihybrid experiment
further demonstrated that rat Serinc1 protein could also interact
with SER3 and YPG1, indicating the possible conservation of these
interactions. Moreover, it was also shown that Serinc proteins
(Serinc1, 2 and 5) could additionally enhance incorporation of
serine into the membrane lipid phosphatidylserine both in
prokaryotes and eukaryotes. On the other hand, sphingolipids are
another class of membrane lipids that could be synthesized by all
eukaryotic cells from serine and palmitoyl-CoA. The condensation of
these two molecules is catalyzed by serine palmitoyltransferase
(SPT), the most key limiting factor in the sphingolipid de
novo biosynthetic pathway. Inuzuka et al also showed
that rat Serinc1 protein facilitated the cellular sphingolipid
biosynthesis in both yeast and mammalian cells, probably via
interacting with and enhancing the activity of SPT. The same
property was also demonstrated for Serinc2 and 5. Results of the
biochemical assays performed by Inuzuka et al indicated that
the Serinc family members might play pivotal roles in the
biosynthesis of membrane lipids. Indeed, besides the upregulation
of Serincs observed in carcinoma tissues, it was postulated that
rat Serinc5/TPO1 might be involved in myelin biogenesis based on
its expression pattern (9).
Furthermore, in the hippocampus of rats with seizures induced by
kainite, Serinc1, 2 and 5 were found to be differentially expressed
compared to that of the control rats (7). These findings suggested that Serinc
proteins might participate in the plasticity of the central nervous
system, perhaps affecting membrane lipid biogenesis.
The conservative nature of the TDE/Serinc proteins
suggests an important biological role for this family. Indeed,
their overexpression was shown to correlate with carcinogenesis. In
addition to the known overexpression of mouse TDE gene in
testicular tumors from PyLT-Ag transgenic mice, human TDE1 (hTDE1)
was also shown to be highly expressed in lung cancer tissues
(10). Moreover, Player et
al (11) cloned and localized
human Serinc2 to chromosome 1 in 2003. Results of both in
situ hybridization and real-time PCR showed that its mRNA was
also upregulated in human non-small cell lung cancer tissue.
However, besides the biochemical function of these proteins
postulated by Inuzuka et al, the molecular mechanism
underlying their oncogenic property remains to be elucidated.
Cell growth is regulated by several factors. In
addition to duplication of DNA, protein, and other cellular
components, recent studies have provided insight into the
involvement of lipid metabolism, including membrane lipid
biogenesis (therefore affecting plasma membrane, or, more
importantly, secretory vesicles), in cell-cycle progression
(12–16). On the one hand, Kurat et al
(15) discovered that Tgl4, the
yeast triacylglycerol lipase, was directly activated by
Cdk1/Cdc28-dependent phosphorylation; on the other hand, sterol
regulatory element-binding proteins (SREBPs), which are
transcriptional factors involved in cholesterol and fatty acid
synthesis, have also been shown to affect cell proliferation
through accumulating cdk inhibitors such as p21 (17–19).
hTDE2/Serinc1 located on chromosome 6, was first cloned by
our group in 1998 during an investigation of genes differentially
expressed in liver cancer tissues (GenBank access number AF087902;
unpublished data). hTDE2/Serinc1 has a broad expression
profile and is significantly upregulated in the hepatocarcinoma
tissues examined, which is consistent with the oncogenic behaviors
of other Serinc members such as Serinc2 and Serinc3. In the present
study, we showed that knockdown of hTDE2/Serinc1 expression
resulted in cell cycle arrest and cell growth inhibition. Moreover,
downregulation of TDE2/Serinc1 upregulated SREBPs, and, eventually,
expression of p21 was elevated causing cell cycle retardation.
Materials and methods
Cell lines, tissue samples and other
materials
The human HCC cell lines L-02, YY-8103, SMMC-7721,
QGY-7703 and Huh-7 were from Fudan University (Shanghai, China).
FOCUS was from the Molecular Hepatology Laboratories, MGH Cancer
Center. All other cell lines used in the present study were
obtained from the American Type Culture Collection (ATCC, Manassas,
VA, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium
or RPMI-1640 supplemented with 10% heat-inactivated fetal bovine
serum (FBS), accordingly. The cell cultures were incubated at 37°C
in a humidified incubator with 5% CO2. The human
multiple tissue cDNA panel was purchased from Clontech.
Paired hepatoma and non-hepatoma tissue samples were
obtained from 32 hepatoma patients who had surgery at Eastern
Hepatobiliary Surgery Hospital, Second Military Medical University,
Shanghai, China. The hospital’s Ethics Committee had approved the
specimen collection procedures, and written informed consent was
obtained from each patient or their relatives.
To perform hTDE2 knockdown, three gene-specific
siRNA fragments (460, 895 and 1394, targeting different regions of
hTDE2) as well as one non-specific fragment (N.C.) were employed to
transfect the HCC cell lines. The sequences were:
5′-CUGCAGCAAUUGCAAUUAUTT-3′ (sense) and 5′-AUAAUUGCAAUUGCUGCAGTT-3′
(antisense) for fragment 460; 5′-GGUCAGCUAUGACCAAUGATT-3′ (sense)
and 5′-UCAUUGGUCAUAGCUGACCTT-3′ (antisense) for fragment 895;
5′-GGCACCACUUGUUCUUACATT-3′ (sense) and 5′-UGUAAGAACAAGUGGUGCCTT-3′
(antisense) for fragment 1394; and 5′-UUCUCCGAACGUGU CACGUTT-3′
(sense) and 5′-ACGUGACACGUUCGGAGAATT-3′ (antisense) for fragment
N.C. All fragments were synthesized at Shanghai GenePharma
(Shanghai, China).
Real-time quantitative PCR and
semi-quantitative PCR
First, total RNA was extracted from tissues or cells
using TRIzol (Invitrogen) and was reverse transcribed into cDNA
(Gibco-BRL) following the manufacturer’s instructions. Then,
real-time PCR was performed with an iCycler iQ (Bio-Rad
Laboratories) to analyze the level of target mRNAs, using the
dsDNA-specific binding dye SYBR-Green Premix (Toyobo). After
normalizing to GAPDH, target mRNA levels were quantified with the
ΔΔCT method. Primer sets used were:
5′-AGATAATGAAAGGGATGGTGTC-3′ (sense) and
5′-ACAGCACGATGCCAATCCAACT-3′ (antisense) for hTDE2;
5′-CGGCCGGGGGAACCCAGTT-3′ (sense) and 5′-CGCAGCCGCCTCC-3′
(antisense) for hSREBP1a; 5′-GAAGGCTGGAGACCAGGAAGA-3′ (sense) and
5′-CGTCCACCACCGACAGATGA-3′ (antisense) for hSREBP2;
5′-TGGAGACTCTCAGGGTCGAAA-3′ (sense) and 5′-AGGACTGCAGGCTTCCTGTG-3′
(antisense) for p21; and 5′-AGGGCTGCTTTTAACTCTGGT-3′ (sense) and
5′-CCCCACTTGATTTTGGAGGGA-3′ (antisense) for GAPDH.
To compare the expression of hTDE2 in
hepatocarcinoma and pericancerous tissue samples, semi-quantitative
PCR was performed. Total RNA was extracted and reverse transcribed
into cDNA as described above. According to the PCR signal generated
from the internal standard β2-MG, the template amount of each cDNA
pool was adjusted to give the same exponential phase signal
strength after 24 cycles. PCR reactions were then performed with
appropriate conditions. The final PCR samples were then subjected
to electrophoresis on 1.5% agarose gels and ethidium bromide
staining. After capturing digital images under UV, the densitometry
value of each PCR-generated DNA fragment was measured with UVP
Gelworks ID Advanced software (Version 2.51). DR (dosage ratio) was
used to describe the relative expression difference between the
tumor and its pericancerous tissues. DR is calculated with the
following formula: DR =
(Densitytumor-hTDE2/Densitytumor-β2-MG)/(Densitypericancerous-hTDE2/Densitypericancerous-β2-MG).
For semi-quantitative PCR, primer set 5′-TCTCTTCCAGTTGGATTGGCATC-3′
(sense) and 5′-CCATAACCTACACTATTGTCCAC-3′ (antisense) was used for
hTDE2, while primer set 5′-ATGAGTATGCCTGCCGTGTGAAC-3′ (sense) and
5′-TGTGGAGCAACCTGCTCAGATAC-3′ (antisense) was used for β2-MG.
Cell cycle, growth curve and colony
formation assay
Cells were transfected with appropriate siRNA
fragments and were collected 48 h after transfection. Prior to flow
cytometric analysis, cells were fixed with pre-chilled (at −20°C)
75% ethanol for 2 h at 4°C, followed by washing with PBS. PI (final
concentration, 50 μg/ml) and RNAase (final concentration, 100
μg/ml) were then added to the cell suspension and the mixture was
kept for 20 min in the dark. Analysis was completed within 3 h
after PI staining.
MTS assay was performed according to the
manufacturer’s instructions. The experiments were carried out in
triplicate and the average absorbance was calculated to generate
the growth curve.
For the colony formation assay, cells were seeded in
6-well plates as 400 cells/well, followed by culturing at 37°C with
proper medium for 6–10 days. Every other day, old medium was
replaced with fresh one. At the end of the incubation, cells were
washed and fixed with 4% paraformaldehyde for 15 min. The fixed
cells were washed again and subjected to Giemsa (Sigma) staining at
room temperature for 2 h. Images of the colonies were captured
after the plates were washed and dried.
Western blot analysis
Cells seeded in 6-well plates were transfected with
appropriate siRNA fragments (50 pmole/well) using Lipofectamine™
2000 (Invitrogen) according to the manufacturer’s instructions.
Forty-eight hours after transfection, cells were lysed and
processed for SDS-PAGE electrophoresis. Proteins were then
transferred to nitrocellular membranes (Bio-Rad Laboratories).
After blocking with 5% milk in TBST buffer, the membranes were
subjected to primary and secondary antibody incubation with washing
in between and after. Membranes were then incubated in enhanced
chemiluminescence (ECL) solution (Amersham) for 1 min followed by
exposure to Hyperfilm. The primary antibodies used were: anti-human
SREBP-1 polyclonal antibody (C-20, sc-366, 1:200; Santa Cruz
Biotechnology), anti-human SREBP-2 polycolonal antibody (H-164,
sc-5603, 1:200; Santa Cruz Biotechnology), anti human p21
monoclonal antibody (1:1,000; Abcam), anti-human p53 monoclonal
antibody (1:200; Sigma) and anti-human β-actin monoclonal antibody
(1:5,000; Sigma). The corresponding secondary antibodies used were
all diluted as 1:3,000.
Luciferase constructs and luciferase
assay
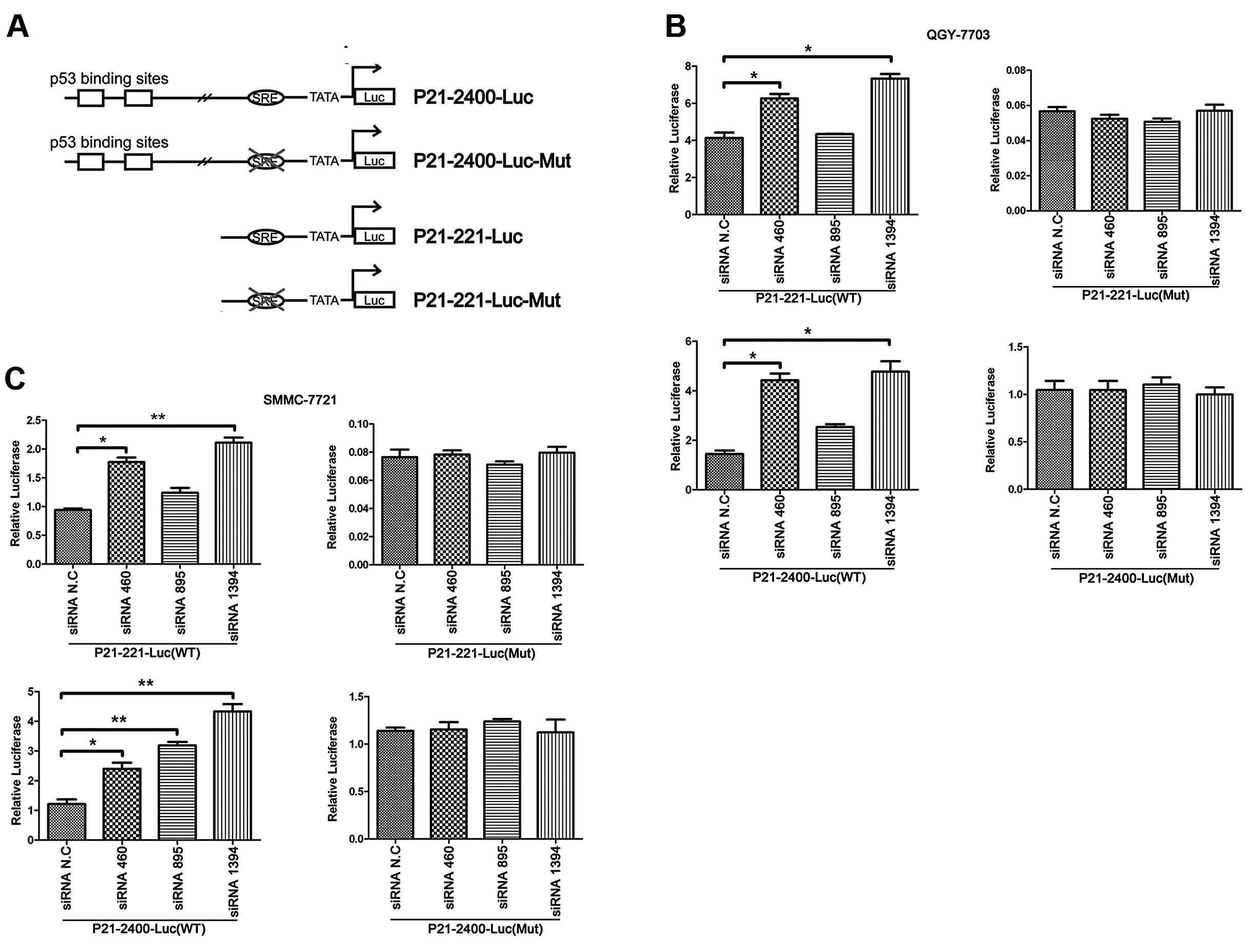
p21-2400-Luc and p21-221-Luc reporter plasmids were
kind gifts from Dr Huang Haojie from Mayo Clinics (Rochester, MN,
USA) with a pGL3-Basic backbone (Promega). In the reporter
plasmids, nucleotide sequences from +73 to −221 and/or −2400 of the
p21 gene were cloned in front of the luciferase reporter
gene. p21-2400-Luc, but not p21-221-Luc, contains two p53-binding
sites. Both plasmids harbor the SRE element which is located
between positions −90 and −98 of the p21 promoter. The other two
luciferase constructs, p21-2400-Luc-Mut and p21-221-Luc-Mut, were
generated by PCR with a site-directed mutagenesis kit (Toyobo). As
a result, the wild-type SRE sequence, TGGGCCGAG, was
replaced by TACAAAATG (20).
To perform the luciferase assay with the
dual-luciferase reporter system (Promega), cells were seeded on
24-well plates one day before transfection. Luciferase reporter
plasmid (0.1 μg) and pRL-SV40, the Renilla luciferase
internal control plasmid (0.01 μg; Promega), were co-transfected
together with appropriate siRNA fragments into the cells using
Lipofectamine 2000 kit (Invitrogen). Luciferase activity in each
transfected sample was examined and normalized to that of the
Renilla luciferase activity.
Statistical analysis
The values are expressed as means ± SD. The
Student’s t-test was used to assess the differences. P<0.05 was
considered to indicate a statistically significant result.
Results
hTDE2 gene is upregulated in
hepatocarcinoma tissues
hTDE2 (hSerinc1) belongs to the Serinc protein
family which was highly conserved from fungi to vertebrates.
Previous research indicated that high expression of these family
members was correlated with carcinogenesis (4,10,11).
However, the physiological function of this family remains unclear.
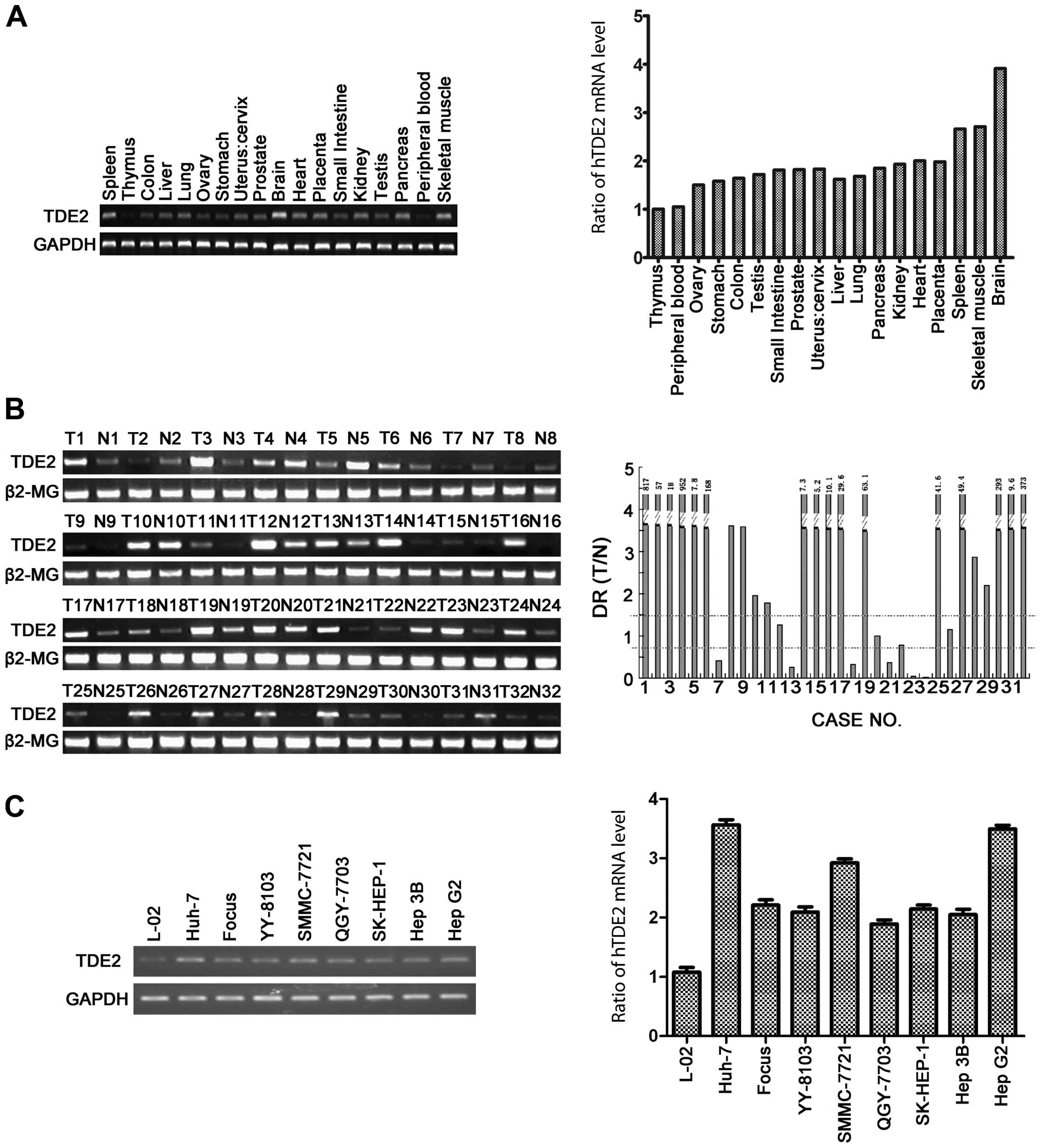
In the present study, we first assessed the expression profile of
hTDE2 in different human tissues with a human multiple
tissue cDNA panel. The quantitative PCR results indicated that the
tissue expression profile of hTDE2 is broad, with the
highest expression in the brain and the lowest in the thymus
(Fig. 1A).
To further analyze whether hTDE2 was also correlated
with hepatocarcinogenesis, we measured hTDE2 expression in 32
paired HCC and corresponding non-HCC neighboring tissue samples
with semi-quantitative PCR. In 6/32 paired samples, hTDE2 was
downregulated in the HCC tumor tissues; in 23 paired samples (23/32
= 71.9%), clear upregulation of hTDE2 was observed in the tumor
tissues. The remaining 3 paired samples showed a similar expression
of hTDE2 in the tumor and normal tissues (Fig. 1B). This result is consistent with
the previous findings that a correlation exists between
upregulation of TDE proteins and carcinogenesis.
We also measured the hTDE2 mRNA level in
different live cell lines commonly used in our laboratory, and
found that all the cell lines expressed this gene and its
expression was lower in L-02 compared to other cell lines
(hepatocarcinoma lines) examined here (Fig. 1C). Since L-02 was generally
considered to be a normal liver cell line, this result was
consistent with previous ones with human tissue cDNA panel and HCC
samples. We selected two of the hepatocarcinoma cell lines,
QGY-7703 and SMMC-7721, to carry out the following study.
Knockdown of hTDE2 hinders the growth of
hepatocarcinoma cells and arrests the cell cycle at G2
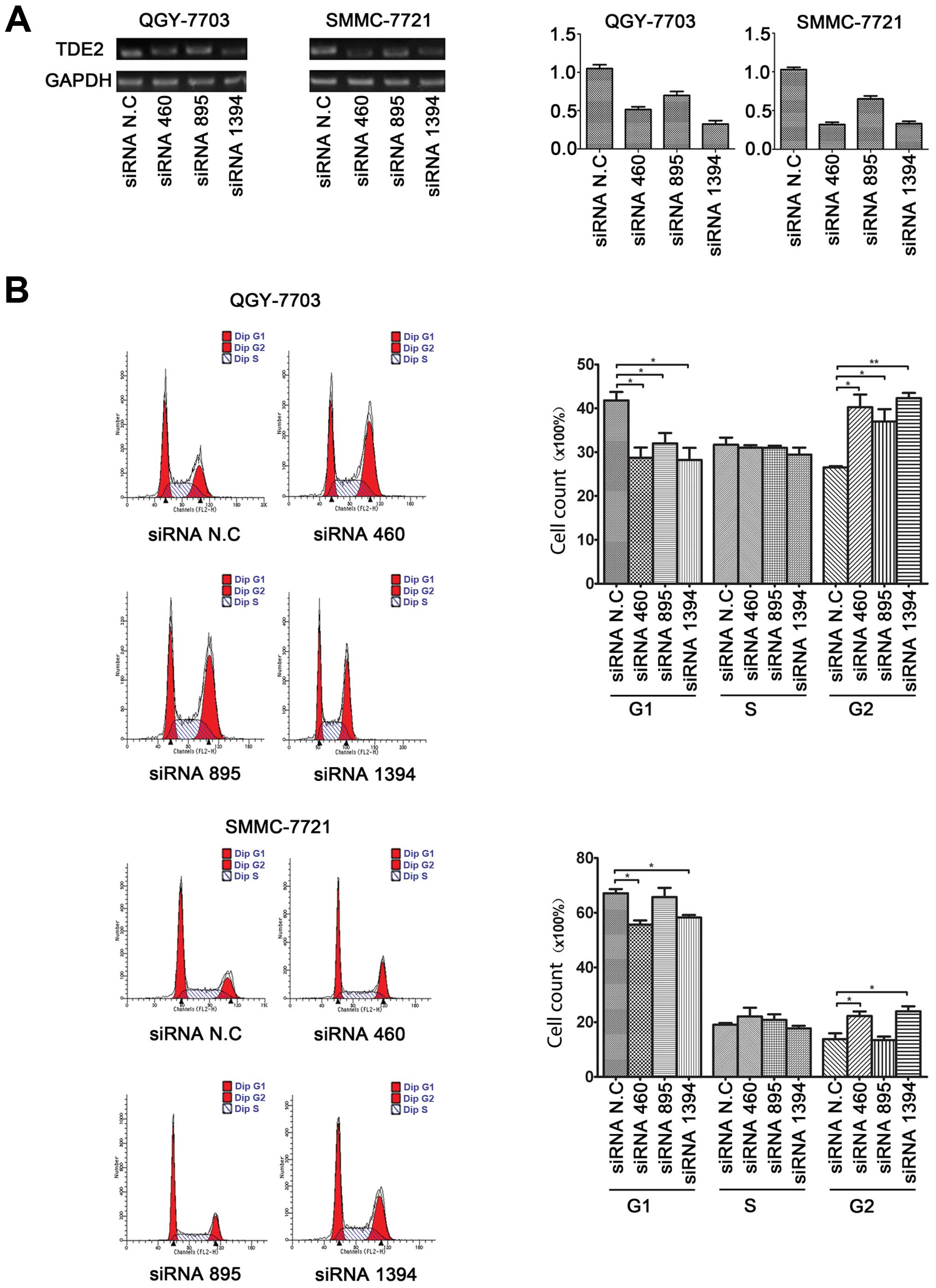
Since hTDE2 was significantly upregulated in
the HCC specimens, we investigated whether knocking down its
expression could have any effect on tumor cell growth. Therefore,
we designed three siRNA fragments specifically targeting
hTDE2 expression (siRNA 460, 895 and 1394) and analyzed
their efficacy with quantitative PCR (Fig. 2A). All three siRNA fragments reduced
hTDE2 mRNA levels in both QGY and SMMC cells with siRNA 895
being relatively less efficient. Then, we utilized FACS assay to
detect the cell cycle progression of both HCC cell lines 48 h after
transient transfection of these siRNA fragments. We found that, in
both QGY and SMMC, the number of cells in G1 phase decreased and
that of G2 cells increased upon hTDE2 knockdown (Fig. 2B). This verified our postulation
that reduction of hTDE2 expression may affect HCC tumor cell
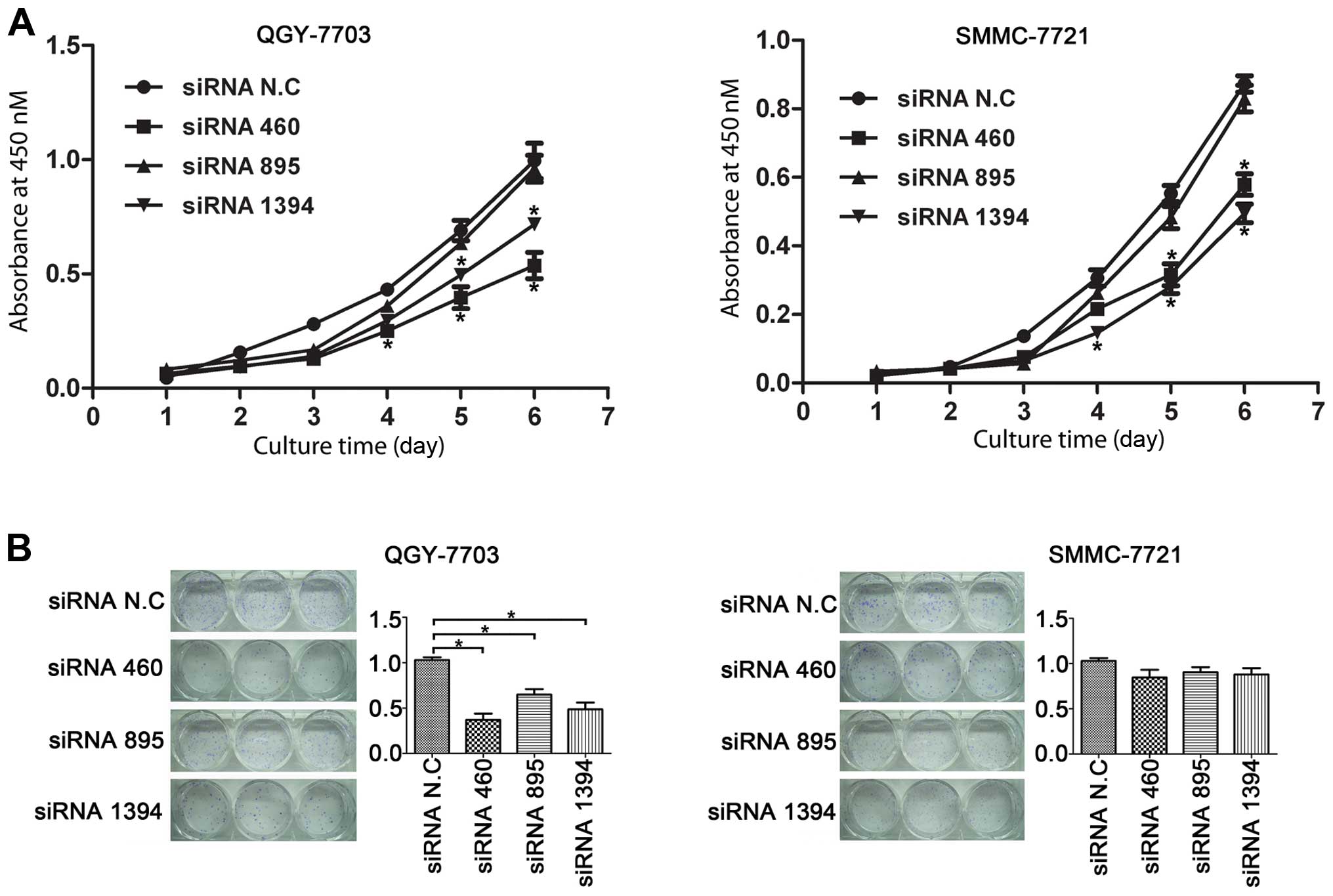
proliferation. Furthermore, our cell growth curve by MTT assay also
confirmed that downregulation of hTDE2 indeed hindered the
growth of tumor cells (Fig. 3A).
Three days after siRNA fragment transfection, growth curve of the
testing cells started to deviate from that of the controls.
We also performed a colony formation test to further
determine the effect of hTDE2 knockdown. With QGY, we found
that the colony numbers decreased significantly when the expression
of hTDE2 was reduced. This effect was not statistically
significant in SMMC cells (Fig.
3B). Our unpublished data further demonstrated that knockdown
of hTDE2 increased apoptosis in QGY, but not in SMMC, which
may explain the above inconsistency within the two lines with the
colony formation test.
In conclusion, we showed that downregulation of
hTDE2 hampered the growth of HCC cells QGY and SMMC, at
least partly, via cell cycle retardation.
Knockdown of hTDE2 upregulates p21 and
SREBP
We then investigated through which signaling pathway
knockdown of TDE2 caused cell cycle arrest. Studies by
Inuzuka et al indicated that the transmembrane Serinc/TDE
proteins might be involved in the biosynthesis of membrane lipids
by facilitating SPT, a rate-limiting enzyme involved in the very
early step of multiple membrane lipid biogenesis (7). Therefore, it is reasonable to
postulate that reduction of TDE2 expression may affect
membrane lipid biogenesis, and this would probably hinder cell
cycle progression. p21 functions as a regulator of cell cycle
progression at G1 and G2, as well as S, and is a transcriptional
target of p53. Moreover, p21 transcription is regulated by factors
other than p53. There are multiple cis-acting elements
residing in the 5′ UTR of the p21 gene, one of which is the
sterol regulatory element (SRE). Combining the biochemical
results from Inuzuka et al and our cell cycle results, as
well as that SREBPs are pivotal regulators of cellular and membrane
lipid homeostasis, we examined whether the expression of SREBPs and
p21 could be affected upon TDE2 knockdown.
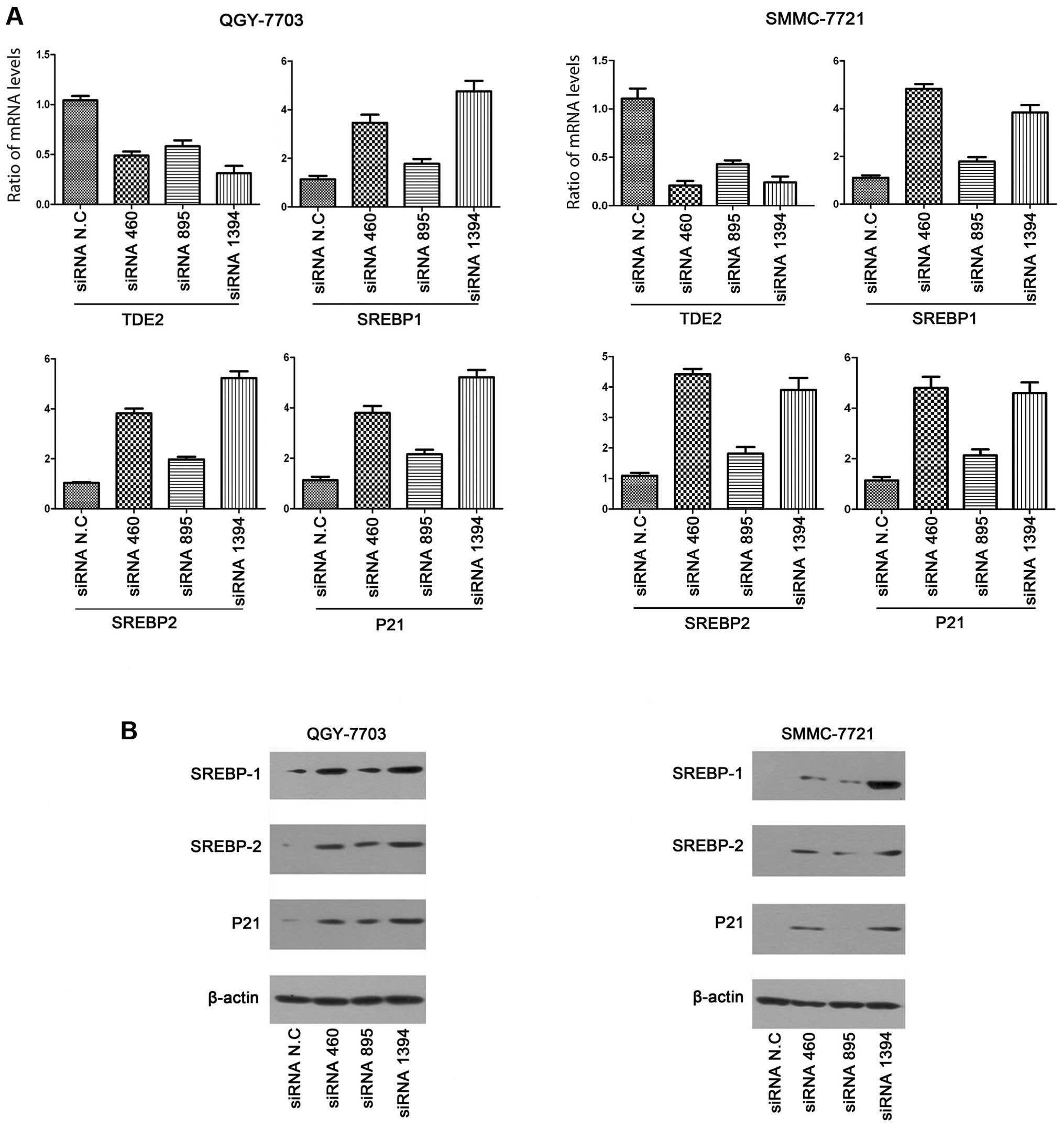
We first transiently transfected the three
TDE2-specific siRNA fragments, as well as the control siRNA,
into both QGY and SMMC cell lines. p21 mRNA levels were
first analyzed with real-time PCR (Fig.
4A). In both cell lines, p21 was indeed upregulated upon
TDE2 knockdown. We also performed western blot analysis to
check p21 expression at the protein level, and similar results were
reached (Fig. 4B). As mentioned
above, we hypothesized the transactivation of p21 here may
be through the SRE element. Therefore, expression of the SRE
binding factors, SREBPs, was also detected with real-time PCR and
western blot analysis. In humans, there are total three SREBP
proteins encoded by two genes, SREBP1 for SREBP-1a and -1c
(resulting from alternative splicing) and SREBP2 for SREBP2
protein. In our experiments, the expression of SREBP-1c (mRNA and
protein) in both QGY and SMMC was very low and undetectable.
Therefore, only results for SREBP-1a and SREBP2 are shown. We found
that SREBPs (-1a and 2) were upregulated at both the mRNA and the
protein level upon TDE2 knockdown, which is consistent with our
speculation.
Moreover, the activation of p21 transcription
here was p53-independent, with p53 protein level in both QGY and
SMMC being unaffected (data not shown). Consistent with this, we
also knocked down TDE2 in H2199, a lung cancer cell line
with p53 deficiency, and upregulation of p21 was detected as
expected (data not shown).
Knockdown of TDE2 transcriptionally
activates p21 promoter via SRE element
To further test whether the transcriptional
activation of p21 is mediated via the SRE element located in
its promoter, we constructed several luciferase reporter plasmids
with p21 promoter sequences (full or partial) inserted in
front of the luciferase gene (as described in Materials and
methods). As shown in Fig. 5A, the
promoter sequence of luciferase gene in p21-2400-Luc harbors both
p53 binding site and SRE element, while that in p21-221-Luc
contains only SRE but not the p53-binding site. For both
reporter plasmids, we also further mutated its SRE element
specifically.
With the dual luciferase report assay, we found that
specific knockdown of TDE2 indeed activated p21 promoter.
Consistent with previous results, this activation was independent
of the p53-binding site. Mutation of SRE sequence completely
abolished this transcriptional activation (Fig. 5B and C). We performed the
experiments in both QGY and SMMC cells, and obtained similar
results.
Discussion
Serinc/TDE proteins belong to a new transmembrane
protein family that is generally tumor differentially expressed. We
found that Serinc1/TDE2 was significantly upregulated in
hepatocarcinoma tissues and cell lines. However, its precise
physiological function remains to be elucidated.
In the present study, we showed that p21 was
upregulated upon knockdown of Serinc1/TDE2 expression, and
this was likely due to SREBP, but not p53. We knocked down
Serinc1/TDE2 expression in H1299 cells (p53-deficient), and
upregulation of p21 was still detected at both the mRNA and
protein levels (unpublished data). Our conclusion was further
supported by results of the dual luciferase report assay. Moreover,
the independence of p53 and SREBP on p21 activation was
previously also shown in the p53-deficient Saos-2 cells (20).
In humans, three isoforms of SREBP (-1a, -1c and 2)
exist. SREBP2 plays a vital role in the regulation of cholesterol
synthesis. While SREBP-1a is involved in the transcription of a
wide scope of genes involved in cholesterol, fatty acids, and
phospholipid synthesis, SREBP-1c has a strong transcriptional
activity for enzyme genes involved in fatty acids and triglycerides
in lipogenic organs (17,18). Other studies showed that both
SREBP-1a and 2 could activate p21 transcription and cause
cell growth inhibition. In addition, SREBP-1a could regulate
p21 transcription by directly binding to SRE
identified in its promoter (20).
Although the physiological significance of SREBPs to p21 activation
still needs to be clarified, it is postulated that fast growing
cells, which require active (membrane) lipid synthesis, may briefly
hold cell growth via activating p21 by SREBPs in case of
lipid deficiency. Results from the biochemical study of
Serinc1/TDE2 (7) and from the
present study support the above postulations.
Furthermore, we exogenously expressed hTDE2 in both
QGY and SMMC and examined its effect on cell cycle progression.
However, compared to the control groups, the experimental groups
did not show significant differences in the cell cycle analysis.
This may be due to the fact that, under normal conditions, cell
cycle progression is limited by factors other than lipid synthesis.
Therefore, the overexpression of Serinc1/TDE2 protein will not show
considerable effects on cell cycle progression. If we culture the
cells in lipid-deficient medium or add drugs to the medium (give
cells pressure), the cells may benefit from over-supply of TDE2.
Indeed, Bossolasco et al (8)
showed that cell apoptosis induced by starvation or drug treatment
could be partially rescued by TDE1 overexpression.
In conclusion, the present study showed that in both
hepatocarcinoma cell lines, downregulation of hSerinc1/hTDE2
clearly arrested cell cycle progression. We also observed an
increase of both SREBPs and p21 expression upon Serinc1/TDE2
knockdown. However, the molecular mechanism underlying this
observation still requires detailed investigation. Moreover,
although we speculated that the cell cycle arrest observed in the
present study might be caused by p21 upregulation via SREBP, we
could not exclude other signaling pathways.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (30700468).
Abbreviations:
|
hTDE2
|
human tumor differentially expressed
2
|
|
serinc
|
serine incorporator
|
|
SREBP
|
sterol regulatory element-binding
protein
|
|
SPT
|
serine palmitoyltransferase
|
References
|
1
|
Lebel M and Mes-Masson AM: Establishment
and characterization of testicular cell lines from MT-PVLT-10
transgenic mice. Exp Cell Res. 213:12–19. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liang P and Pardee AB: Differential
display of eukaryotic messenger RNA by means of the polymerase
chain reaction. Science. 257:967–971. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liang P, Averboukh L and Pardee AB:
Distribution and cloning of eukaryotic mRNAs by means of
differential display: refinements and optimization. Nucleic Acids
Res. 21:3269–3275. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lebel M and Mes-Masson AM: Sequence
analysis of a novel cDNA which is overexpressed in testicular
tumors from polyomavirus large T-antigen transgenic mice. DNA Seq.
5:31–39. 1994.PubMed/NCBI
|
|
5
|
Grossman TR, Luque JM and Nelson N:
Identification of a ubiquitous family of membrane proteins and
their expression in mouse brain. J Exp Biol. 203:447–457.
2000.PubMed/NCBI
|
|
6
|
Aoki S, Su O, Li H, Nishikawa K, Ayukawa
K, et al: Identification of an axotomy-induced glycosylated
protein, AIGP1, possibly involved in cell death triggered by
endoplasmic reticulum-golgi stress. J Neurosci. 22:10751–10760.
2002.PubMed/NCBI
|
|
7
|
Inuzuka M, Hayakawa M and Ingi T: Serinc,
an activity-regulated protein family, incorporates serine into
membrane lipid synthesis. J Biol Chem. 280:35776–35783. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bossolasco M, Veillette F, Bertrand R and
Mes-Masson AM: Human TDE1, a TDE1/TMS family member, inhibits
apoptosis in vitro and stimulates in vivo
tumorigenesis. Oncogene. 25:4549–4558. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Krueger WH, Gonye GE, Madison DL, Murray
KE, Kumar M, et al: TPO1, a member of a novel protein family, is
developmentally regulated in cultured oligodendrocytes. J
Neurochem. 69:1343–1355. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bossolasco M, Lebel M, Lemieux N and
Mes-Masson AM: The human TDE gene homologue: localization to
20q13.1-13.3 and variable expression in human tumor cell lines and
tissue. Mol Carcinog. 26:189–200. 1999.
|
|
11
|
Player A, Gillespie J, Fujii T, Fukuoka J,
Dracheva T, et al: Identification of TDE2 gene and its
expression in non-small cell lung cancer. Int J Cancer.
107:238–243. 2003.
|
|
12
|
Mizuta K and Warner JR: Continued
functioning of the secretory pathway is essential for ribosome
synthesis. Mol Cell Biol. 14:2493–2502. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Athenstaedt K and Daum G: Tgl4p and Tgl5p,
two triacylglycerol lipases of the yeast Saccharomyces
cerevisiae are localized to lipid particles. J Biol Chem.
280:37301–37309. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kurat CF, Natter K, Petschnigg J, Wolinski
H, Scheuringer K, et al: Obese yeast: triglyceride lipolysis is
functionally conserved from mammals to yeast. J Biol Chem.
281:491–500. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kurat CF, Wolinski H, Petschnigg J,
Kaluarachchi S, Andrews B, et al: Cdk1/Cdc28-dependent activation
of the major triacylglycerol lipase Tgl4 in yeast links lipolysis
to cell-cycle progression. Mol Cell. 33:53–63. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Futcher B: Tgl4 Lipase: a big fat target
for cell-cycle entry. Mol Cell. 33:143–144. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shimano H: Sterol regulatory
element-binding proteins (SREBPs): transcriptional regulators of
lipid synthetic genes. Prog Lipid Res. 40:439–452. 2001. View Article : Google Scholar
|
|
18
|
Horton JD, Goldstein JL and Brown MS:
SREBPs: activators of the complete program of cholesterol and fatty
acid synthesis in the liver. J Clin Invest. 109:1125–1131. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nakakuki M, Shimano H, Inoue N, Tamura M,
Matsuzaka T, et al: A transcription factor of lipid synthesis,
sterol regulatory element-binding protein (SREBP)-1a causes
G1 cell-cycle arrest after accumulation of
cyclin-dependent kinase (cdk) inhibitors. FEBS J. 274:4440–4452.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Inoue N, Shimano H, Nakakuki M, Matsuzaka
T, Nakagawa Y, et al: Lipid synthetic transcription factor SREBP-1a
activates p21WAF1/CIP1, a universal cyclin-dependent
kinase inhibitor. Mol Cell Biol. 25:8938–8947. 2005. View Article : Google Scholar : PubMed/NCBI
|