Introduction
Esophageal cancer is the sixth most prevalent cancer
worldwide and ranks fifth as the most common cause of
cancer-related mortality in males. Among all histological subtypes,
esophageal squamous cell carcinoma (ESCC) is the predominant type
in Asia while adenocarcinoma occurs frequently in Western countries
(1). In our center and around the
world, the current management of ESCC which incorporates
chemotherapy with or without radiation into mainstay esophagectomy
has been proven to greatly improve the survival of patients
(2,3). Yet, not all patients can benefit from
such medical strategies, especially those who are intrinsically
resistant or acquire resistance over the course of therapy.
Similar to most solid tumors, ESCC involves the
imbalance of oncogenes and tumor suppressors and deregulation of
tumorigenic pathways. Among these causes, a number of studies have
suggested the role of iron in the progression of esophageal cancers
(4–6). Iron participates in many cellular
processes related to energy metabolism, respiration and DNA
synthesis by being a cofactor or an enzyme component. Notable
involvement includes the functionality of iron-containing enzyme,
ribonucleotide reductase, in catalyzing the conversion of
ribonucleotides to deoxyribonucleotides for DNA synthesis (7). On the molecular level, iron interacts
with iron regulatory proteins to post-transcriptionally modulate
the expression of mRNAs containing iron-responsive elements (IREs)
at 5′- or 3′-untranslated regions, for which these include genes
responsible for iron metabolism and cell cycle regulation (6,8).
Cancer cells on the other hand require an extra iron supply for
sustaining a rapid growth rate. Positive findings have illustrated
the role of iron in tumorigenesis based on the effects of
intracellular iron on modulating the tumorigenic Wnt signaling
pathway in colorectal and leukemic cancer cells (9,10). In
addition, the iron level was also found to influence the
cancer-related MEK/ERK pathway in neuroblastoma, head and neck
squamous carcinoma and hepatocellular carcinoma (11–13).
Apart from its direct effect on individual genes and selected
tumorigenic pathways, surplus iron can also generate reactive
oxygen species, thereby triggering oxidative damage to lipids,
proteins and DNA. This type of DNA damage is detrimental because of
the potentiality of the subsequent advent of gene mutations
inducing cancer. Based on all these cellular effects associated
with iron, a homeostatic level of iron must be tightly maintained
by a team of molecules for import, export and storage of iron (e.g.
transferrin receptor, ferroportin and H-ferritin) (6).
In order to cope with the high iron demand, cancer
cells overexpress a panel of molecules related to iron metabolism
(14–16). Among them, transferrin receptor CD71
was chosen for the present study since it is an important regulator
for controlling cellular iron level and is frequently upregulated
in different cancer types (15,17).
CD71 or transferrin receptor 1 (TFR1) is a 95-kDa homodimeric
transmembrane glycoprotein. It facilitates iron uptake via its
binding with transferrin, the major iron-carrying protein in
systemic circulation (18–20). During the uptake step, the complex
of CD71/transferrin is internalized by endocytosis before the
release of iron in the cellular content. An increase in the
expression of CD71 is readily detected when cells turn malignant.
Immunohistochemical study has associated intense CD71 with active
cell growth (21). Despite the
correlative study of CD71 in other cancer types suggesting its
diagnostic and prognostic value, to our knowledge there is only a
single study on CD71 and ESCC (22). Here, we address this by examining
the expression of CD71 in ESCC tissues before revealing its
clinical association in our population. The tumorigenic properties
of CD71 were studied using transient RNA interference (RNAi)
approach, and the effects of CD71 suppression on cell growth, cell
cycle and the MEK/ERK tumorigenic pathway were also analyzed.
Materials and methods
Clinical specimens
Tumors and adjacent non-tumor tissues from 26 ESCC
patients who had undergone esophagectomy without any prior
chemotherapy and/or radiotherapy at the Department of Surgery,
Queen Mary Hospital, Hong Kong were used in the present study.
Cancer stage was defined according to the TNM system of the
American Joint Committee on Cancer (AJCC) 7th edition, and the
relevant information is summarized in Table I. Consent regarding the use of
clinical specimens for research was obtained from patients, and the
study was approved by the Institutional Review Board of the
University of Hong Kong/Hospital Authority Hong Kong West Cluster
(HKU/HA HKW IRB).
 | Table IClinicopathological parameters of the
study cohort and their correlation with CD71 expression. |
Table I
Clinicopathological parameters of the
study cohort and their correlation with CD71 expression.
| Variables | N | Correlation with
CD71 expression (P-value) |
|---|
| Age (years) |
| ≤65 | 11 | 0.575 |
| >65 | 15 | |
| Gender |
| Male | 15 | 0.8687 |
| Female | 11 | |
| Tumor
differentiation |
| Poor | 8 | 0.6657 |
| Moderate | 16 | |
| Well | 2 | |
| R category |
| R0 | 14 | 0.1793 |
| R1/R2 | 12 | |
| T-stage |
| T1 | 1 | 0.1751 |
| T2 | 3 | |
| T3 | 13 | |
| T4 | 9 | |
| N-stage |
| N0 | 11 | 0.7201 |
| N1 | 8 | |
| N2 | 2 | |
| N3 | 5 | |
| M-stage |
| M0 | 24 | 0.5980 |
| M1 | 2 | |
| Pathological
stage |
| I/II | 9 | 0.4621 |
| III/IV | 17 | |
Cell culture
In-house human cell lines of four ESCCs (HKESC-1,
HKESC-2, HKESC-3 and SLMT-1) and one immortalized non-neoplastic
esophageal epithelial cell line NE-1 were previously developed by
our research team (23–26). Cultures were maintained as
previously described (27,28).
RNAi-mediated CD71 suppression of ESCC
cells
CD71 suppression was achieved by transfecting
HKESC-1, HKESC-2 and SLMT-1 cells using short interfering RNA
(siRNA) against different regions of human CD71. Cultured cells
were plated at 30–50% confluence on 6-well plates overnight before
transfection using 1.5 ml Opti-MEM (Life Technologies, Carlsbad,
CA, USA) with 5 μl Lipofectamine RNAiMAX reagent (Life
Technologies) and 60 pmol of either scramble siRNA (Stealth RNAi
siRNA; Life Technologies; as negative control) or CD71 siRNAs (CD71
Stealth Select RNAi siRNA; Life Technologies). Transfection
efficiency was assessed at both the mRNA and protein levels.
Immunohistochemistry (IHC)
IHC was performed as previously described (28,29).
Paraffin-embedded clinical sections (5 μm) were deparaffinized with
xylene and rehydrated with graded ethanol to water. Antigen
retrieval was performed by heating the rehydrated sections in 10 mM
citric buffer (pH 6.0) for 20 min. After quenching the endogenous
peroxidase activity with hydrogen peroxide and blocking with 3%
bovine serum albumin (BSA), the sections were incubated with 1:50
monoclonal mouse anti-human CD71 (10F11; Abcam, Cambridge, MA, USA)
or 1:100 monoclonal mouse anti-human Ki-67 antigen (MIB-1; Dako,
Carpinteria, CA, USA). Primary antibody binding was detected using
EnVision+ System-HRP labelled polymer anti-mouse (Dako), and the
signals were visualized with Liquid DAB+ substrate chromogen system
(Dako) before counterstaining with hematoxylin. Negative controls
were performed using the same concentration of mouse IgG2b and IgG1
instead of the anti-human CD71 and anti-human Ki-67 antibodies,
respectively. Images of stained sections were captured using a
Nikon DXM1200F digital camera (Nikon, Melville, NY, USA).
Quantitative polymerase chain reaction
(qPCR)
Total RNA from cultured cells and clinical specimens
were extracted using TRIzol reagent (Life Technologies) and
converted to cDNA using SuperScript III First-Strand Synthesis
system for RT-PCR kit (Life Technologies). qPCR was performed as
previously described (28,30) with CD71 primers (forward, 5′-GAG GAG
CCA GGA GAG GAC TT-3′ and reverse, 5′-ACG CCA GAC TTT GCT GAG
TT-3′) and Platinum SYBR-Green qPCR SuperMix-UDG (Life
Technologies). GAPDH was used as an internal control for
normalization. The reactions were performed using the ABI PRISM
7900HT sequence detection system (Life Technologies). In parallel,
semi-quantitative PCR was performed using Platinum Taq DNA
polymerase (Life Technologies), and the endpoint PCR products were
resolved on a 1.5% (w/v) agarose gel.
Flow cytometry for CD71
After trypsinization of cultured ESCC and NE-1
cells, the cells were stained with mouse anti-human CD71 antibody
(BD Biosciences, Franklin Lakes, NJ, USA), followed by the
FITC-labeled secondary antibody. Isotype control was performed in
parallel for each cell line.
Western blotting
Five days post-transfection, the cultured cells were
rinsed with phosphate-buffered saline (PBS) before cell lysis.
Protein lysate (15 μg) from the control and experimental groups was
resolved using 10% SDS-polyacrylamide gel electrophoresis and
subjected to western blot analysis with antibodies specific for
CD71 (BD Biosciences), MEK1/2 (Cell Signaling Technology, Danvers,
MA, USA), phospho-MEK1/2 (Cell Signaling Technology), ERK1/2 (Cell
Signaling Technology) and phospho-ERK1/2 (Cell Signaling
Technology). Signals were visualized with the ECL Plus Western
blotting detection reagent (GE Healthcare Biosciences, Pittsburgh,
PA, USA).
Colony formation assay
Colony formation ability of the control and
transfected cells was examined using a colony formation assay as
previously described (31,32). Briefly, 1,000 cells/well were seeded
on a 6-well plate 24 h after transfection. Colonies were fixed with
4% paraformaldehyde after culturing for 8 days and stained with
0.5% crystal violet for visualization.
Flow cytometry for cell cycle
analysis
Cultured cells were seeded on a 6-well plate at
1×105 cells/well and trypsinized for flow cytometric
analysis 6 days after cell seeding. After washing twice with PBS,
cells were fixed in 70% ethanol on ice before staining with 50
mg/ml propidium iodide (PI) (Sigma-Aldrich, Munich, Germany) and
100 mg/ml PureLink RNase (Life Technologies) in PBS at room
temperature. Stained cells were analyzed using the Cytomics FC500
flow cytometer (Beckman Coulter, Danvers, MA, USA).
Statistical analyses
Data in the bar charts are expressed as means ± SD,
and the significance of difference was calculated by the Student’s
t-test. Unpaired Student’s t-test or one-way ANOVA, where
appropriate, was used to assess the clinical correlation of CD71
expression. Kaplan-Meier method was employed for analyzing
survival. A P-value of <0.05 was considered to indicate a
statistically significant result. All the statistical analyses were
performed using GraphPad Prism 5 for Mac (GraphPad Software, La
Jolla, CA, USA).
Results
Overexpression of CD71 correlates with
advanced ESCC
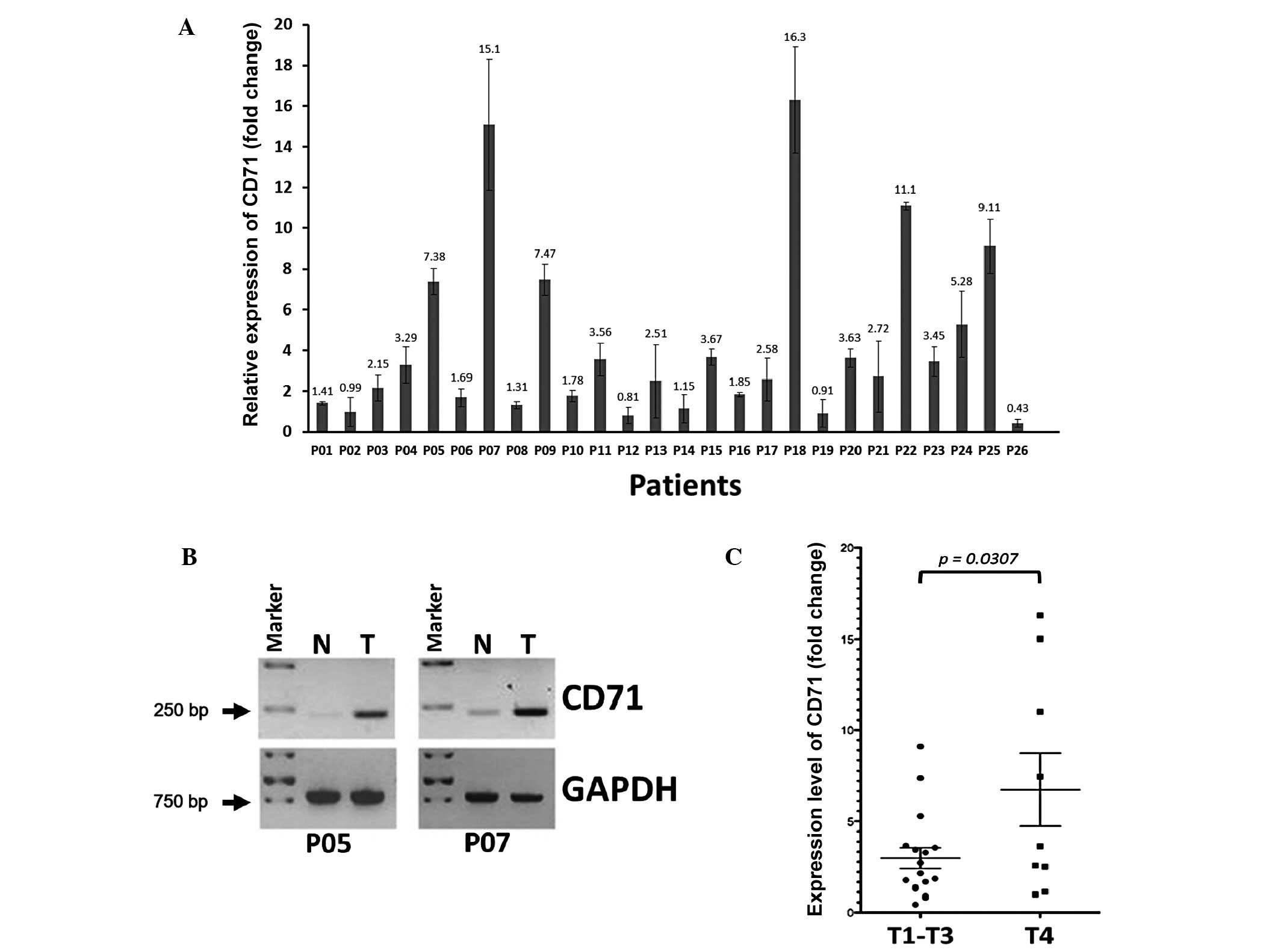
qPCR analysis revealed a >2-fold upregulation of
CD71 transcripts in 61.5% (16/26) of the frozen ESCC tissues when
compared to their adjacent non-tumor tissues. The increase in
expression level ranged from 2.15- to 5-fold in 34.6% (9/26) of
patients, >5- to 10-fold in 15.4% (4/26) of patients, and
>10-fold in 11.5% (3/26) of patients (Fig. 1A). When these tissue pairs were run
for conventional semi-quantitative RT-PCR, an obvious increase in
CD71 expression was found in all tumors exhibiting a >5-fold
mRNA upregulation as shown in Fig.
1A, while only slight expression of CD71 was detected in the
adjacent non-tumor tissues (Fig.
1B). When the fold-change of CD71 expression (tumor/non-tumor)
was correlated with clinicopathological characteristics, we did not
find significant correlation with tumor differentiation, degree of
tumor invasion ‘T-stage’, status of tumor local regional lymph node
involvement ‘N-stage’, the presence of systemic metastases
‘M-stage’, and the ‘R-category’ (Table
I). However, when the CD71 expression level of patients in
T1–T3 stages was compared with that in the T4 stage patients, the
CD71 mRNA expression level was correlated with advanced T4 stage of
the primary tumor (P=0.0307; Fig.
1C), suggesting that patients in advanced T4 stage have tumors
with high expression of the CD71 transcript. In the analysis
concerning survival time after surgery, no significant difference
in survival was noted between patients having tumors with high or
low expression of CD71 when 2.65-fold (this is the median fold
ratio of CD71 upregulation in our patient cohort) of tumor vs.
non-tumor CD71 expression was used as a cut-off (P=0.2861).
However, 3 patients (P07, P18 and P22) with >10-fold
upregulation of the CD71 transcript in tumors, had a reduced
survival time after surgery (P07, 2.95 months; P18, 8.59 months and
P22, 11.01 months) when compared to the average survival rate of
the study cohort (28.46 months).
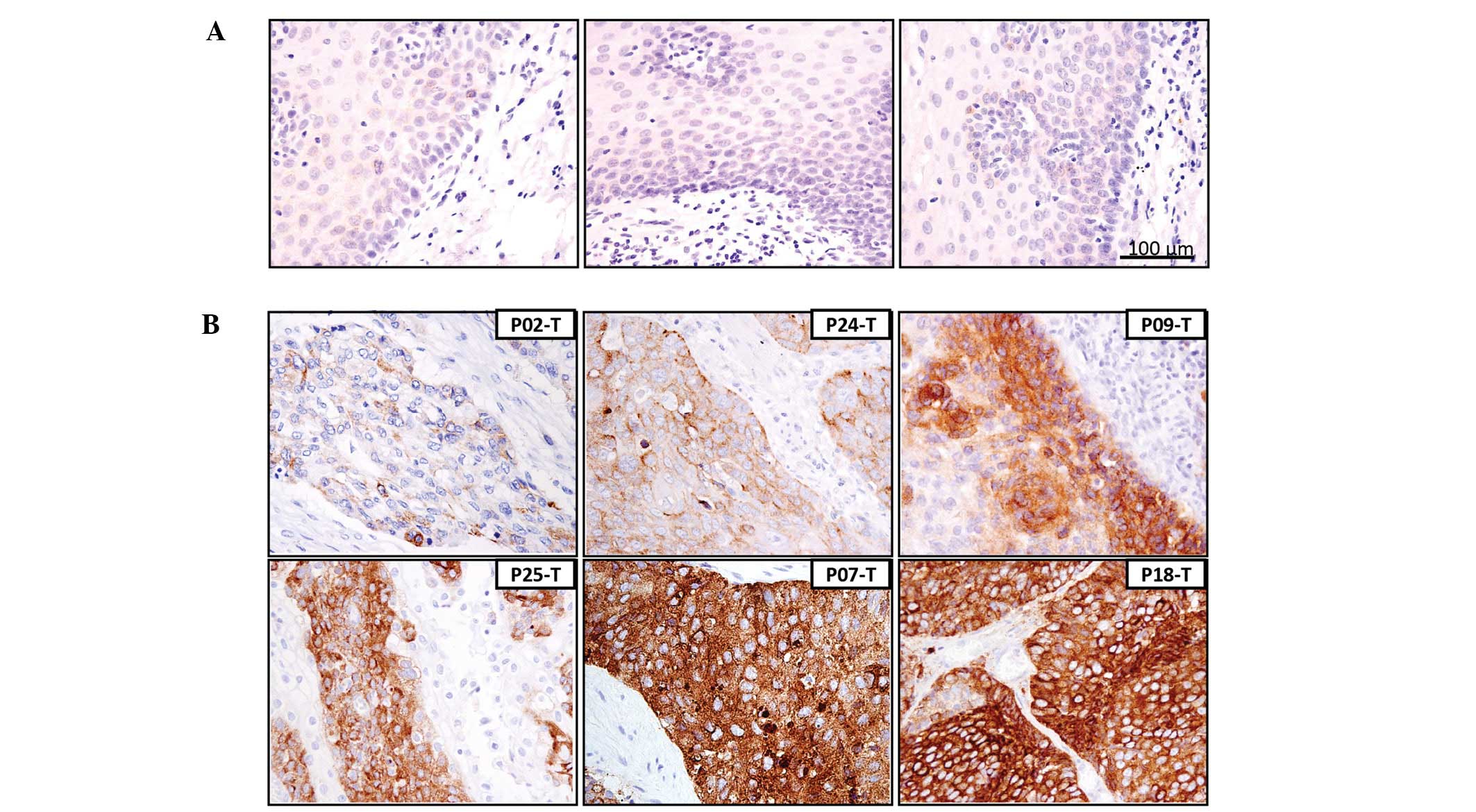
When we examined the protein level of CD71 and its
localization in paraffin-embedded tumor tissues and adjacent
non-tumor tissues using IHC from the same patient cohort, CD71
staining patterns were mainly membranous and cytoplasmic in the
ESCC tissues (Fig. 2). In the
frozen non-tumor tissues and formalin-fixed non-neoplastic
esophageal epithelium cells, a weak CD71 signal was barely detected
using RT-PCR (Fig. 1B) and IHC
(Fig. 2A), respectively. Faded CD71
staining was detected in the tumor tissues from patients with ~1-
to 2-fold CD71 transcript induction as shown in the case of P02
(0.99-fold) (P02-T; Fig. 2B).
Representative IHC images of the cases with a medium to high level
of CD71 in tumors (based on their respective transcript level as in
Fig. 1A) are shown in Fig. 2B. The highest IHC signal intensity
was observed in patients P07 and P18 with >15-fold mRNA
upregulation (P07-T and P18-T), while a medium IHC signal was noted
in patients P24, P09 and P25 with 5.28-, 7.47- and 9.11-fold mRNA
upregulation, respectively (P24-T, P09-T and P25-T). Of note, a
discrepancy between real-time PCR and IHC results existed in 2
cases (P05 and P23). In P05, the paraffin-embedded tumor sections
were weakly stained regardless of the high level of CD71 mRNA
expression, while strong CD71 staining was found in P23 although
the level of CD71 induction was relatively low (data not shown). In
general, the level of mRNA in the frozen tissues matched well with
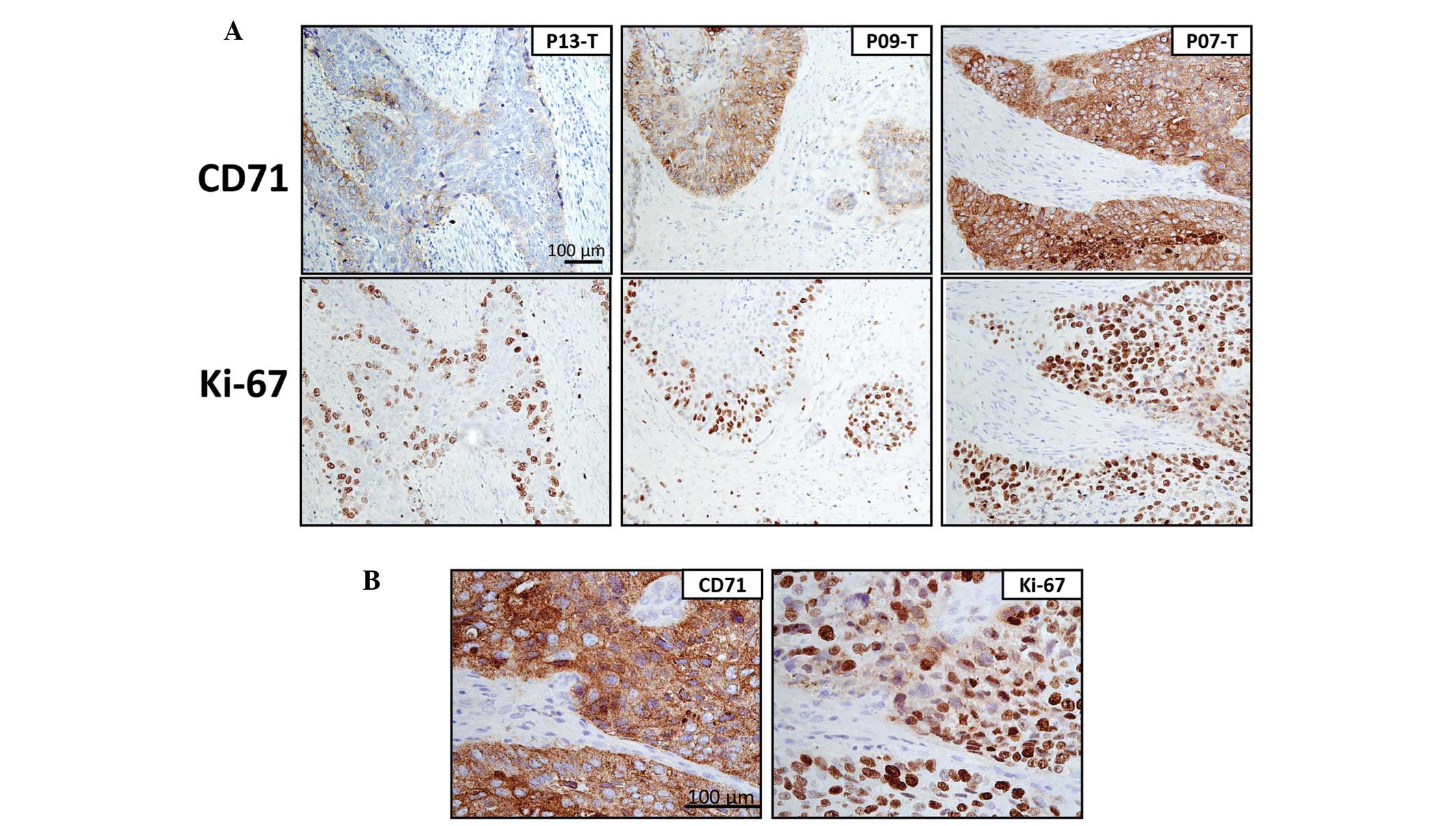
the protein level in the tissue sections. To ascertain whether
overexpression of CD71 is related to active cell proliferation in
ESCC, we stained the paraffin sections for cell proliferation
marker Ki-67. Apart from the presence of Ki-67-positive cells at
the basal epithelial layer of the non-tumor tissues (data not
shown), co-localized staining patterns of CD71 and Ki-67 were
clearly shown in the tumor tissues with CD71 overexpression
(Fig. 3). This observation suggests
a link between CD71 overexpression and active cell
proliferation.
Knockdown of CD71 inhibits tumor
phenotypes of ESCC cells
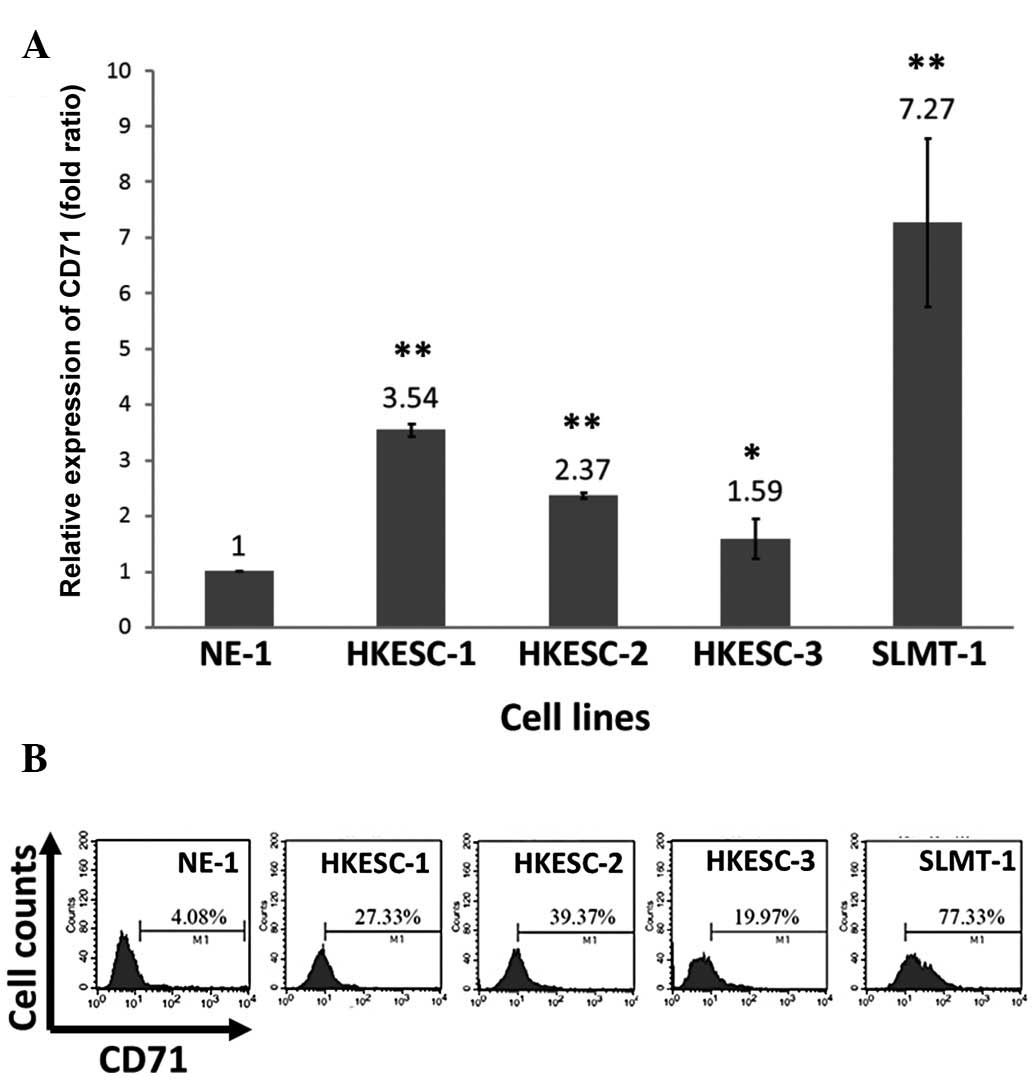
As CD71 was overexpressed in more than half of the
ESCC cases examined, it was expected that higher expression of the
CD71 transcript should be detected in cultured ESCC cell lines when
compared to the non-neoplastic esophageal epithelial cell line
NE-1. Using real-time PCR, a >2-fold upregulation of the CD71
transcript was found in 3 (HKESC-1, HKESC-2 and SLMT-1) out of the
4 examined ESCC cell lines (Fig.
4A). This transcript expression data was highly correlated with
the flow cytometry-derived protein data, which showed the highest
CD71 protein level in the SLMT-1 cells and lowest in the NE-1 cells
(Fig. 4B). A slight deviation in
CD71 mRNA and protein levels were noted for HKESC-1 and HKESC-2
cells (Fig. 4).
To ascertain the role of CD71 in ESCC tumorigenesis,
siRNA-mediated RNAi was used to suppress CD71 in CD71-expressing
HKESC-2 cells before assessing the effects on tumor phenotypes.
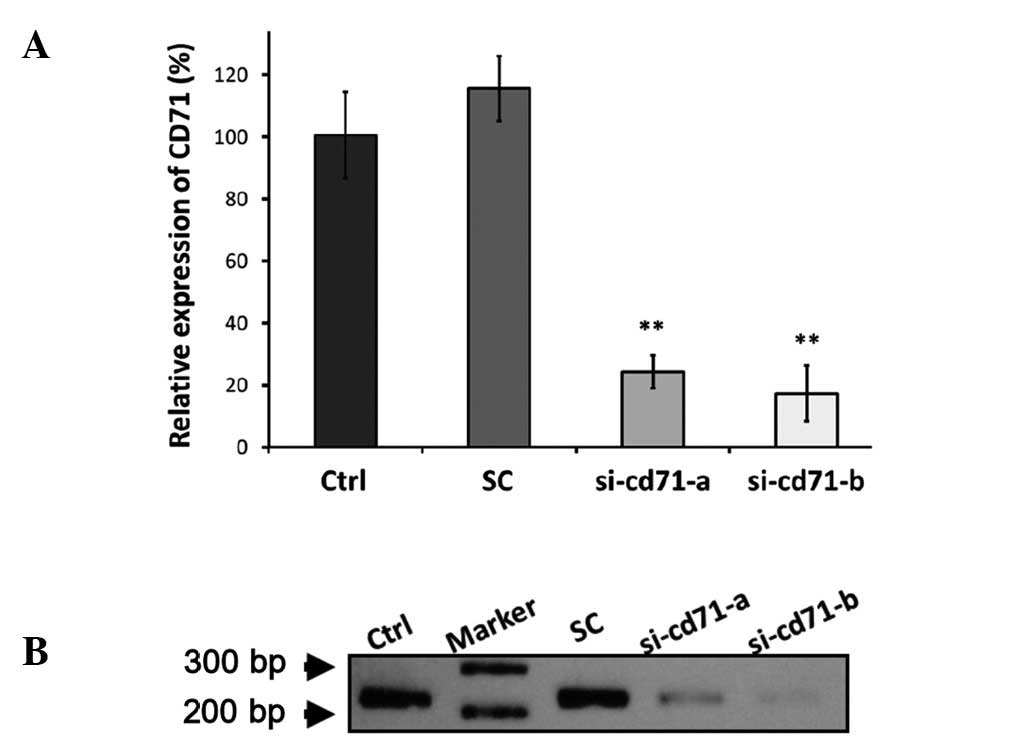
Si-cd71-a and si-cd71-b were two siRNAs targeting different regions
of the CD71 transcript and both caused significant reduction in the
CD71 mRNA level in the HKESC-2 cells by >75% (si-cd71-a,
76.7±5.4%; si-cd71-b, 82.7±9%) compared to the parental line as
revealed using real-time PCR (Fig.
5A). Consistent with the real-time and conventional RT-PCR
results (Fig. 5A and B),
siRNA-mediated suppression of CD71 also reduced the levels of CD71
protein (Fig. 6E). These results
confirmed a better suppression efficiency of si-cd71-b when
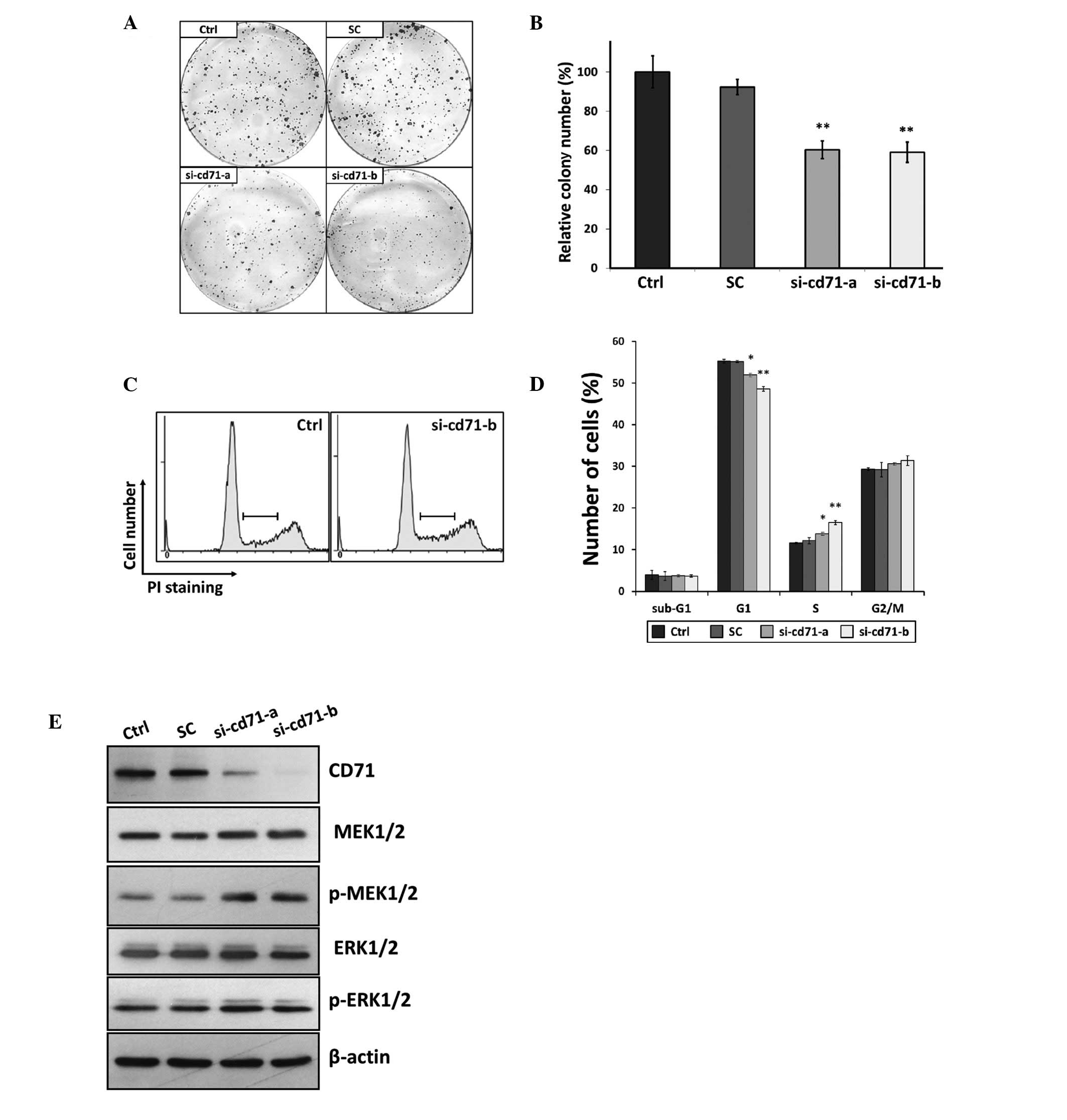
compared with that of si-cd71-a. In the colony formation assay,
suppression of CD71 using si-cd71-a and si-cd71-b significantly
reduced the size and number of colonies to 60.3±4.5 and 59.0±4.5%,
respectively (P<0.01) when compared to the parental cells
(Fig. 6A and B). Analysis of cell
cycle distribution after suppression of CD71 showed cell cycle
arrest at S phase; the percentage of cells in the S phase was
significantly increased from 11.56±0.28% in untreated cells to
13.81±0.36% (P<0.05) and 16.46±0.36% (P<0.01) by si-cd71-a
and si-cd71-b, respectively (Fig. 6C
and D). However, no change in the level of apoptosis was found
between the untreated and CD71-suppressed cells as indicated in the
sub-G1 phase population (Fig. 6D).
Similar results for reducing colony formation ability and arresting
cells in the S phase were obtained following suppression of CD71 by
transfection of HKESC-1 and SLMT-1 with si-cd71-b (data not shown).
No alteration in examined parameters was noted between the
untreated parental cells and cells transfected with the GC
content-matched scramble control siRNA (Fig. 6).
Activation of the MEK/ERK pathway in
CD71-suppressed cells
An obvious increase in the level of phospho-MEK1/2
was found in both the si-cd71-a- and si-cd71-b-transfected cells as
revealed in the immunoblot assay using an antibody against
activated MEK1/2 with phosphorylation at Ser217/221. This
activation of phospho-MEK1/2 upon CD71 suppression was coupled with
an increase in its downstream factor phospho-ERK1/2 (Fig. 6E), suggesting an association between
CD71 suppression and activation of the MEK/ERK pathway in ESCC. In
addition to the effect on the tumorigenic pathway by CD71
suppression, we also attempted to ascertain whether suppression of
CD71 leads to any alteration associated with iron metabolism. Using
real-time PCR, we did not detect any changes in the expression of
several components related to iron metabolism such as iron storage
factor H-ferritin, iron import factor divalent metal transporter 1,
and iron export factor ferroportin after CD71 knockdown (data not
shown). Therefore, the effects on CD71 knockdown-induced cellular
phenotypes were not related to changes in the above iron metabolic
factors.
Discussion
Iron is circulated in the form of iron-bound
transferrin in the body and its cellular uptake is mediated via a
transferrin cell surface receptor named CD71 (15). Similar to surplus iron having a link
with cancer, overexpression of CD71 is frequently observed in
cancers and is correlated with carcinogenesis and several
clinicopathological parameters in tumors originating in brain,
colon, breast and lung (14,16,17,21,33–35).
To date, only a limited number of studies have defined the role of
CD71 in ESCC. In 2006, Wada et al (22) reported expression of CD71 mRNA in
22.4% of paraffin-embedded ESCC tissues using conventional RT-PCR.
Although this study provided valuable information on CD71 in ESCC,
paraffin-embedded tissue is not an ideal source for RNA extraction
due to the long fixation and embedding process that may affect RNA
quality. In the present study, we supplemented this earlier study
by providing real-time PCR results generated from frozen tumors. A
higher percentage of patients (61.5%) was shown to have a
>2-fold increase in CD71 mRNA expression. In support of this
finding, ESCC cell lines derived from tumors resected from
different regions of the esophagus were also noted to have elevated
expression of CD71. Both the clinical and cell line data
unequivocally suggest the importance of this iron transport
receptor in ESCC. Explanation for CD71 overexpression in ESCC was
provided by Wada et al (22), who stated that amplification of
chromosome 3q is one way that leads to this observation. Although
we did not perform a similar experiment in our patient cohort, 2 of
our studied cell lines, HKESC-1 and HKESC-2, with CD71
overexpression indeed harbored chromosomal gain in the 3q region
(23,24), which might account for the induced
expression. Even considering this finding, chromosomal gain appears
not to be the only mechanism for the upregulation of CD71
expression, since SLMT-1 cells that lack chromosomal gain at the 3q
region also exhibit a drastic induction of CD71 (25). It is plausible that SLMT-1 cells
might be subjected to post-transcriptional regulation as a number
of microRNA binding sites can be found in the 2.5 kb (NM_003234.2:
2567–5241) 3′ untranslated region of CD71 mRNA (36–38).
Alternatively, other epigenetic mechanisms such as promoter
demethylation may also take part in the regulation of CD71
expression. Additional regulatory mechanisms leading to CD71
activation in ESCC require further investigation.
CD71 overexpression has been correlated with tumor
stage in several cancer types (15,21),
thereby suggesting CD71 as a marker for tumor diagnosis and
progression (15,17). Here, no significant correlation was
found between expression of the CD71 transcript and several tumor
parameters, such as lymph node involvement and degree of tumor
differentiation. Yet, a high level of CD71 was correlated with
advanced T4 tumor stage. In particular for cases P07, P18 and P22
with >10-fold upregulation of the CD71 transcript, all had T4
stage disease and none of the cases survived longer than 1 year
after surgery. Collectively, this suggests the prognostic value of
CD71 in indicating advanced T4 stage disease. In esophageal cancer,
T-stage defines the depth of tumor invasion into the esophageal
wall and T4 denotes tumors that have infiltrated through the
esophageal adventitia into adjacent structures. High expression of
CD71 in the late tumor stage implicates the involvement of CD71 in
tumor invasion. It is believed that iron overload is one factor
that increases the expression of matrix metalloproteinase, which is
an enzyme facilitating tumor invasion by breaking down
extracellular matrix in head and neck squamous cell carcinoma
(13). At present, no concrete
evidence is available correlating CD71 overexpression and ESCC
invasiveness, which warrants further investigation.
As shown, CD71 is mainly localized at the membrane
and cytoplasm in ESCC tumor cells, which is consistent with reports
of other cancer types (15) and is
in line with the functional roles of CD71 for transferrin binding
and internalization. Most intense CD71 signals were found in 2
cases (P07 and P18), for which their tumors had a >15-fold CD71
mRNA upregulation. In these 2 cases, strong CD71 staining occupied
the entire cytoplasmic region in some cells. In most cases
examined, the staining intensity of CD71 was in agreement with the
mRNA expression data, except for 2 cases (e.g. P05) with weak CD71
staining in the presence of high mRNA expression. This discrepancy
between the mRNA and protein level in a minority subgroup of tumors
might be due to other undefined mechanisms regulating CD71 at the
post-transcriptional or translational level, and the protein
turnover rate might be taken into account for further
investigation.
In the present study, when we concomitantly analyzed
the expression of CD71 and Ki-67 using adjacent tissue sections
less than 30 μm apart, both of these stains were found in similar
localizations. In non-neoplastic esophageal squamous tissue,
Ki-67-stained cells were restricted to the proliferating layer of
the esophageal epithelium weakly expressing CD71. While in tumor
tissues, Ki-67 staining increased with CD71 expression. As Ki-67 is
a proliferative marker, the IHC results indicate that CD71 may
contribute to rapid cell growth in ESCC. Having demonstrated this,
we next tested whether suppression of CD71 inhibits tumor
phenotypes of ESCC cells. The HKESC-2 ESCC cell line was chosen for
siRNA transfection due to its high competence of transfection based
on our prior experience. Although cancer cells might evolve other
mechanisms to compensate for the loss of CD71 such as production of
other iron importers, application of siRNA against CD71 was
sufficient to inhibit ESCC cell growth as revealed by the small
size and reduced number of colonies formed as detected in the
colony formation assay. Cell cycle analysis further showed that
treatment of CD71 siRNA induced cell accumulation in the S phase
coupled with cell depletion in the G1 phase. Unexpectedly, this
cell cycle arrest resulted in no change in the percentage of
apoptotic cells. From the CD71 knockdown experiments and the fact
that iron is an important element for carcinogenesis, targeting
CD71 appears to bear therapeutic potential in CD71-expressing
tumors. Based on this concept, preclinical cancer therapeutic
research has focused on reducing the systemic iron level by
chelating agents. Recently Ford et al (39) reported the possible application of
an iron chelator deferasirox in inhibiting esophageal cancer
growth. Indeed, we provided evidence to support the targeting of
the iron importer CD71 as a way to achieve antitumorigenesis. To
the best of our knowledge, this is the first report detailing the
growth inhibiting effect of targeting CD71 on ESCC tumorigenesis.
This study provides valuable insight into the potential therapeutic
value of CD71 in ESCC. CD71 is an important candidate for study
regarding its therapeutic potential for other cancer types. An
investigation focusing on cancer immunotherapy has revealed the
antiproliferative effect of the anti-CD71 antibody against lymphoma
cells (40). In view of these
findings, targeting iron-related molecules such as CD71 seems to be
a plausible way to counteract tumorigenesis of different
origins.
Previous studies have provided clues on how the iron
level can modulate tumorigenesis. In colon cancer cells, elevation
of intracellular iron enhances tumorigenic Wnt signaling as
indicated by increased transcription of its downstream targets of
this pathway (9). In contrary,
application of iron chelating agents to reduce the iron level
abrogated Wnt signaling and inhibited cell growth in colorectal and
leukemic cells (10). Moreover, the
MEK/ERK pathway is activated by iron in PC12 neuroblastoma cells
and neck squamous cell carcinoma cells (11,13).
Other iron-sensitive molecules, such as cell division cycle 14A
(cdc14A), are cell cycle regulators under IRE control (8). Given the fact that CD71 is responsible
for iron binding and internalization, it is reasonable to believe
that the mentioned signaling pathways and molecules might be
affected upon CD71 manipulation in ESCC. However, no deviation in
the level of Wnt pathway downstream targets and cell cycle
regulator cdc14A was noted after CD71 suppression in the present
study (data not shown). An unexpected increase in the
phospho-MEK1/2 coupled with elevation of its downstream target
phosphor-ERK1/2 was detected when CD71 was suppressed, indicating
that knockdown of CD71 activates this specific pathway. The MEK/ERK
pathway exerts its effect through phosphorylation of many
downstream targets related to cell growth and apoptosis, and it is
generally believed that activation of the MEK/ERK signaling pathway
promotes cell proliferation and malignant transformation. In line
with our data, this finding fails to explain the entire scenario.
MEK/ERK signaling is composed of a complicated network with other
signaling molecules, such as those belonging to the PI3K/PTEN/AKT
pathway and p53 (41,42). Moreover, this pathway is also
capable of activating members of the kinases, transcription factors
and apoptotic regulators. Emerging evidence has suggested that
activation of the MEK/ERK pathway leads to cell growth inhibition.
In a colon cancer cell line, activation of the MEK/ERK pathway
resulted in p14ARF-induced cell growth arrest (41). Benzyl isothiocyanate-induced cell
growth arrest and apoptosis were mediated by ERK activation in
human pancreatic cancer (43). The
decisive role of the MEK/ERK pathway in ESCC is still unclear. With
the presented results as a background, further study of the
molecular carcinogenesis of ESCC should focus on the link between
CD71 suppression and MEK/ERK activation.
Based on the results derived from the present study,
we demonstrated the overexpression of CD71 in ESCC and that
suppression of CD71 in cultured ESCC cells leads to reduced
tumorigenic properties. The results presented in the present study
also indicate the therapeutic potential of targeting CD71 in ESCC,
which may contribute to the development of novel anticancer
agents.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Law S, Kwong DL, Kwok KF, et al:
Improvement in treatment results and long-term survival of patients
with esophageal cancer: impact of chemoradiation and change in
treatment strategy. Ann Surg. 238:339–348. 2003.PubMed/NCBI
|
|
3
|
Law S and Wong J: The current management
of esophageal cancer. Adv Surg. 41:93–119. 2007. View Article : Google Scholar
|
|
4
|
Chen X, Yang G, Ding WY, Bondoc F, Curtis
SK and Yang CS: An esophagogastroduodenal anastomosis model for
esophageal adenocarcinogenesis in rats and enhancement by iron
overload. Carcinogenesis. 20:1801–1808. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cross AJ, Freedman ND, Ren J, et al: Meat
consumption and risk of esophageal and gastric cancer in a large
prospective study. Am J Gastroenterol. 106:432–442. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boult J, Roberts K, Brookes MJ, et al:
Overexpression of cellular iron import proteins is associated with
malignant progression of esophageal adenocarcinoma. Clin Cancer
Res. 14:379–387. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Desoize B: Metals and metal compounds in
cancer treatment. Anticancer Res. 24:1529–1544. 2004.PubMed/NCBI
|
|
8
|
Sanchez M, Galy B, Dandekar T, et al: Iron
regulation and the cell cycle: identification of an iron-responsive
element in the 3′-untranslated region of human cell division cycle
14A mRNA by a refined microarray-based screening strategy. J Biol
Chem. 281:22865–22874. 2006.
|
|
9
|
Brookes MJ, Boult J, Roberts K, et al: A
role for iron in Wnt signalling. Oncogene. 27:966–975. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song S, Christova T, Perusini S, et al:
Wnt inhibitor screen reveals iron dependence of β-catenin signaling
in cancers. Cancer Res. 71:7628–7639. 2011.PubMed/NCBI
|
|
11
|
Munoz P, Zavala G, Castillo K, Aguirre P,
Hidalgo C and Nunez MT: Effect of iron on the activation of the
MAPK/ERK pathway in PC12 neuroblastoma cells. Biol Res. 39:189–190.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu Y and Richardson DR: Cellular iron
depletion stimulates the JNK and p38 MAPK signaling transduction
pathways, dissociation of ASK1-thioredoxin, and activation of ASK1.
J Biol Chem. 286:15413–15427. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kaomongkolgit R, Cheepsunthorn P, Pavasant
P and Sanchavanakit N: Iron increases MMP-9 expression through
activation of AP-1 via ERK/Akt pathway in human head and neck
squamous carcinoma cells. Oral Oncol. 44:587–594. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kukulj S, Jaganjac M, Boranic M, Krizanac
S, Santic Z and Poljak-Blazi M: Altered iron metabolism,
inflammation, transferrin receptors, and ferritin expression in
non-small-cell lung cancer. Med Oncol. 27:268–277. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Habashy HO, Powe DG, Staka CM, et al:
Transferrin receptor (CD71) is a marker of poor prognosis in breast
cancer and can predict response to tamoxifen. Breast Cancer Res
Treat. 119:283–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang XP, Elliott RL and Head JF:
Manipulation of iron transporter genes results in the suppression
of human and mouse mammary adenocarcinomas. Anticancer Res.
30:759–765. 2010.PubMed/NCBI
|
|
17
|
Magro G, Cataldo I, Amico P, et al:
Aberrant expression of TfR1/CD71 in thyroid carcinomas identifies a
novel potential diagnostic marker and therapeutic target. Thyroid.
21:267–277. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Testa U, Pelosi E and Peschle C: The
transferrin receptor. Crit Rev Oncog. 4:241–276. 1993.
|
|
19
|
Aisen P: Transferrin receptor 1. Int J
Biochem Cell Biol. 36:2137–2143. 2004. View Article : Google Scholar
|
|
20
|
Sargent PJ, Farnaud S and Evans RW:
Structure/function overview of proteins involved in iron storage
and transport. Curr Med Chem. 12:2683–2693. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Prutki M, Poljak-Blazi M, Jakopovic M,
Tomas D, Stipancic I and Zarkovic N: Altered iron metabolism,
transferrin receptor 1 and ferritin in patients with colon cancer.
Cancer Lett. 238:188–196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wada S, Noguchi T, Takeno S and Kawahara
K: PIK3CA and TFRC located in 3q are new prognostic factors in
esophageal squamous cell carcinoma. Ann Surg Oncol. 13:961–966.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu Y, Lam KY, Wan TS, et al: Establishment
and characterization of HKESC-1, a new cancer cell line from human
esophageal squamous cell carcinoma. Cancer Genet Cytogenet.
118:112–120. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu YC, Lam KY, Law SY, et al:
Establishment, characterization, karyotyping, and comparative
genomic hybridization analysis of HKESC-2 and HKESC-3: two newly
established human esophageal squamous cell carcinoma cell lines.
Cancer Genet Cytogenet. 135:120–127. 2002. View Article : Google Scholar
|
|
25
|
Tang JC, Wan TS, Wong N, et al:
Establishment and characterization of a new xenograft-derived human
esophageal squamous cell carcinoma cell line SLMT-1 of Chinese
origin. Cancer Genet Cytogenet. 124:36–41. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang H, Jin Y, Chen X, et al: Cytogenetic
aberrations in immortalization of esophageal epithelial cells.
Cancer Genet Cytogenet. 165:25–35. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hui MK, Chan KW, Luk JM, et al:
Cytoplasmic Forkhead Box M1 (FoxM1) in esophageal squamous cell
carcinoma significantly correlates with pathological disease stage.
World J Surg. 36:90–97. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hui MK, Lai KK, Chan KW, et al: Prognostic
significance of phosphorylated RON in esophageal squamous cell
carcinoma. Med Oncol. 29:1699–1706. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee NP, Leung KW, Wo JY, Tam PC, Yeung WS
and Luk JM: Blockage of testicular connexins induced apoptosis in
rat seminiferous epithelium. Apoptosis. 11:1215–1229. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee NP, Tsang FH, Shek FH, et al:
Prognostic significance and therapeutic potential of eukaryotic
translation initiation factor 5A (eIF5A) in hepatocellular
carcinoma. Int J Cancer. 127:968–976. 2010.PubMed/NCBI
|
|
31
|
Chan KT and Lung ML: Mutant p53 expression
enhances drug resistance in a hepatocellular carcinoma cell line.
Cancer Chemother Pharmacol. 53:519–526. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu LX, Lee NP, Chan VW, et al: Targeting
cadherin-17 inactivates Wnt signaling and inhibits tumor growth in
liver carcinoma. Hepatology. 50:1453–1463. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hanninen MM, Haapasalo J, Haapasalo H, et
al: Expression of iron-related genes in human brain and brain
tumors. BMC Neurosci. 10:362009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ryschich E, Huszty G, Knaebel HP, Hartel
M, Buchler MW and Schmidt J: Transferrin receptor is a marker of
malignant phenotype in human pancreatic cancer and in
neuroendocrine carcinoma of the pancreas. Eur J Cancer.
40:1418–1422. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sciot R, Paterson AC, van Eyken P, Callea
F, Kew MC and Desmet VJ: Transferrin receptor expression in human
hepatocellular carcinoma: an immunohistochemical study of 34 cases.
Histopathology. 12:53–63. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cmejla R, Petrak J and Cmejlova J: A novel
iron responsive element in the 3′ UTR of human MRCKα. Biochem
Biophys Res Commun. 341:158–166. 2006.
|
|
37
|
Cmejla R, Ptackova P, Petrak J, et al:
Human MRCKα is regulated by cellular iron levels and interferes
with transferrin iron uptake. Biochem Biophys Res Commun.
395:163–167. 2010.
|
|
38
|
Schaar DG, Medina DJ, Moore DF, Strair RK
and Ting Y: miR-320 targets transferrin receptor 1 (CD71) and
inhibits cell proliferation. Exp Hematol. 37:245–255. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ford SJ, Obeidy P, Lovejoy DB, et al:
Deferasirox (ICL670A) effectively inhibits oesophageal cancer
growth in vitro and in vivo. Br J Pharmacol.
168:1316–1328. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Loisel S, Andre PA, Golay J, et al:
Antitumour effects of single or combined monoclonal antibodies
directed against membrane antigens expressed by human B cells
leukaemia. Mol Cancer. 10:422011. View Article : Google Scholar
|
|
41
|
Du H, Yao W, Fang M and Wu D: ARF triggers
cell G1 arrest by a P53 independent ERK pathway. Mol Cell Biochem.
357:415–422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
McCubrey JA, Steelman LS, Chappell WH, et
al: Roles of the Raf/MEK/ERK pathway in cell growth, malignant
transformation and drug resistance. Biochim Biophys Acta.
1773:1263–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sahu RP, Zhang R, Batra S, Shi Y and
Srivastava SK: Benzyl isothiocyanate-mediated generation of
reactive oxygen species causes cell cycle arrest and induces
apoptosis via activation of MAPK in human pancreatic cancer cells.
Carcinogenesis. 30:1744–1753. 2009. View Article : Google Scholar
|