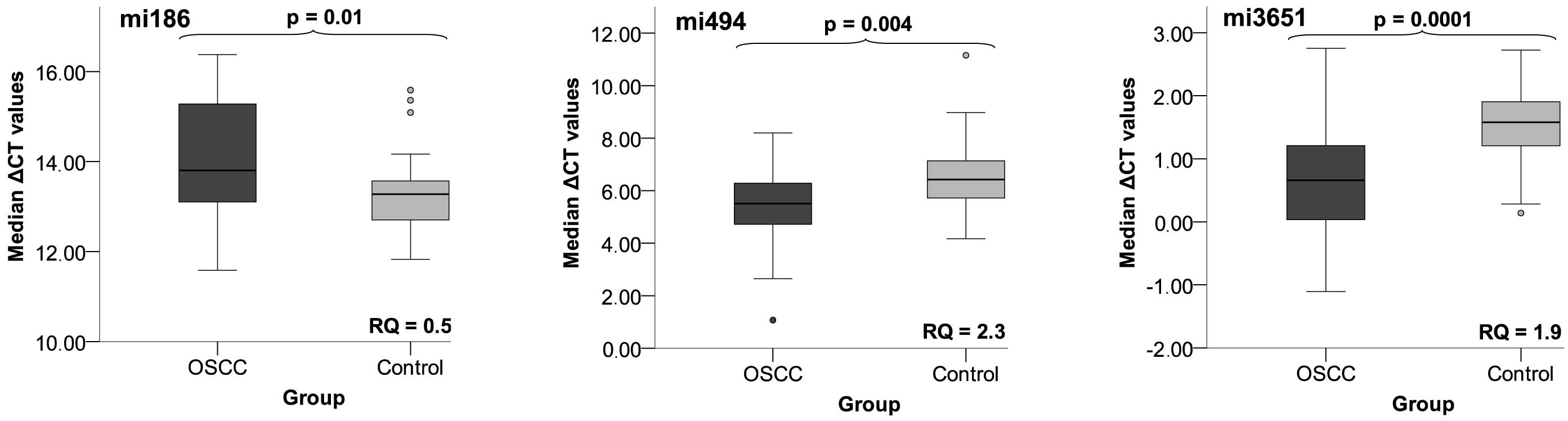
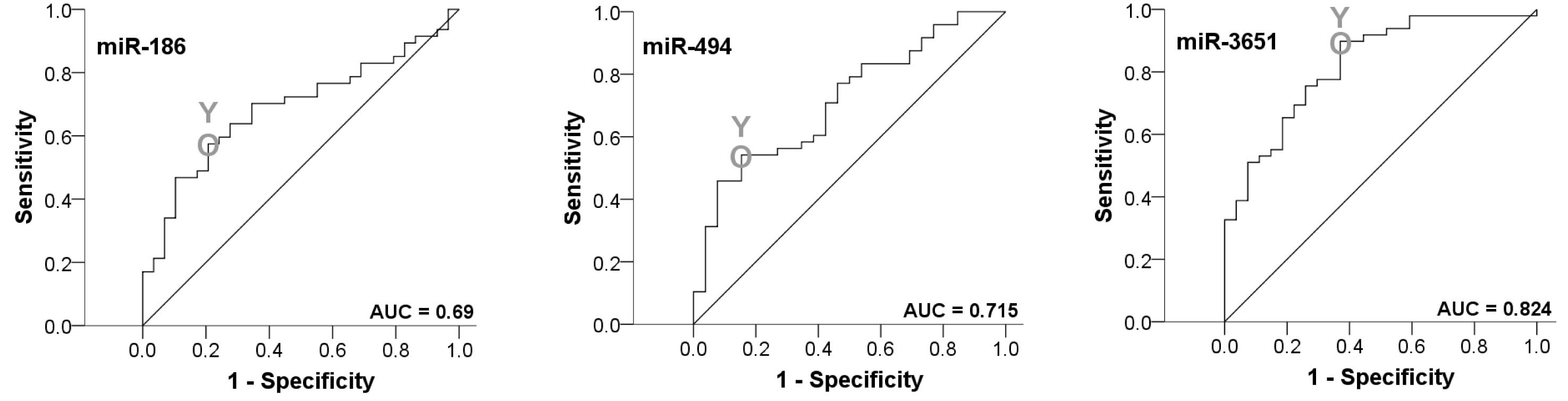
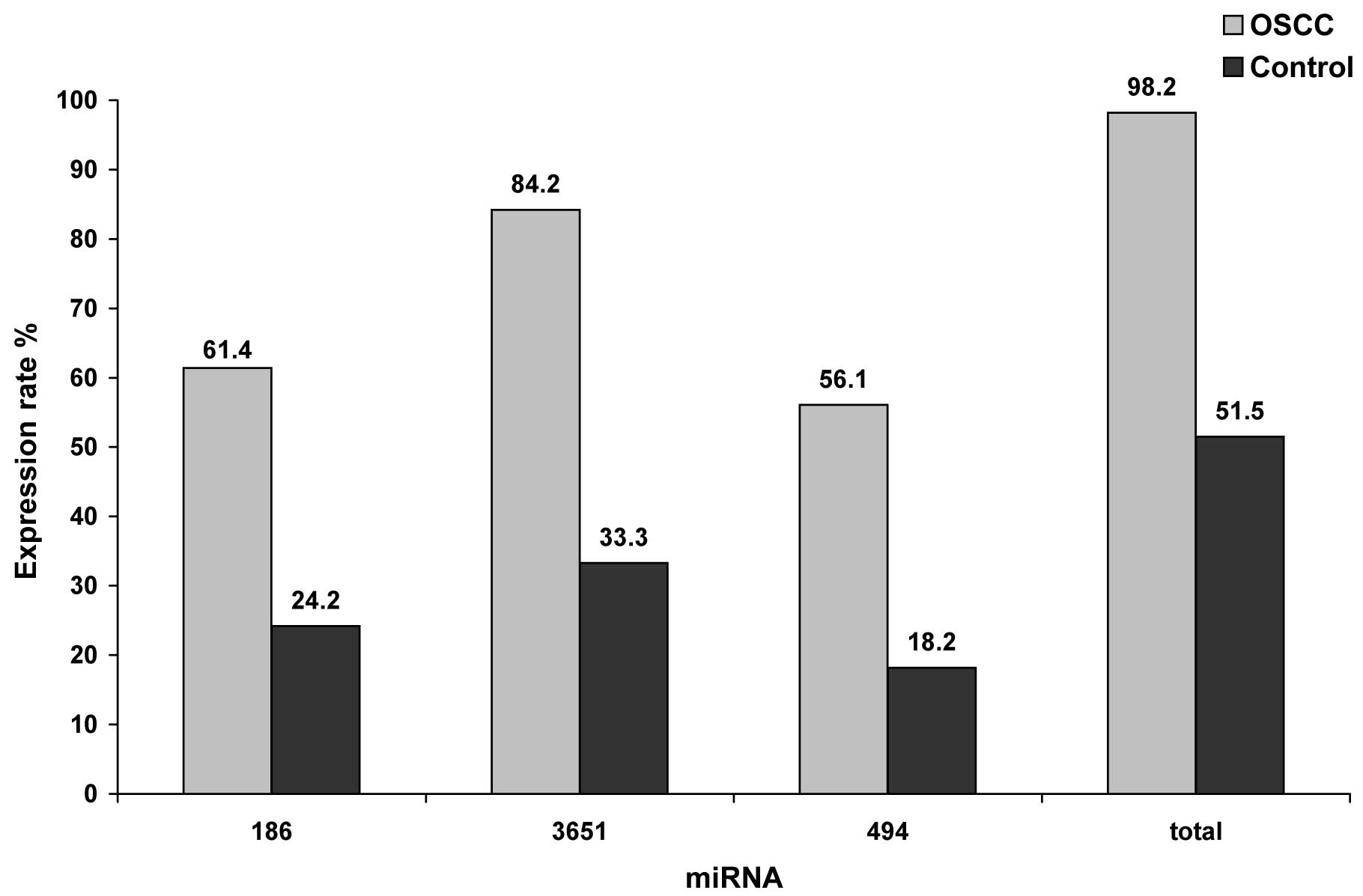
|
1
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gomes CC, de Sousa SF and Gomez RS:
microRNAs: small molecules with a potential role in oral squamous
cell carcinoma. Curr Pharm Des. 19:1285–1291. 2013.PubMed/NCBI
|
|
3
|
Gomes CC and Gomez RS: MicroRNA and oral
cancer: future perspectives. Oral Oncol. 44:910–914. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu BH, Xiong XP, Jia J and Zhang WF:
MicroRNAs: new actors in the oral cancer scene. Oral Oncol.
47:314–319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lajer CB, Nielsen FC, Friis-Hansen L, et
al: Different miRNA signatures of oral and pharyngeal squamous cell
carcinomas: a prospective translational study. Br J Cancer.
104:830–840. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Scapoli L, Palmieri A, Lo Muzio L, et al:
MicroRNA expression profiling of oral carcinoma identifies new
markers of tumor progression. Int J Immunopathol Pharmacol.
23:1229–1234. 2010.PubMed/NCBI
|
|
9
|
Chang SS, Jiang WW, Smith I, et al:
MicroRNA alterations in head and neck squamous cell carcinoma. Int
J Cancer. 123:2791–2797. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Soga D, Yoshiba S, Shiogama S, et al:
microRNA expression profiles in oral squamous cell carcinoma. Oncol
Rep. 30:579–583. 2013.PubMed/NCBI
|
|
12
|
Mitchell PS, Parkin RK, Kroh EM, et al:
Circulating microRNAs as stable blood-based markers for cancer
detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen X, Ba Y, Ma L, et al:
Characterization of microRNAs in serum: a novel class of biomarkers
for diagnosis of cancer and other diseases. Cell Res. 18:997–1006.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Häusler SF, Keller A, Chandran PA, et al:
Whole blood-derived miRNA profiles as potential new tools for
ovarian cancer screening. Br J Cancer. 103:693–700. 2010.PubMed/NCBI
|
|
15
|
Heegaard NH, Schetter AJ, Welsh JA, et al:
Circulating micro-RNA expression profiles in early stage nonsmall
cell lung cancer. Int J Cancer. 130:1378–1386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kosaka N, Iguchi H and Ochiya T:
Circulating microRNA in body fluid: a new potential biomarker for
cancer diagnosis and prognosis. Cancer Sci. 101:2087–2092. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin SC, Liu CJ, Lin JA, et al:
miR-24 up-regulation in oral carcinoma: positive association
from clinical and in vitro analysis. Oral Oncol. 46:204–208. 2010.
View Article : Google Scholar
|
|
18
|
Liu CJ, Kao SY, Tu HF, et al: Increase of
microRNA miR-31 level in plasma could be a potential marker
of oral cancer. Oral Dis. 16:360–364. 2010.
|
|
19
|
Maclellan SA, Lawson J, Baik J, et al:
Differential expression of miRNAs in the serum of patients with
high-risk oral lesions. Cancer Med. 1:268–274. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wong TS, Liu XB, Wong BY, et al: Mature
miR-184 as potential oncogenic microRNA of squamous cell carcinoma
of tongue. Clin Cancer Res. 14:2588–2592. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wittmann J and Jäck HM: Serum microRNAs as
powerful cancer biomarkers. Biochim Biophys Acta. 1806:200–207.
2010.PubMed/NCBI
|
|
22
|
Liu CJ, Lin SC, Yang CC, et al: Exploiting
salivary miR-31 as a clinical biomarker of oral squamous
cell carcinoma. Head Neck. 34:219–224. 2012.PubMed/NCBI
|
|
23
|
Yang CC, Hung PS, Wang PW, et al:
miR-181 as a putative biomarker for lymph-node metastasis of
oral squamous cell carcinoma. J Oral Pathol Med. 40:397–404. 2011.
View Article : Google Scholar
|
|
24
|
Kosaka N, Iguchi H, Yoshioka Y, et al:
Competitive interactions of cancer cells and normal cells via
secretory microRNAs. J Biol Chem. 287:1397–1405. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Heneghan HM, Miller N, Lowery AJ, et al:
Circulating microRNAs as novel minimally invasive biomarkers for
breast cancer. Ann Surg. 251:499–505. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Patel SG and Shah JP: TNM staging of
cancers of the head and neck: striving for uniformity among
diversity. CA Cancer J Clin. 55:242–258. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Benjamini Y, Drai D, Elmer G, et al:
Controlling the false discovery rate in behavior genetics research.
Behav Brain Res. 125:279–284. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fluss R, Faraggi D and Reiser B:
Estimation of the Youden Index and its associated cutoff point.
Biom J. 47:458–472. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gorenchtein M, Poh CF, Saini R and Garnis
C: MicroRNAs in an oral cancer context - from basic biology to
clinical utility. J Dent Res. 91:440–446. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li H, Yin C, Zhang B, et al: PTTG1
promotes migration and invasion of human non-small cell lung cancer
cells and is modulated by miR-186. Carcinogenesis. 34:2145–2155.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao BS, Liu SG, Wang TY, et al: Screening
of microRNA in patients with esophageal cancer at same tumor node
metastasis stage with different prognoses. Asian Pac J Cancer Prev.
14:139–143. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cai J, Wu J, Zhang H, et al: miR-186
downregulation correlates with poor survival in lung
adenocarcinoma, where it interferes with cell-cycle regulation.
Cancer Res. 73:756–766. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim SY, Lee YH and Bae YS: MiR-186,
miR-216b, miR-337-3p, and miR-760 cooperatively induce cellular
senescence by targeting alpha subunit of protein kinase CKII in
human colorectal cancer cells. Biochem Biophys Res Commun.
429:173–179. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Myatt SS, Wang J, Monteiro LJ, et al:
Definition of microRNAs that repress expression of the tumor
suppressor gene FOXO1 in endometrial cancer. Cancer Res.
70:367–377. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Diakos C, Zhong S, Xiao Y, et al: TEL-AML1
regulation of survivin and apoptosis via miRNA-494 and miRNA-320a.
Blood. 116:4885–4893. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yamanaka S, Campbell NR, An F, et al:
Coordinated effects of microRNA-494 induce G2/M arrest
in human cholangiocarcinoma. Cell Cycle. 11:2729–2738. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ohdaira H, Sekiguchi M, Miyata K and
Yoshida K: MicroRNA-494 suppresses cell proliferation and induces
senescence in A549 lung cancer cells. Cell Prolif. 45:32–38. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Duttagupta R, Jiang R, Gollub J, et al:
Impact of cellular miRNAs on circulating miRNA biomarker
signatures. PLoS One. 6:e207692011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Heneghan HM, Miller N and Kerin MJ:
Circulating microRNAs: promising breast cancer biomarkers. Breast
Cancer Res. 13:402–403. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Friedman EB, Shang S, de Miera EV, et al:
Serum microRNAs as biomarkers for recurrence in melanoma. J Transl
Med. 10:1552012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schwarzenbach H, Hoon DS and Pantel K:
Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev
Cancer. 11:426–437. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zen K and Zhang CY: Circulating microRNAs:
a novel class of biomarkers to diagnose and monitor human cancers.
Med Res Rev. 32:326–348. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Osaki M, Takeshita F and Ochiya T:
MicroRNAs as biomarkers and therapeutic drugs in human cancer.
Biomarkers. 13:658–670. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tong AW and Nemunaitis J: Modulation of
miRNA activity in human cancer: a new paradigm for cancer gene
therapy? Cancer Gene Ther. 15:341–355. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang B and Farwell MA: microRNAs: a new
emerging class of players for disease diagnostics and gene therapy.
J Cell Mol Med. 12:3–21. 2008. View Article : Google Scholar : PubMed/NCBI
|