Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of
the most difficult-to-treat types of cancer and has an extremely
poor prognosis (1,2). The only effective treatment for PDAC
is surgical resection. Research and development of chemotherapy and
immunotherapy are progressing, but their clinical utility remains
insufficient (1,2). To classify the poor prognosis group of
PDAC, an immunohistological analysis using paraffin sections from
surgical specimens was developed as a new molecular biological
method (3,4). Endothelin B receptor (ETBR) is a
glycoprotein that consists of 442 amino acid residues and
penetrates through the cell membrane seven times (5). It has been reported that ETBR of tumor
endothelial cells prevents antitumor immunity. Specifically, ETBR
overexpression prevents the endothelial barrier to T cell homing to
tumors in human ovarian cancer (6).
It was previously reported in our facilities that the existence of
tumor-infiltrating T lymphocytes in PDAC tissue served as a
significant factor related to good prognosis (3). The role of the endothelin axis in a
cancer domain other than ETAR of prostate cancer has rarely been
studied (7,8). Several studies have examined breast
cancer (9), melanoma (10,11),
glioblastoma (12), non-small cell
lung (13), colon (14–16),
gastric (17) and squamous cell
carcinoma (18). Therefore, we
sought to examine ETBR expression in PDAC tissue. However, no
studies have reported the conditions needed for
immunohistochemistry (IHC) to detect ETBR expression. Therefore,
the aim of the present study was to confirm the appropriate
conditions required for IHC of ETBR using ETBR cDNA and
transfectant cells, as well as to assess ETBR expression in PDAC
patients.
Materials and methods
Human tissue samples, culture cells and
xenografts
Frozen normal human tissues were received from the
Department of Gastroenterological Surgery II, Hokkaido University
Graduate School of Medicine, Sapporo, Japan. PBMCs were obtained
from healthy Japanese adult volunteers. HEK293 and 293FT cells were
purchased from Invitrogen Corporation (Carlsbad, CA, USA). The
human pancreatic cancer cell line PANC-1 was provided by RIKEN
(Tsukuba, Japan), PK-9 and PK-45P were from Tohoku University
(Sendai, Japan), and SUIT-2 was from Health Science Research
Resources Bank (Osaka, Japan). Human pancreatic cancer xenografts
were established by subcutaneous injection of these cell lines into
female BALB/c-SCID mice.
ETBR subcloning
Total RNA from normal human adrenal gland was
extracted by the RNeasy Mini kit (Qiagen, Tokyo, Japan). The cDNA
synthesis reaction was performed as previously described (19). ETBR-cDNA was amplified by PCR.
Briefly, each 50-μl reaction mixture contained 1 μl of reverse
transcription reaction products, 1 unit of KOD-Plus-DNA polymerase,
5 μl PCR buffer, 5 μl of 2 mM deoxynucleotide triphosphate, 2 μl of
25-mM MgSO4 (all from Toyobo, Osaka, Japan), and 1.5 μl
of each 10-μM 3′ and 5′ primer specific for ETBR (sense,
5′-CGGCTAGCCCTTCTGGAGCAGGTA-3′ and antisense,
5′-CGCGGATCCTCAAGATGAGCTGTA-3′). ETBR cDNA was amplified for 30
cycles. Conditions for ETBR PCR were: 94°C for 15 sec, 40°C for 30
sec and then 68°C for 3 min. All PCR products were electrophoresed
in a 2.0% agarose gel and visualized by ethidium bromide staining.
Plasmids expressing ETBR were generated by the PCR amplification of
ETBR cDNA and cloning into the NheI and BamHI sites
of pcDNA3.1(+) (Invitrogen).
Transfection
Subsequently, pcDNA-3.1(+)-ETBR-IRES-GFP (pETBR) was
transfected into 293FT or HEK293 cells using
Lipofectamine® 2000 (Invitrogen). Transfected cells were
incubated at 37°C in a 5% CO2 incubator for 20 h prior
to harvest.
Semi-quantitative reverse
transcription-polymerase chain reaction (RT-PCR)
Total RNA extraction and cDNA synthesis of normal
human tissues were performed as described above. Multiplex PCR was
performed using primers specific for ETBR or
glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Briefly, each
20-μl reaction mixture contained 1 μl reverse transcription
reaction products, 0.2 μl Taq DNA polymerase, 4 μl reaction
buffer (both from Promega, Madison, WI, USA), 0.5 μl of 10-mM
deoxynucleotide triphosphate, and 0.5 μl of each 10-μM 3′ and 5′
primer specific for ETBR (as described above) and GAPDH (sense,
5′-ACCCCTTCATTGACCTCAACT-3′ and antisense,
5′-TGAGTCCTTCCACGATACCAA-3′). ETBR and GAPDH cDNA was amplified for
25, 30, 35 and 40 cycles. Conditions for ETBR and GAPDH PCR were:
95°C for 30 sec, 50°C for 80 sec, and then 72°C for 5 min. All PCR
products were electrophoresed in a 2.0% agarose gel and visualized
by ethidium bromide staining.
Western blot analysis
Western blot analysis was performed to analyze ETBR
and GFP protein expression in transfected 293FT and HEK293 cells,
normal human tissues, human pancreatic cancer cell lines and
xenografts. Briefly, lysates from cells, tissues and xenografts
were resolved using 15% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and then transferred to polyvinylidene fluoride
microporous membranes (Millipore, Billerica, MA). Anti-ETBR rabbit
polyclonal antibodies (Ab1, 3, 4), mouse monoclonal antibody (Ab2),
and anti-GFP and anti-β-actin mouse monoclonal antibodies were used
as the primary antibodies. Peroxidase-conjugated goat anti-rabbit
or mouse IgG was used as the secondary antibody (1:10,000
dilution). The detection of bound antibodies was performed using
the electrogenerated chemiluminescence (ECL) system (Amersham,
Aylesbury, UK).
Flow cytometry
Flow cytometry was performed by FACSCalibur (BD
Biosciences, Franklin Lakes, NJ, USA). Briefly, pETBR-transfected
293FT cells [293FT (pETBR)] were harvested by 0.25% trypsin, and
then anti-ETBR antibodies (Ab1–4) were used as the primary
antibody. PE-conjugated goat anti-rabbit or mouse IgG (Beckman
Coulter, Fullerton, CA, USA) was used as the secondary
antibody.
Patients and tissue specimens
Tumor specimens were obtained from the same 80
patients reported previously (3).
This examination was carried out with the approval of the Hokkaido
University Ethics Committee.
IHC
Immunohistochemical reactions were carried out using
the universal immunoenzyme polymer method. Sections were
deparaffinized with xylene, rehydrated through a graded series of
ethanol/water and treated in a pressure cooker for 20 min.
Endogenous peroxidase activity was blocked by 30-min incubation
with 0.3% hydrogen peroxide in methanol. After washing in TBS-T,
specimens were saturated with 10% normal goat serum [Histofine
MAX-PO (Multi) kit; Nichirei Corp., Tokyo, Japan] for 30 min.
Sections were then incubated overnight with anti-ETBR antibodies
(Ab1–4) at 4°C. After three additional washes, the sections were
incubated with Histofine Simple Stain MAX-PO (Multi) (Nichirei
Corp.) for 20 min at room temperature. Reaction products were
observed by incubation for ~5 min with 3,30-diaminobenzidine
tetrahydrochloride (Nichirei Corp.). Sections were counterstained
in hematoxylin for 1 min and then mounted in Marinol (micro slides;
Muto-Glass, Tokyo, Japan).
Results
Establishment of ETBR-expressing
cells
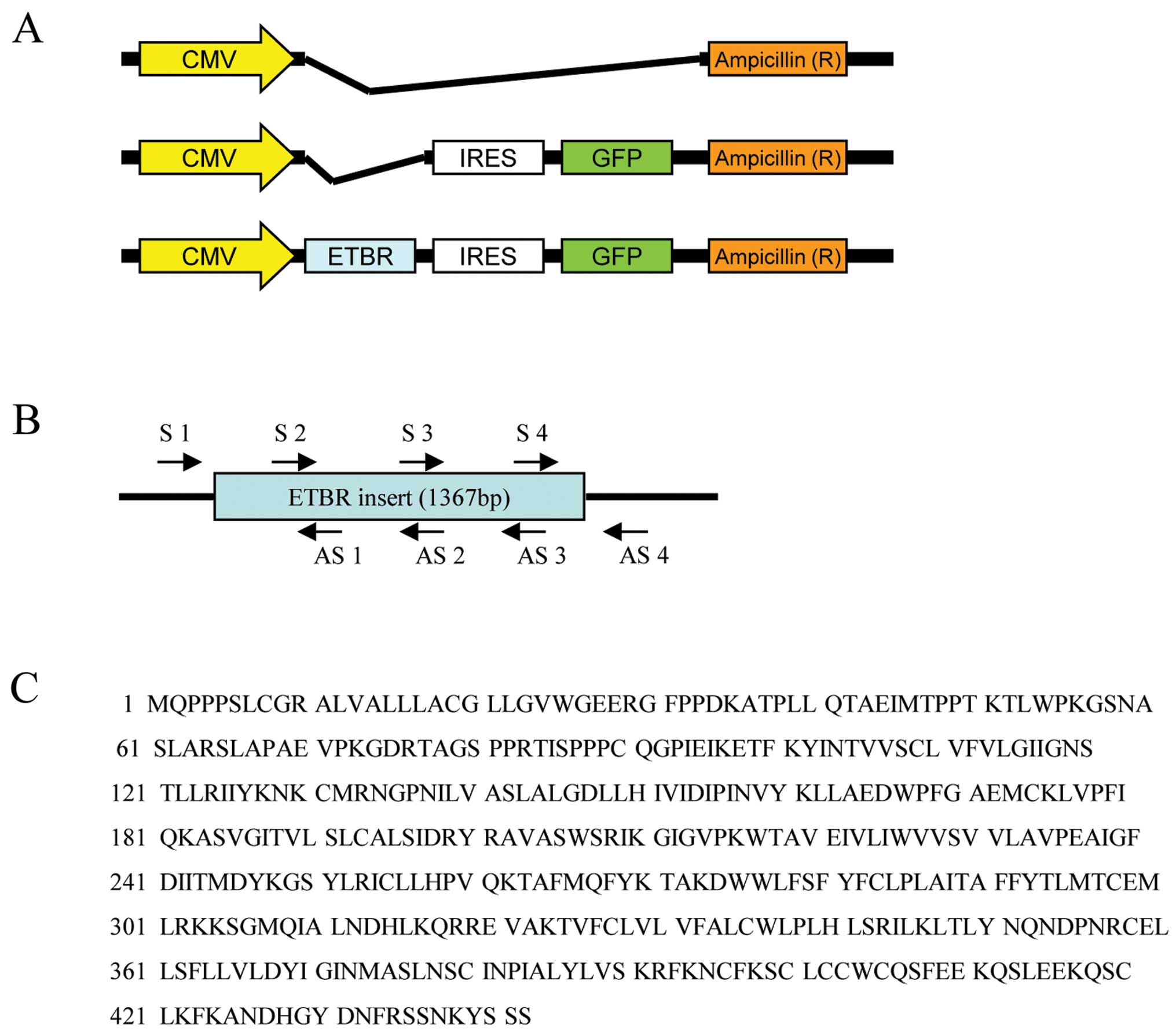
Three vectors were constructed as follows (Fig. 1A): pcDNA3.1(+) (pEmpty Vector),
pcDNA3.1(+)-IRES-GFP (pGFP), and pcDNA3.1(+)-ETBR-IRES-GFP (pETBR).
Four sense and antisense primers were synthesized (Fig. 1B), and the sequence after subcloning
was confirmed (data not shown). The amino acid sequence
corresponding to the ETBR base sequence was equal to the ETBR
sequence described in The National Center for Biotechnology
Information (NCBI) (Fig. 1C).
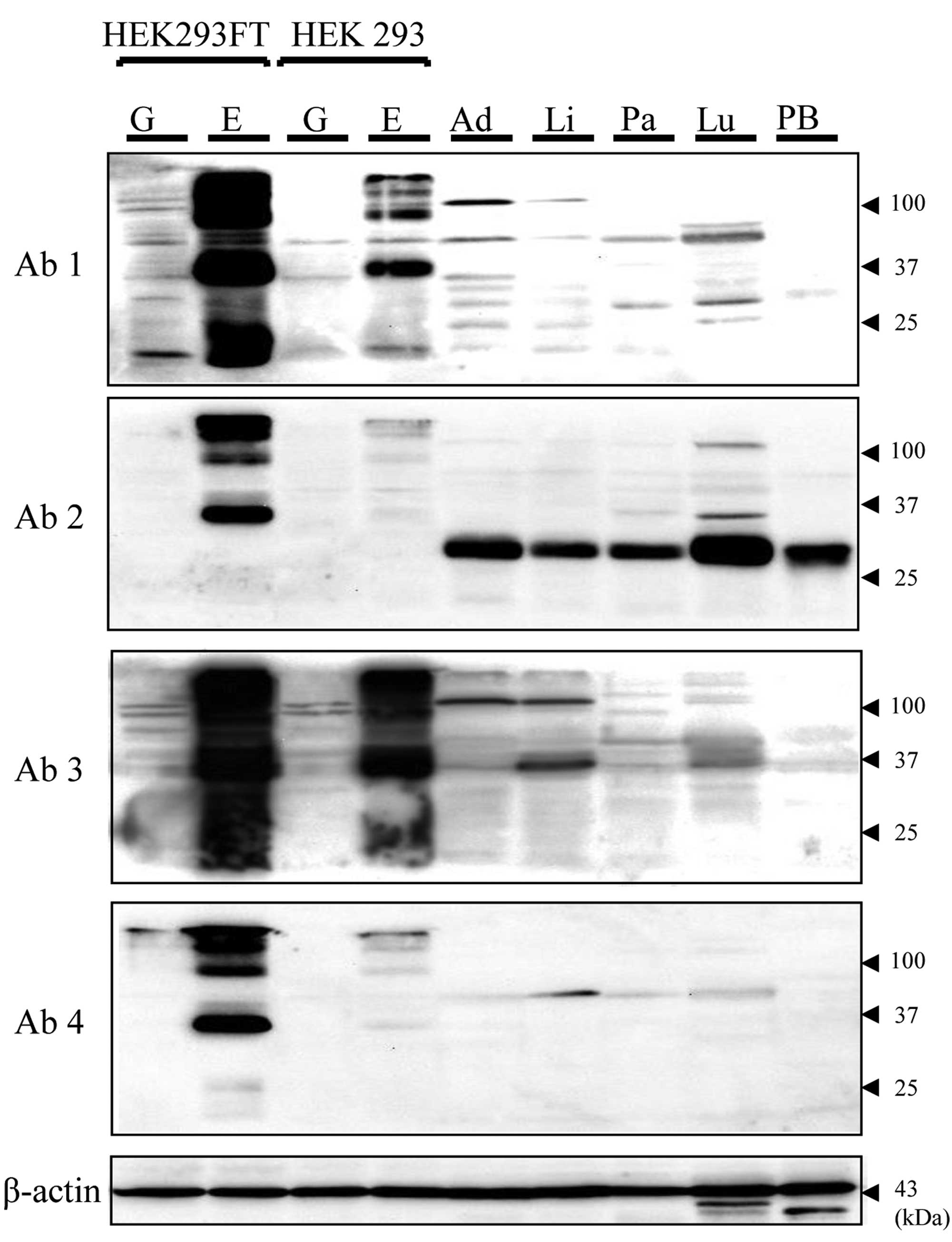
Transfection was then performed, and the detectability of ETBR
expression by western blot analysis (WB), flow cytometry and IHC
was evaluated. On WB, the expression of GFP protein was confirmed
in both 293FT (pGFP) and 293FT (pETBR) lysates. For the ETBR
protein, bands (100, 37, 25 kDa) were shown only in the 293FT
(pETBR) lysate by all four ETBR antibodies. The density of the
25-kDa band appeared different with each antibody (Fig. 2). HEK293 cell lysates showed lower
density bands of the same size as compared with 293FT. In the
normal human tissue sample, specific bands of the same size as the
positive control [PC; 293FT (pETBR)] were not observed with all
tissue samples. On the other hand, different size bands from PC
were found. This tendency was different with each antibody
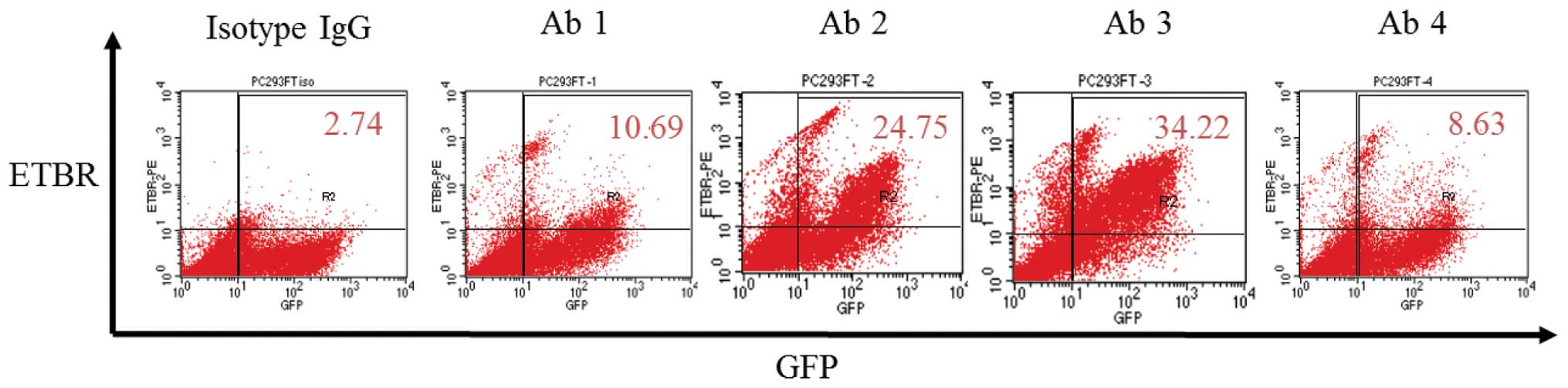
(Fig. 2). On flow cytometry, ETBR
expression of 293FT (pETBR) was observed with all antibodies.
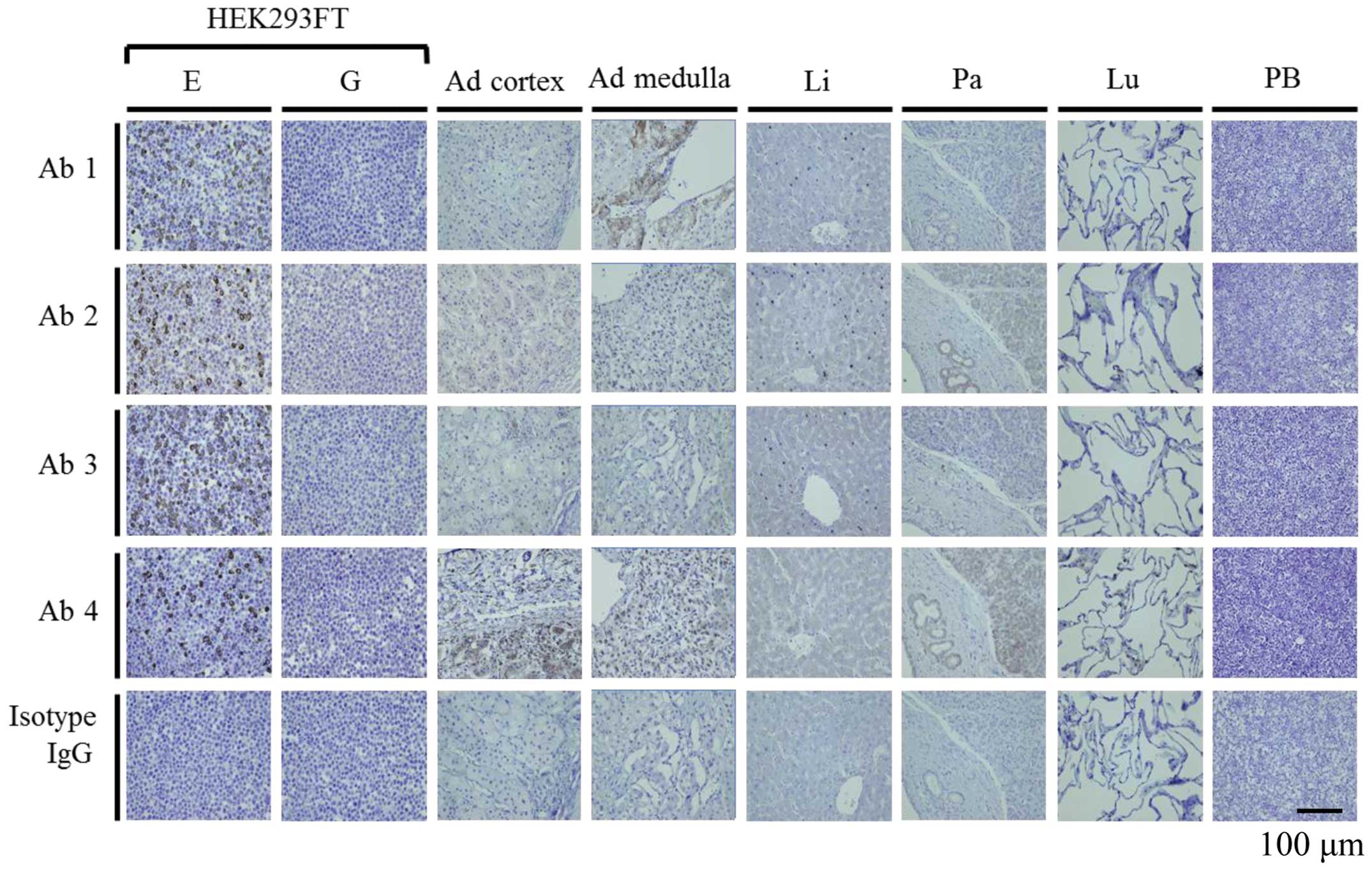
However, the ratio of ETBR positivity varied by antibody (Fig. 3). On IHC, no immunostaining was
observed with the negative control [NC; 293FT (pGFP)] by all
antibodies, and immunostaining was found on the membrane surface
with the PC by all antibodies. No immunostaining was seen with
isotype IgG (Fig. 4). This result
remained the same under different pH conditions (data not shown).
In normal human tissues, excluding the adrenal gland, obvious
immunostaining such as 293FT (pETBR) was not shown. In the adrenal
gland, the staining region (cortex or medulla) and intensity were
different with each antibody (Fig.
4). These staining results changed under different pH
conditions (data not shown).
Analysis of ETBR mRNA expression
The mRNA of normal human tissues was extracted, and
semi-quantitative RT-PCR was performed with the ETBR primer. ETBR
mRNA expression was weak only in the adrenal gland sample (Fig. 5).
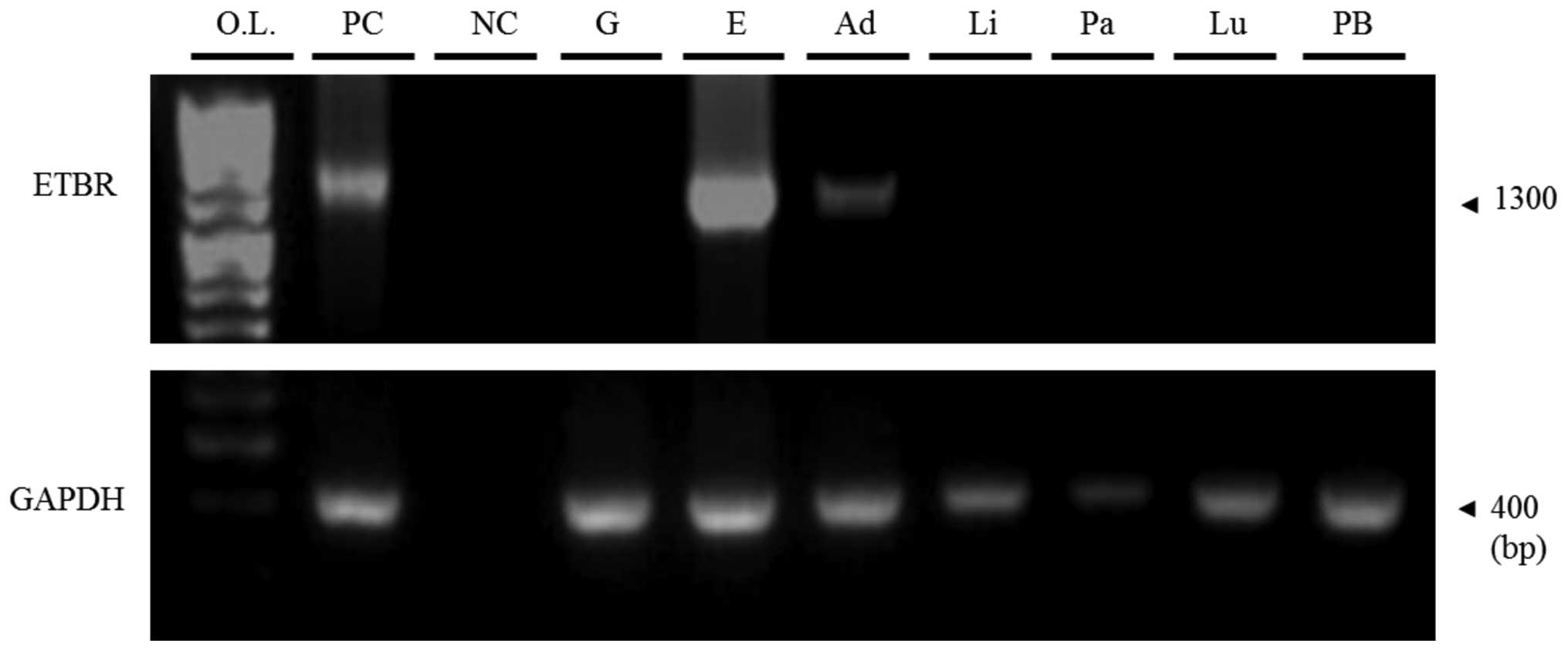 | Figure 5Semi-quantitative reverse
transcription-polymerase chain reaction (RT-PCR) of HEK293FT
transfectants or normal human tissues by ETBR primers for 35
cycles. Positive control (PC), pcDNA3.1(+)-IRES-GFP-ETBR; negative
control (NC), DDW; O.L., one step ladder (marker); G, 293FT (pGFP);
E, 293FT (pETBR); Ad, adrenal gland; Li, liver; Pa, pancreas; Lu,
lung; PB, PBMCs. |
Evaluation of ETBR expression in human
PDAC cell lines and tumor specimens
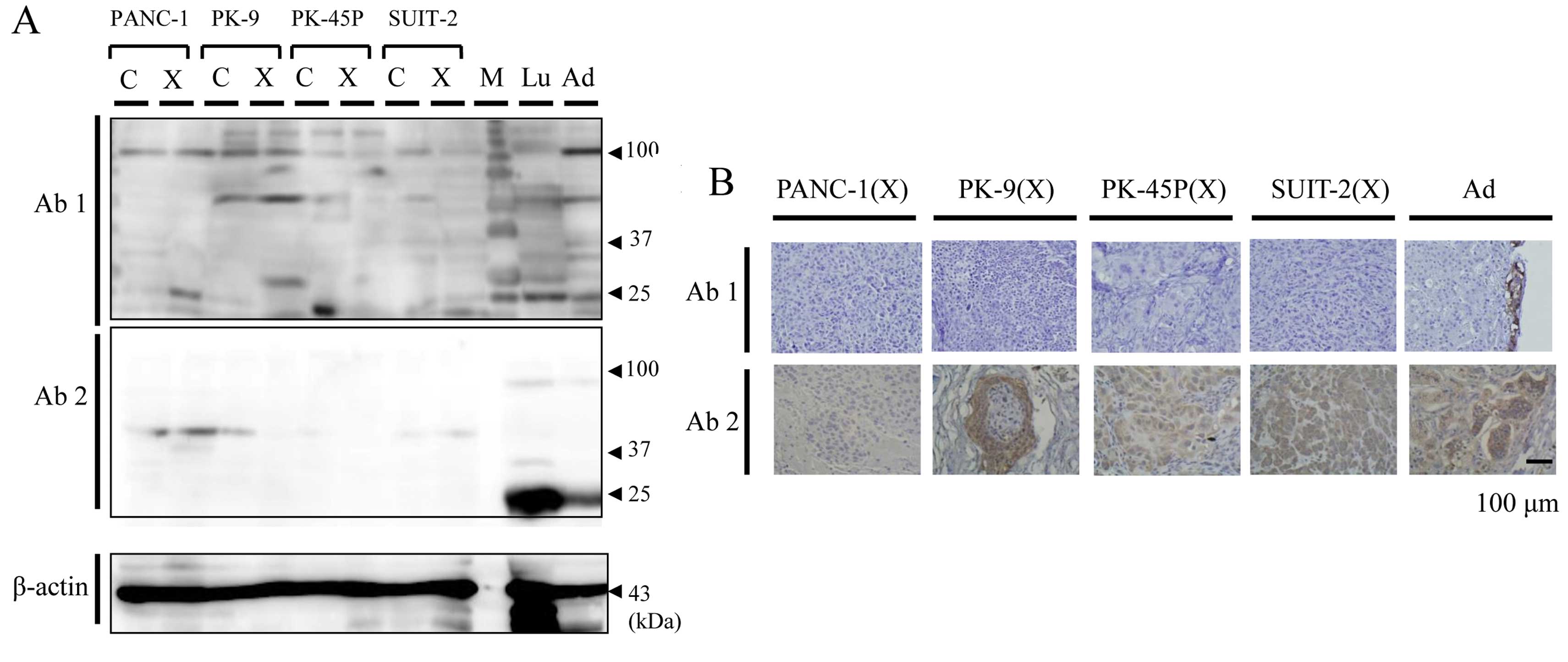
On WB, no specific band of ETBR was shown by two
antibodies (Fig. 6A). On IHC,
immunostaining was shown only in the adrenal medulla section by
Ab1, maintaining consistency between cell cultures and xenografts
(Fig. 6B). Thus, it was considered
that Ab1 was more suitable for IHC than Ab2 as almost all
immunostaining by Ab2 was shown to be non-specific. Ab1 was then
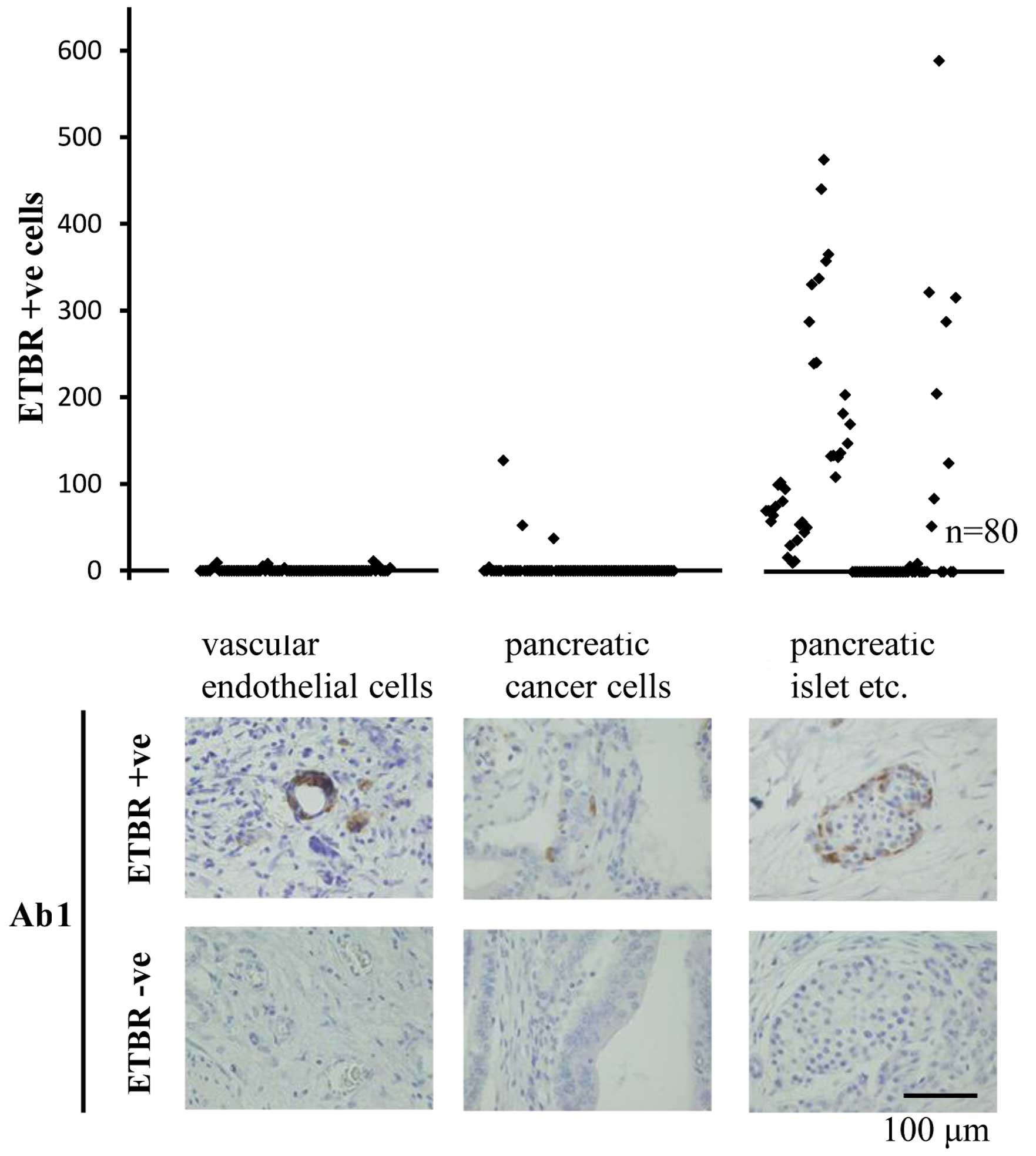
used for IHC of PDAC tumor specimens. No biologically significant
staining was shown in the vascular endothelial cells and the
pancreatic cancer cells. Some staining was shown in the pancreatic
acinar cells and pancreatic islets (Fig. 7).
Discussion
An appropriate ETBR gene expression system was
created, with 293FT (pETBR) as the PC and 293FT (pGFP) as the NC.
Although the theoretical molecular weight of ETBR protein is 49.64
kDa, the results of WB revealed several bands of different sizes.
Although all the bands (25, 37, 100, 200 kDa) of PC differed from
the theoretical molecular weight of ETBR, it is appropriate to
consider all these bands are ETBR. The PC was the NC with the
addition of the ETBR gene. Therefore, all bands seen in the PC that
were not seen in the NC must be ETBR. Therefore, it is logical that
these bands did not vary with the different antibodies.
It has been reported that ETBR can form a homodimer
(20,21). In this case, the homodimer shows a
size of 100 kDa, and polymerization of the homodimer or a
combination of the homodimer and other membrane proteins can show a
size exceeding 200 kDa. If one assumes that although ETBR homodimer
is comparatively stable biochemically, monomer is unstable and is
cleaved by a specific site during protein extraction, the band of
ETBR may also appear with sizes of 25 and 37 kDa. Splicing variants
(22,23) or glycosylation modification may
explain the band size variation from the theoretical molecular
weight of ETBR. Since the band of the PC differed from the
theoretical molecular weight, examination of the type and
concentration of reagent in the case of protein extraction was also
performed. It was found that the band of the PC was similarly
different from the theoretical figure.
The adrenal gland can serve as a PC of IHC under
appropriate conditions due to ETBR-mRNA expression. Of the four
ETBR antibodies, Ab1 was considered to be the most suitable
antibody for IHC, as adrenal gland was stained and other tissues
were not stained as much (Fig. 4).
The examination results for PDAC cell lines suggest that both
cultured cells and xenograft tumors may have no biologically
significant expression of ETBR. The assessment data on the PDAC
tumor specimens demonstrate that both pancreatic cancer cells and
vascular endothelial cells have no overexpression of ETBR.
Furthermore, it is possible that some intracellular enzymes may
contribute to the non-specific staining observed in gland tissue
such as pancreatic islets.
Based on the present results, ETBR appears to have
no biological significance in PDAC tissue, as non-staining of IHC
was regarded as low expression of ETBR.
In IHC, if positive and negative controls with high
objectivity are not used, there can be a risk of misleading
non-specific staining for the expression of target molecules.
In conclusion, ETBR expression in human PDAC tissues
may not be detected by IHC based on the strict objective controls
we have established and, therefore, it was not considered to
reflect the grade of malignancy of PDAC.
Acknowledgements
The authors thank Dr Satoshi Kondo and Dr Masaki
Miyamoto (Department of Gastroenterological Surgery II, Division of
Surgery, Hokkaido University Graduate School of Medicine, Sapporo,
Japan) for their high level of contribution to this research. The
authors would also like to thank Hikaru Shida and Naomi Saito for
their technical support in immunohistochemical analyses.
References
|
1
|
Hidalgo M: Pancreatic cancer. N Engl J
Med. 362:1605–1617. 2010. View Article : Google Scholar
|
|
2
|
Vincent A, Herman J, Schulick R, Hruban RH
and Goggins M: Pancreatic cancer. Lancet. 378:607–620. 2011.
View Article : Google Scholar
|
|
3
|
Fukunaga A, Miyamoto M, Cho Y, et al:
CD8+ tumor-infiltrating lymphocytes together with
CD4+ tumor-infiltrating lymphocytes and dendritic cells
improve the prognosis of patients with pancreatic adenocarcinoma.
Pancreas. 28:e26–e31. 2004.
|
|
4
|
Uehara H, Miyamoto M, Kato K, et al:
Expression of pigment epithelium-derived factor decreases liver
metastasis and correlates with favorable prognosis for patients
with ductal pancreatic adenocarcinoma. Cancer Res. 64:3533–3537.
2004. View Article : Google Scholar
|
|
5
|
Sakurai T, Yanagisawa M, Takuwa Y, et al:
Cloning of a cDNA encoding a non-isopeptide-selective subtype of
the endothelin receptor. Nature. 348:732–735. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Buckanovich RJ, Facciabene A, Kim S, et
al: Endothelin B receptor mediates the endothelial barrier to T
cell homing to tumors and disables immune therapy. Nat Med.
14:28–36. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nelson JB, Fizazi K, Miller K, et al:
Phase 3, randomized, placebo-controlled study of zibotentan
(ZD4054) in patients with castration-resistant prostate cancer
metastatic to bone. Cancer. 118:5709–5718. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Miller K, Moul JW, Gleave M, et al: Phase
III, randomized, placebo-controlled study of once-daily oral
zibotentan (ZD4054) in patients with non-metastatic
castration-resistant prostate cancer. Prostate Cancer Prostatic
Dis. 16:187–192. 2013.PubMed/NCBI
|
|
9
|
Clézardin P: Therapeutic targets for bone
metastases in breast cancer. Breast Cancer Res. 13:2072011.
|
|
10
|
Asundi J, Reed C, Arca J, et al: An
antibody-drug conjugate targeting the endothelin B receptor for the
treatment of melanoma. Clin Cancer Res. 17:965–975. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cruz-Muñoz W, Jaramillo ML, Man S, et al:
Roles for endothelin receptor B and BCL2A1 in spontaneous CNS
metastasis of melanoma. Cancer Res. 72:4909–4919. 2012.PubMed/NCBI
|
|
12
|
Liu Y, Ye F, Yamada K, et al: Autocrine
endothelin-3/endothelin receptor B signaling maintains cellular and
molecular properties of glioblastoma stem cells. Mol Cancer Res.
9:1668–1685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Knight LJ, Burrage J, Bujac SR, et al:
Epigenetic silencing of the endothelin-B receptor gene in non-small
cell lung cancer. Int J Oncol. 34:465–471. 2009.PubMed/NCBI
|
|
14
|
Puglisi MA, Barba M, Corbi M, et al:
Identification of Endothelin-1 and NR4A2 as
CD133-regulated genes in colon cancer cells. J Pathol. 225:305–314.
2011.PubMed/NCBI
|
|
15
|
Sørby LA, Kleiveland CR, Andersen SN,
Bukholm IR and Jacobsen MB: The endothelin axis in the metastatic
process of colon carcinoma. Anticancer Res. 31:861–869.
2011.PubMed/NCBI
|
|
16
|
Wang R, Löhr CV, Fischer K, et al:
Epigenetic inactivation of endothelin-2 and endothelin-3 in colon
cancer. Int J Cancer. 132:1004–1012. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tao K, Wu C, Wu K, et al: Quantitative
analysis of promoter methylation of the EDNRB gene in gastric
cancer. Med Oncol. 29:107–112. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ishimoto S, Wada K, Tanaka N, et al: Role
of endothelin receptor signalling in squamous cell carcinoma. Int J
Oncol. 40:1011–1019. 2012.PubMed/NCBI
|
|
19
|
Hase R, Miyamoto M, Uehara H, et al:
Pigment epithelium-derived factor gene therapy inhibits human
pancreatic cancer in mice. Clin Cancer Res. 11:8737–8744. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Evans NJ and Walker JW: Endothelin
receptor dimers evaluated by FRET, ligand binding, and calcium
mobilization. Biophys J. 95:483–492. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Klammt C, Srivastava A, Eifler N, et al:
Functional analysis of cell-free-produced human endothelin B
receptor reveals transmembrane segment 1 as an essential area for
ET-1 binding and homodimer formation. FEBS J. 274:3257–3269. 2007.
View Article : Google Scholar
|
|
22
|
Elshourbagy NA, Adamou JE, Gagnon AW, Wu
HL, Pullen M and Nambi P: Molecular characterization of a novel
human endothelin receptor splice variant. J Biol Chem.
271:25300–25307. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shyamala V, Moulthrop TH, Stratton-Thomas
J and Tekamp-Olson P: Two distinct human endothelin B receptors
generated by alternative splicing from a single gene. Cell Mol Biol
Res. 40:285–296. 1994.PubMed/NCBI
|