Introduction
Hepatocellular carcinoma (HCC), the fifth in
incidence among malignant tumors, accounts for 70–85% of primary
liver cancer cases. Each year, 748,000 cases are newly diagnosed as
HCC worldwide, and over half of them are in China (1), where HCC is the second most frequent
cause for cancer-related mortality since the 1990s (2). HBV infection is the primary etiology
that leads to liver cirrhosis-related carcinoma (3). The reason for the high mortality in
HCC is that the tumor is always detected at advanced stages when
curative therapy cannot be carried out due to intrahepatic or
extrahepatic metastases. Although serum α-fetoprotein (AFP) has
been widely accepted and used as a serous biomarker for screening
HCC in a high risk population for years, the sensitivity and
specificity of serum AFP only ranged from 40–65 and 76–96%,
respectively (4), suggesting it may
not be an ideal indicator to identify HCC from other diseases.
Therefore, discovering new biomarkers for early diagnosis of HCC
are still needed in clinical practice.
Mature microRNAs (miRNAs) are 19- to 25-nt
transcripts of small non-coding RNA family processed from 70- to
100-nt hairpin-shaped precursors. The sequences of some miRNAs are
conserved in various biological species, suggesting that these tiny
molecules participate in essential processes of development,
proliferation, differentiation and/or apoptosis in organisms
(5). Depending on certain cellular
functions of their targets, dysregulation of miRNAs may play
oncogenic roles, such as Myc, or tumor suppressor roles, such as
p53, to induce or inhibit tumorigenesis (6). Meanwhile, the importance of miRNAs in
cancer progression has been reported, as miRNAs can both influence
the effect of chemotherapy (7) as
well as the development of drug resistance (8). Although the precise biological
functions of miRNAs are not yet fully understood, some studies
demonstrated that miRNA expression profile is distinguished in
diverse diseases, indicating that miRNAs may be used as biomarkers
for cancer diagnosis and prognosis prediction (9,10).
Whether those dysregulated miRNAs are related to HCC and/or as a
general mechanism in disease progression to cancer is an important
concern for utilization of miRNAs as biomarkers for HCC risk,
treatment response and clinical outcome prediction (11).
In the present study, we investigated comprehensive
miRNA expression profiling of HCC using miRNA microarray in
hepatocellular cancerous tissue and peritumoral non-cancerous
tissue, and found that microRNA-139 (miRNA-139) was significantly
downregulated in hepatocellular cancerous tissue. Furthermore, we
detected and verified the miRNA-139 expression level in tissue and
blood to investigate the correlation between miRNA-139 and clinical
characteristics to identify the diagnostic and prognostic values of
miRNA-139 in HCC patients.
Materials and methods
Patients, samples and data
collection
The study population was enrolled at the general
surgery department of Tangdu Hospital affiliated to the Fourth
Military Medical University (Xi’an, China). Among the study
population, 31 patients were newly diagnosed and histologically
confirmed HCC, and subsequently received curative hepatectomy
according to the National Comprehensive Cancer Network (NCCN)
guidelines for hepatobiliary cancer. Cancerous tissue samples (CT
group) were collected from surgery and pathologically confirmed as
HCC; peritumoral non-cancerous tissue samples (NT group) were
collected from normal liver tissue at 3 cm beyond the tumor margin.
Plasma samples (PS group) were obtained from patients prior to
surgery, while the matched plasma samples (MPS group) were obtained
from 31 age- and gender-matched chronic HBV-hepatitis (CH)
patients. The clinical characteristics of all subjects are
summarized in Table I. All study
subjects provided informed consent, and the present study was
approved by the Review Board of Tangdu Hospital Ethics
Committee.
 | Table IClinicopathological characteristics of
hepatocellular carcinoma (HCC) patients and chronic HBV-hepatitis
(CH) patients. |
Table I
Clinicopathological characteristics of
hepatocellular carcinoma (HCC) patients and chronic HBV-hepatitis
(CH) patients.
| Patients with
different diseases | |
|---|
|
| |
|---|
| HCC (n=31) | CH (n=31) | P-value |
|---|
| Clinical factors |
| Age (years) | 49±11 | 49±10 | 0.981 |
| Gender
(male/female) | 26/5 | 26/5 | 0.999 |
| WBC
(×109/l) | 6.06±2.48 | 5.51±2.21 | 0.358 |
| RBC
(×1012/l) | 4.54±0.78 | 4.49±0.81 | 0.830 |
| PLT
(×109/l) | 128.90±59.10 | 131.84±60.26 | 0.847 |
| Hb (g/l) | 139.87±20.48 | 140.74±21.30 | 0.870 |
| ALB (g/l) | 40.25±4.32 | 40.96±8.78 | 0.684 |
| TB (μmol/l) | 20.17±7.88 | 22.17±12.66 | 0.458 |
| DB (μmol/l) | 5.71±4.15 | 7.05±4.35 | 0.220 |
| IB (μmol/l) | 14.14±5.75 | 15.13±9.40 | 0.620 |
| ALT (U/l) | 45.42±22.53 | 45.39±29.88 | 0.996 |
| AST (U/l) | 57.23±30.47 | 43.00±33.00 | 0.115 |
| ALP (U/l) | 93.00±61.00 | 81.50±61.75 | 0.134 |
| GGT (U/l) | 67.00±55.00 | 59.00±73.25 | 0.714 |
| Operation time
(min) | 194±42 | | |
| Blood loss (ml) | 740.65±582.33 | | |
| Liver cirrhosis
(+/−) | 20/11 | | |
| Hypersplenism
(+/−) | 16/15 | | |
| Tumor-related
factors |
| AFP (ng/ml) | 17.80±598.60 | 7.10±13.10 |
0.007a |
| CEA (ng/ml) | 2.57±1.81 | 2.15±1.32 | 0.311 |
| CA19-9 (U/ml) | 20.97±15.86 | 16.50±33.00 | 0.749 |
| Child-Pugh grading
(A/B) | 26/5 | 19/12 | 0.086 |
| Tumor no.
(single/multiple) | 26/5 | | |
| Tumor size
(cm) | 8.24±3.20 | | |
| Vascular invasion
(+/−) | 2/29 | | |
| Edmondson-Steiner
grading (I/II+III) | 5/26 | | |
| TNM staging
(I/II/III) | 12/3/16 | | |
| Okuda staging
(1/2) | 14/17 | | |
| BCLC staging
(A/B/C) | 14/8/9 | | |
| CLIP scoring
(1/2/3) | 15/13/3 | | |
miRNA microarray
miRNA microarray was performed in three pairs of
tissue specimens which were collected from two male and one female
HCC patients, using a service provider (LC Sciences, USA). The
assay started from 4 to 8 μg total RNA sample and was 3′-extended
with a poly(A) tail using poly(A) polymerase. An oligonucleotide
tag was then liquated to the poly(A) tail for later fluorescent dye
staining. Hybridization was performed overnight on a μParaflo
microfluidic chip using a micro-circulation pump (Atactic
Technologies, USA). On the microfluidic chip, each detection probe
consisted of a chemically modified nucleotide coding segment
complementary to target miRNA or control RNA and a spacer segment
of polyethylene glycol to extend the coding segment away from the
substrate. The detection probes were made by in situ
synthesis using photogenerated reagent chemistry. Hybridization
used 100 l 6X SSPE buffer containing 25% formamide at 34°C. After
RNA hybridization, tag-conjugating Cy3 dye was circulated through
the microfluidic chip for dye staining. Fluorescence images were
collected using a laser scanner (GenePix 4000B; Molecular Devices,
USA) and digitized using Array-Pro image analysis software (Media
Cybernetics, USA). Data were analyzed by first subtracting the
background and then normalizing the signals using a LOWESS filter
(locally-weighted regression).
Total RNA extraction and
quantification
All tissue samples were quick-frozen in liquid
nitrogen and stored at −80°C immediately after removal. The
isolation of tissue total RNA was completed using miRNeasy Mini kit
(Qiagen, Germany). While extracting RNA, 25 mg frozen tissue with
liquid nitrogen was ground to fine powder manually by mortar and
pestle. After transferring the fine powder into an Eppendorf tube
instantly, 700 μl QIAzol lysis reagent and 140 μl chloroform was
added into the tube. The mixture was centrifuged at 12,000 rpm
(revolutions/min) for 15 min at 4°C, and the aqueous phase,
together with 1.5 volumes of 100% ethanol, was pipeted into a fresh
tube. Then, the sample was purified in RNeasy Mini column using 700
μl buffer RWT once and 500 μl buffer RPE twice. Finally, 50 μl
RNase-free ddH2O was used to elute total RNA from the
RNeasy Mini column.
The plasma samples were isolated from whole blood
before storing. In order to completely remove cell debris, the
fresh whole blood was centrifuged at 1,600 rpm for 5 min, followed
by 13,000 rpm for another 15 min. The plasma was stored in a
cryogenic tube at −80°C. The miRNeasy Serum/Plasma kit provided by
Qiagen was used to carry out extraction of total RNA from plasma
according to the manufacturer’s instructions. The purity and
concentration of total RNA preparations were determined by
measuring the absorbance at UV 260 nm (A260) and UV 280 nm (A280)
in a spectrophotometer (BioTek Epoch, USA). Pure RNA solution has
an A260/A280 ratio of 1.9–2.1. All RNA preparations were stored at
−80°C.
Reverse transcription and quantitative
polymerase chain reaction (RT-qPCR)
Quantification of mature miRNAs was accomplished by
a two-step method. Firstly, RNA was 3′-extended with a poly(A) tail
using poly(A) polymerase, then the poly(A) product was reverse
transcribed using oligo(dT)-Universal Tag (Tiangen, China).
Subsequently, real-time quantitative PCR was performed with
miRNA-139 primer and internal normalization primer according to the
procedures of miRcute miRNA SYBR-Green qPCR detection kit (Tiangen)
in Mx3000p sequence detection system (Agilent, USA), using the
following conditions: 94°C for a 2-min cycle, followed by 45 cycles
of 94°C for 20 sec, and 60°C for 34 sec. Briefly, 20 μl PCR system
contained 2 μl of RT product solution, 10 μl of 2X miRcute miRNA
Premix (including SYBR), 0.4 μl forward primer, 0.4 μl reverse
primer and 7.2 μl RNase-free ddH2O. Triplicate PCRs were
carried out for every cDNA sample, including negative controls
without templates. hsa-miR-U6 and hsa-miR-16 were used as the
internal normalization control for tissue and plasma sample
respectively. All primers were designed and provided by Tiangen.
The expression level of miRNA was computed using the comparative
ΔCt method as previously reported (12).
Statistical analysis
Data analysis was performed by software SPSS 21.0
for Windows (IBM SPSS, USA). The difference of miRNA expression
levels between groups was calculated using the t-test or
Mann-Whitney U test. The Pearson’s correlation coefficient was used
to calculate correlations. The diagnostic value for differentiating
HCC patients from chronic HBV-hepatitis patients was evaluated by
receiver operator characteristic curve (ROC curve) and the areas
under ROC curve (AUC). The clinicopathological data were
represented as means ± SD or frequencies, and differences between
groups were calculated by the t-test, Mann-Whitney U test or
Fisher’s exact test. The Kaplan-Meier method was applied to
determine 1-year survival rate, and the statistical difference
between two groups was analyzed using Cox’s proportional hazard
model. A value of p<0.050 was considered to indicate
statistically significant differences.
Results
Patient characteristics
There were 31 HCC patients (26 males and 5 females)
with a mean age of 49 years (SD, 11; range, 27–69 years) while
another 31 age- and gender-matched CH patients with a mean age of
49 years (SD, 10; range, 27–69 years) were enrolled in the present
study. There was no significant difference in terms of demographic
characteristics (age and gender), ALT (p=0.996), AST (p=0.115), TB
(p=0.458), CEA (p=0.311), CA19-9 (p=0.749), and Child-Pugh grading
(p=0.086). There was a significant difference in serum AFP value
(p=0.007) between the two groups (Table
I). Despite the difference in AFP value, the two groups were
completely comparable in the present study. All HCC patients
received curative hepatectomy during 2010–2012 with a median
survival time of 362 days (± 160 days).
Association between miRNA-139 and HCC
risk
Ten miRNAs dysregulated in HCC
We hypothesized that miRNAs may participate in liver
tumorigenesis, and are aberrantly expressed in HCC samples. The
miRNA microarray was performed in 3 hepatocellular cancerous
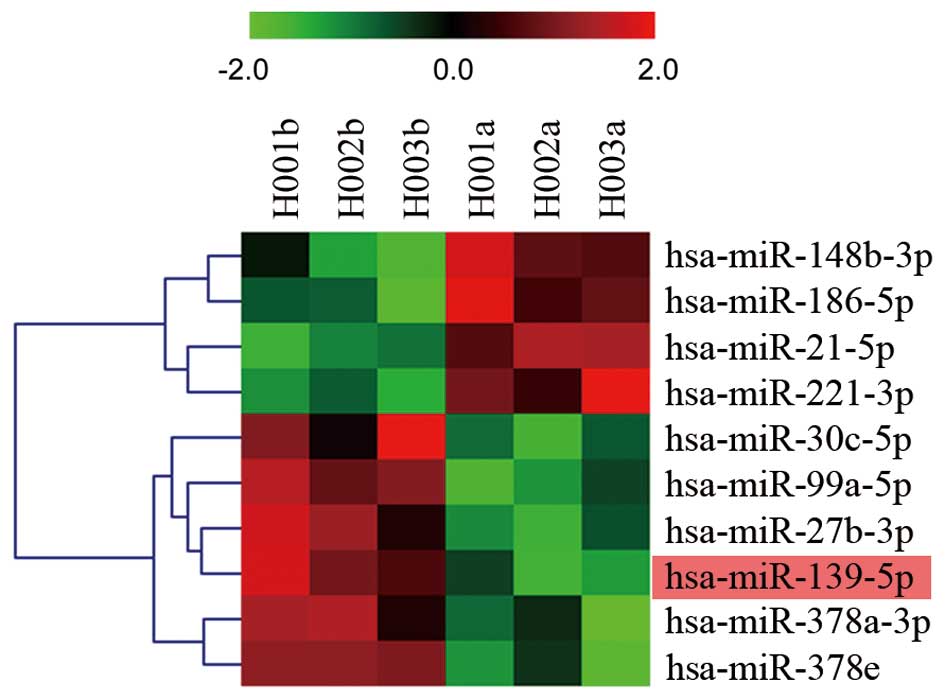
tissues and corresponding paired non-cancerous tissues. Fig. 1 shows the heat map of these
dysregulated targets and defined comprehensive miRNA expression
profiling. The most conspicuous underexpression values were found
for miRNA-139, miRNA-99a, miRNA-27b, miRNA-378a, miRNA-378e and
miRNA-30c, while the overexpression values were found for miRNA-21,
miRNA-221, miRNA-148b and miRNA-186.
miRNA-139 is downregulated in HCC
tissue and plasma samples
Among these dysregulated miRNAs, miRNA-139
expression level was further analyzed in the CT and NT group by
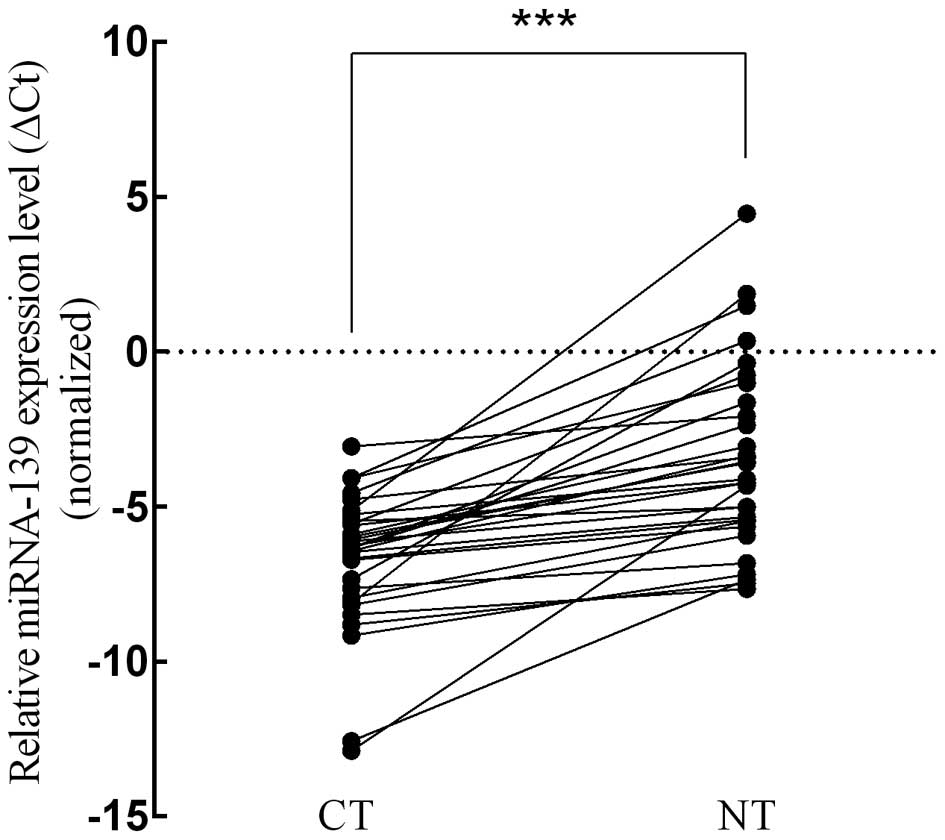
RT-qPCR to confirm the dysregulation in tumoral tissues. We found
that miRNA-139 expression level was significantly lower in the
patients of the CT group, compared to those in the NT group, with
an average ΔCt value of −6.704 vs. −3.490 (p<0.001, t=−6.785;
Fig. 2). Subsequently, we analyzed
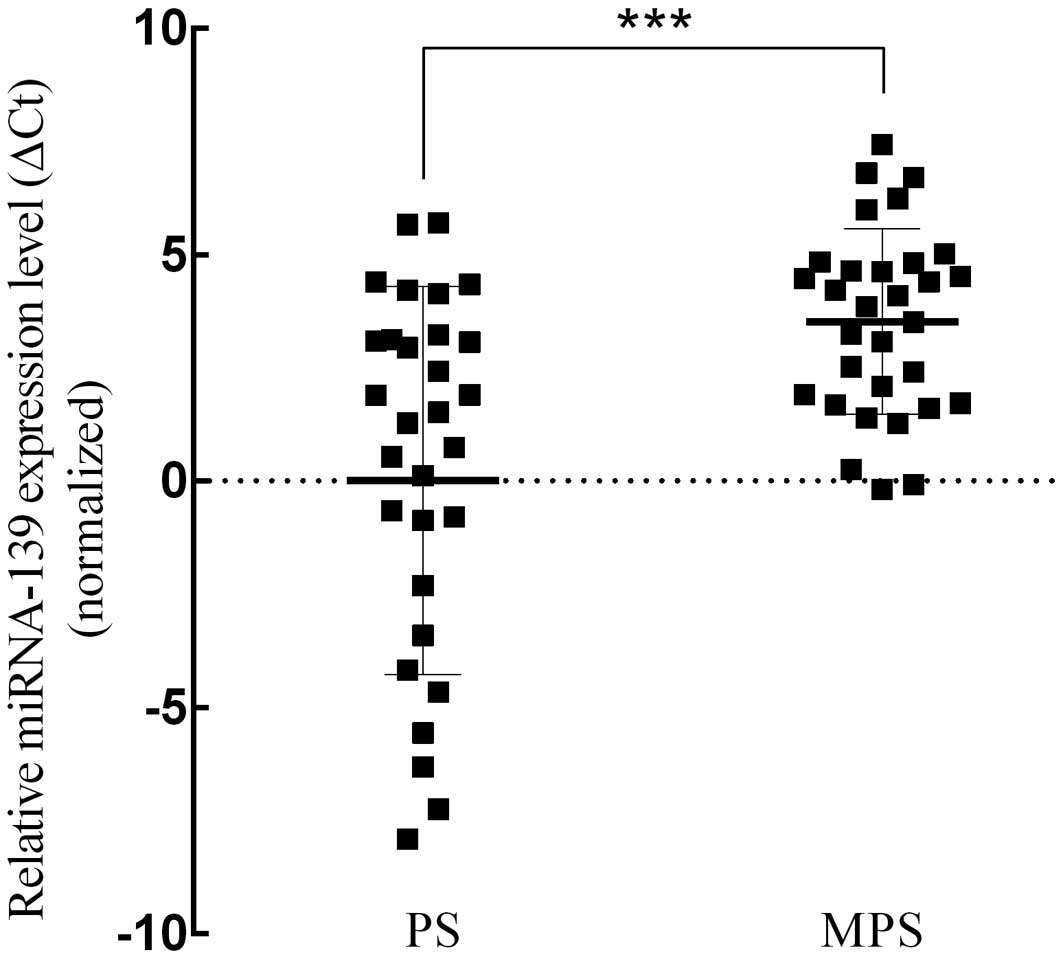
miRNA-139 expression level in the PS and MPS groups. The results
demonstrated that miRNA-139 expression level in the PS group was
significantly lower than that in the MPS group (average ΔCt value:
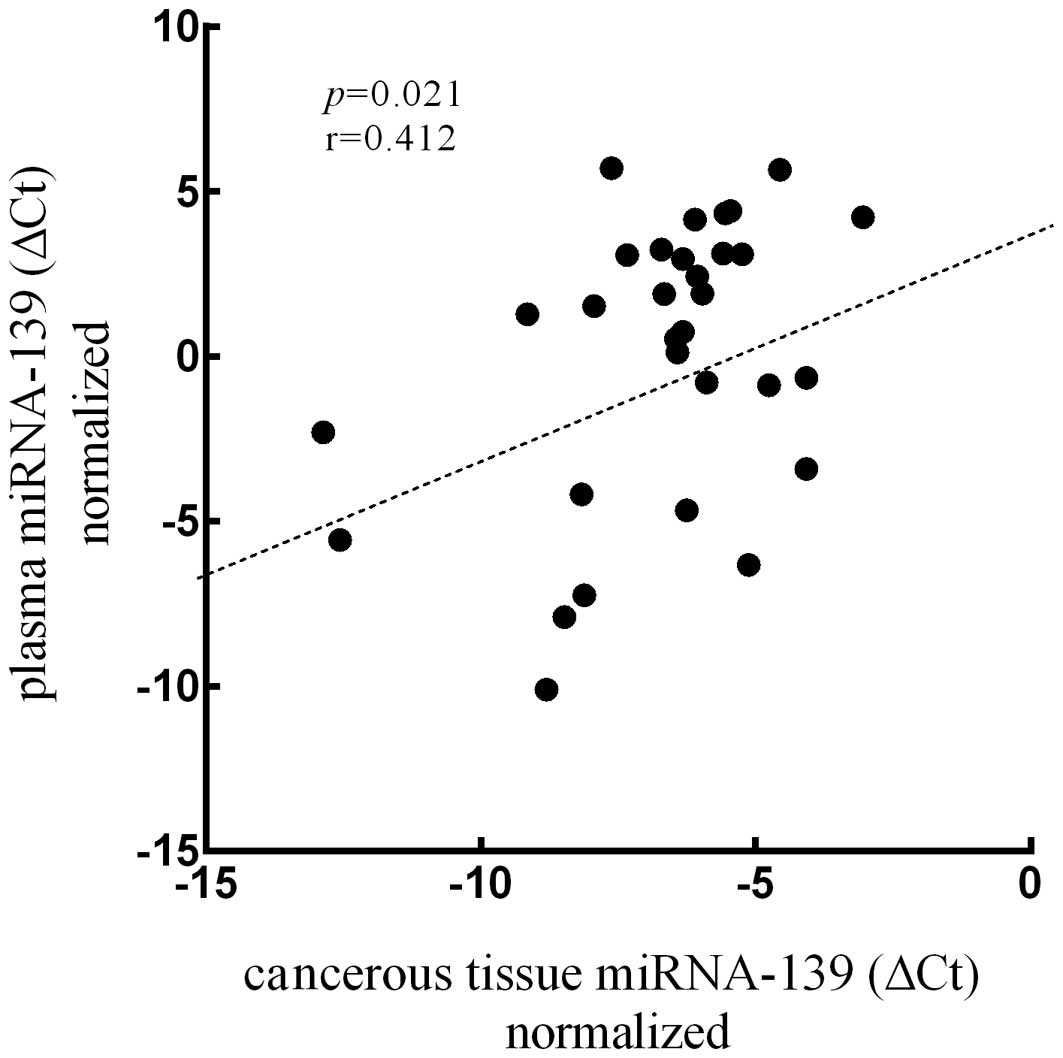
0.009 vs. 3.516, p<0.001, t=−4.117; Fig. 3). Meanwhile, the expression quantity
of miRNA-139 in plasma was positively correlated to that in
cancerous tissues (p=0.021, r=0.412; Fig. 4).
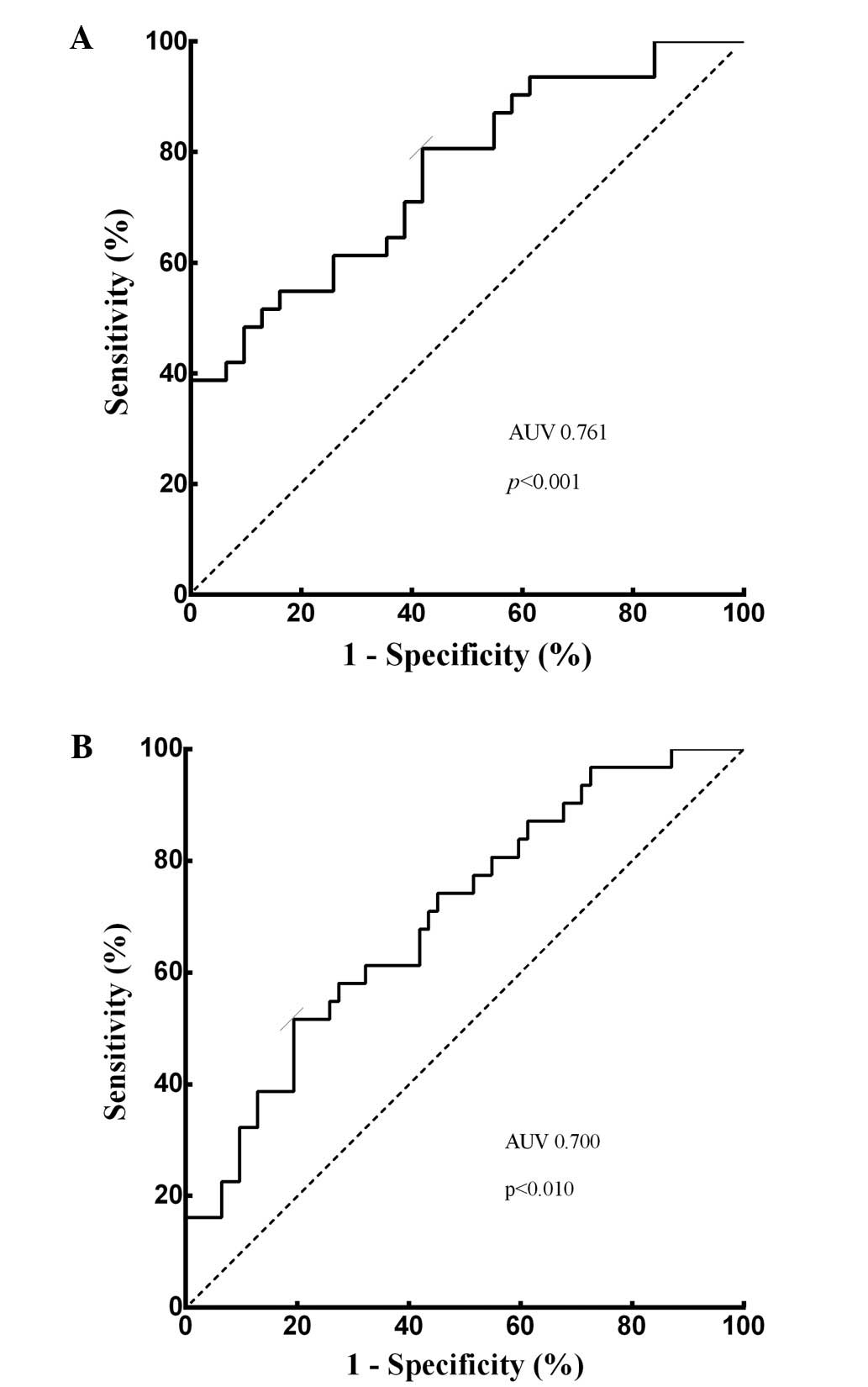
The diagnostic value of miRNA-139,
AFP, and combination of miRNA-139 and AFP for HCC
ROC curve was performed. To evaluate the
differentiating power of miRNA-139, plasma miRNA-139 expression
level was confirmed to be weakly correlated with serum AFP value
(p=0.024, r=0.406). Plasma miRNA-139 was able to identify HCC
patients from CH patients with AUC value of 0.761 (95% CI,
0.643–0.878, p<0.001; Fig. 5A).
At the cut-off value of −3.240 for plasma miRNA-139, the optimal
sensitivity and specificity were 80.6 and 58.1%, respectively.
Subsequently, with the cut-off value of 17.05 ng/ml, the AUC of
serum AFP was 0.700 (p<0.010, 95% CI, 0.571–0.829; Fig. 5B), while the sensitivity and
specificity were 51.6 and 79.3%, respectively, which was consistent
with previous studies (13).
Similarly, the differentiating power for combination of plasma
miRNA-139 with serum AFP was analyzed, and we found that the
combination of these two markers improved the power of screening
HCC. The combination retrieved a significantly higher sensitivity
of 90.3 and specificity of 87.1%, while the AUC increased to 0.770
(p<0.010, 95% CI, 0.654–0.886).
Plasma miRNA-139 correlates with
clinicopathological features and 1-year survival analysis of
HCC
We examined the correlations between plasma
miRNA-139 expression level and some clinical features (Table II). All HCC patients were divided
into two groups according to the expression status of miRNA-139:
the low expression group (n=25), representing the patients with
plasma miRNA-139 level under the optimal cut-off of −3.240, and the
high expression group, representing the remaining 6 patients. The
results revealed that plasma miRNA-139 expression level was
correlated with Edmondson-Steiner grading (p=0.038), serum AFP
value (p=0.043), CEA value (p=0.034) and DB value (p=0.041).
However, there was no correlation between miRNA-139 expression
level and other features, such as age, gender and clinical staging.
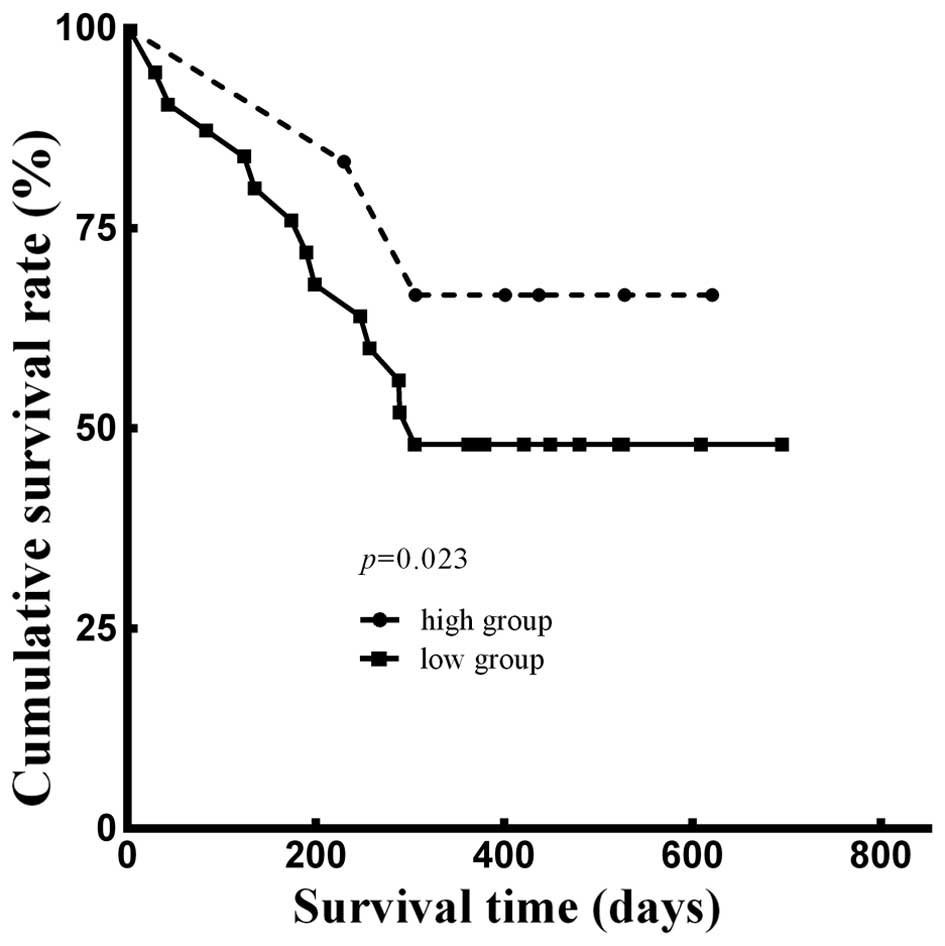
Furthermore, we analyzed the 1-year survival rate using the
Kaplan-Meier method. Adjusted for age, gender, weight loss
percentage, smoking status (14),
Edmondson-Steiner grading (15),
Child-Pugh grading (16) and CLIP
scoring (17), there was a
significant difference between the two groups (p=0.023; Fig. 6). The Kaplan-Meier analysis
indicated that the 1-year cumulative survival rate of patients in
the high expression group was 66.7% with 503 survival days (± 69
days) median survival time, while that in the low expression group
was 48.0%, and the median survival time was 439 days (± 50
days).
 | Table IIClinicopathological characteristics
of HCC patients categorized according to the plasma miRNA-139
expression level. |
Table II
Clinicopathological characteristics
of HCC patients categorized according to the plasma miRNA-139
expression level.
| Plasma miRNA-139
expression level | |
|---|
|
| |
|---|
| Low group (n=25)
[<−3.240 (ΔCt)] | High group (n=6) [≥
−3.240 (ΔCt)] | P-value |
|---|
| Clinical
factors |
| Age (years) | 49±11 | 48±12 | 0.747 |
| Gender
(male/female) | 20/5 | 6/0 | 0.553 |
| WBC
(×109/l) | 5.92±2.53 | 6.63±2.38 | 0.538 |
| RBC
(×1012/l) | 4.61±0.80 | 4.23±0.69 | 0.297 |
| PLT
(×109/l) | 135.44±61.46 | 101.67±41.50 | 0.214 |
| Hb (g/l) | 141.40±20.03 | 133.50±23.05 | 0.405 |
| ALB (g/l) | 40.62±4.22 | 38.70±4.92 | 0.340 |
| TB (μmol/l) | 22.53±7.69 | 19.61±7.97 | 0.423 |
| DB (μmol/l) | 8.78±5.85 | 4.97±3.39 |
0.041a |
| IB (μmol/l) | 13.71±6.92 | 14.24±5.60 | 0.843 |
| ALT (U/l) | 44.28±24.28 | 50.17±13.48 | 0.574 |
| AST (U/l) | 55.04±31.22 | 66.33±27.73 | 0.424 |
| ALP (U/l) | 173.08±309.04 | 115.33±34.00 | 0.655 |
| GGT (U/l) | 104.84±96.05 | 59.33±28.13 | 0.265 |
| Operation time
(min) | 198±44 | 179±45 | 0.355 |
| Blood loss
(ml) | 728±570 | 793±684 | 0.810 |
| Liver cirrhosis
(+/−) | 16/9 | 4/2 | 0.999 |
| Hypersplenism
(+/−) | 13/12 | 3/3 | 0.999 |
| Tumor-related
factors |
| AFP (ng/ml) | 29.50±934.35 | 8.80±13.48 |
0.043a |
| CEA (ng/ml) | 4.18±2.85 | 1.55±0.59 |
0.034a |
| CA19-9 (U/ml) | 22.21±16.89 | 15.82±10.00 | 0.384 |
| Child-Pugh grading
(A/B) | 21/4 | 5/1 | 0.999 |
| Tumor no.
(single/multiple) | 22/3 | 4/2 | 0.241 |
| Tumor size
(cm) | 8.24±3.2 | 7.63±4.22 | 0.698 |
| Vascular invasion
(+/−) | 1/24 | 1/5 | 0.366 |
| Edmondson-Steiner
grading (I/II+III) | 2/23 | 3/3 |
0.038a |
| TNM staging
(I/II/III) | 10/1/14 | 2/2/2 | 0.171 |
| Okuda staging
(1/2) | 11/14 | 3/3 | 0.872 |
| BCLC staging
(A/B/C) | 11/7/7 | 3/1/2 | 0.958 |
| CLIP scoring
(1/2/3) | 11/11/3 | 4/2/0 | 0.823 |
Discussion
Comprehensive out-of-hospital surveillance for
chronic HBV-hepatitis patients has led to an earlier diagnosis of
small lesions being precursors to malignancy (18). However, current existing tumor
markers are insufficient to diagnose HCC at early onset. As a
result, HCC is always defined as one of the most common and
aggressive malignancies and usually with a poor prognosis
worldwide. Although, liver transplantations are considered possible
curative therapies, the strict Milan criteria and the limitations
of donor availability impede patients from receiving liver
transplantations (19). In
addition, surgical resection is feasible only if the patient was
evaluated with adequate functional liver remnant and solitary mass
without major vascular invasion (20). To cure HCC patients at the
relatively early stage, serum AFP value has been mostly applied for
screening HCC, whereas the sensitivity and specificity are not
satisfied.
Previous results revealed that miRNAs in blood
circulation were markedly stable (21) and they may be potential diagnostic
and prognostic factors in diverse diseases, particularly in the
field of malignant neoplasms. Since the serum miRNA-21 was
identified as the first one for diagnosing patients with diffuse
large B cell lymphoma, and was associated with recurrence-free
survival (22), circulating miRNAs
were frequently studied as potential biomarkers for several types
of cancer. To date, some miRNAs have been reported to be associated
with the diagnosis and prognosis of liver cancer (23).
In the present study, we found that miRNA-139
expression was significantly lower in hepatocellular cancerous
tissues than in their peritumoral non-cancerous tissues in miRNA
microarray analysis. Then, the miRNA-139 expression profile was
further detected in the tissue and plasma samples of 31 HCC
patients and another 31 age- and gender-matched CH patients. Our
results demonstrated that the average value of miRNA-139 expression
level in HCC patients was 0.009, which was significantly lower than
the value of 3.516 in CH patients. To our knowledge, the present
study is the first one to identify miRNA-139 expression profiles
both in HCC patients and CH patients. Low miRNA-139 level has been
reported in digestive malignant tumor, such as gastric (24) and colorectal cancer (25), and also in adrenocortical carcinomas
(26), parathyroid cancer (27), and squamous cell carcinoma in tongue
(28). Thus, we consider that the
downregulation of plasma miRNA-139 expression may be a common event
in malignancies. In terms of liver cancer, Wong et al
(29) first reported low-expression
of miRNA-139 in HCC may suppress metastasis and progression of
cancer cells by downregulating Rho-kinase 2. The following year,
Professor Wong’s study team indicated that miRNA-139 is a
tumor-suppressor miRNA, and enhancer of zeste homolog 2 (EZH2) may
be responsible for the downregulation of the miRNAs in human HCCs
(30). Based on the aforementioned
findings, we further hypothesized that miRNA-139 may be excreted
into the extracellular space and it may also be observed in blood.
In the present study, we confirmed the correlation between
miRNA-139 expression in plasma and cancerous tissue. Many studies
have revealed the correlation between plasma miRNAs and cancerous
tissue miRNAs, and have proposed the viewpoint that tissue
intracellular miRNAs would be released into circulation during
tumorigenesis, which was accompanied by pathological injury or
cellular destruction (21,22). However, due to the sample size and
the inconsistent data of certain patients in correlation analysis,
the correlation coefficient of miRNA-139 expression in plasma and
cancerous tissue was only 0.412. Further verification of our
findings is required in a large population.
ROC curves for the diagnostic value of plasma
miRNA-139 yielded an AUC of 0.761 with the sensitivity of 80.6% and
the specificity of 58.1% in HCC diagnosis. Meanwhile, in the same
population, AUC of serum AFP was 0.700 at the cut-off value of
17.05 ng/ml, with the sensitivity of 51.6 and the specificity of
79.3%. However, the combination of miRNA-139 and AFP increased the
sensitivity to 90.3 and the specificity to 87.1%, while the AUC was
0.770, which was considerably better than miRNA-139 or AFP
alone.
For a better understanding of the clinical
implications of plasma miRNA-139, we also examined the correlations
between plasma miRNA-139 expression level and clinical features.
Edmondson-Steiner grading, which classifies HCC according to HCC
cell differentiation, morphology, and mitotic phase (15), was negatively associated with plasma
miRNA-139 expression. The plasma miRNA-139 expression level
decreased with the increase of Edmondson-Steiner grading.
Edmondson-Steiner grade was always considered to be positively
correlated with the invasion and tumor recurrence may account for
the dismal prognosis of patients with poorly differentiated HCC
(31), indicating that miRNA-139
acted as the protective agent for HCC from differentiation.
However, no association was found between miRNA-139 and clinical
staging or scoring system (including TNM staging, Okuda scoring,
BCLC staging, and CLIP scoring). Furthermore, the 1-year survival
analysis showed the HCC patients with lower miRNA-139 expression
presented shorter survival time, indicating miRNA-139 may be a
potential indicator of survival prediction for HCC patients.
In conclusion, miRNA-139 is downregulated in both
cancerous tissue and plasma of HCC. The plasma miRNA-139 is a
possible diagnostic biomarker for identifying HCC patients while
combined with other biomarkers, it is also a prognostic factor for
indicating patient survival. However, the mechanisms of miRNA-139
dysregulation due to primary expression or secondary changes,
require further investigation.
Acknowledgements
This study was supported by grant nos. 81172287 and
30901457 from the National Natural Science Foundation of China.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Srivatanakul P, Sriplung H and Deerasamee
S: Epidemiology of liver cancer: an overview. Asian Pac J Cancer
Prev. 5:118–125. 2004.
|
|
3
|
Thorgeirsson SS and Grisham JW: Molecular
pathogenesis of human hepatocellular carcinoma. Nat Genet.
31:339–346. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marrero JA and Lok AS: Newer markers for
hepatocellular carcinoma. Gastroenterology. 127(Suppl 1):
S113–S119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jannot G and Simard MJ: Tumour-related
microRNAs functions in Caenorhabditis elegans. Oncogene.
25:6197–6201. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Meng F, Henson R, Lang M, et al:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xia L, Zhang D, Du R, et al: miR-15b and
miR-16 modulate multidrug resistance by targeting BCL2 in human
gastric cancer cells. Int J Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Budhu A, Jia HL, Forgues M, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Calin GA, Ferracin M, Cimmino A, et al: A
MicroRNA signature associated with prognosis and progression in
chronic lymphocytic leukemia. N Engl J Med. 353:1793–1801. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo X, Burwinkel B, Tao S and Brenner H:
MicroRNA signatures: novel biomarker for colorectal cancer? Cancer
Epidemiol Biomarkers Prev. 20:1272–1286. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative CT method. Nat
Protoc. 3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marrero JA, Feng Z, Wang Y, et al:
α-fetoprotein, des-γ carboxyprothrombin, and lectin-bound
α-fetoprotein in early hepatocellular carcinoma. Gastroenterology.
137:110–118. 2009.
|
|
14
|
Shih WL, Chang HC, Liaw YF, et al:
Influences of tobacco and alcohol use on hepatocellular carcinoma
survival. Int J Cancer. 131:2612–2621. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Edmondson HA and Steiner PE: Primary
carcinoma of the liver: a study of 100 cases among 48,900
necropsies. Cancer. 7:462–503. 1954. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Child CG and Turcotte JG: Surgery and
portal hypertension. Major Probl Clin Surg. 1:1–85. 1964.
|
|
17
|
No authors listed. A new prognostic system
for hepatocellular carcinoma: a retrospective study of 435
patients: the Cancer of the Liver Italian Program (CLIP)
investigators. Hepatology. 28:751–755. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sherman M: Approaches to the diagnosis of
hepatocellular carcinoma. Curr Gastroenterol Rep. 7:11–18. 2005.
View Article : Google Scholar
|
|
19
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar
|
|
20
|
Maluccio M and Covey A: Recent progress in
understanding, diagnosing, and treating hepatocellular carcinoma.
CA Cancer J Clin. 62:394–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mitchell PS, Parkin RK, Kroh EM, et al:
Circulating microRNAs as stable blood-based markers for cancer
detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lawrie CH, Gal S, Dunlop HM, et al:
Detection of elevated levels of tumour-associated microRNAs in
serum of patients with diffuse large B-cell lymphoma. Br J
Haematol. 141:672–675. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tomimaru Y, Eguchi H, Nagano H, et al:
Circulating microRNA-21 as a novel biomarker for hepatocellular
carcinoma. J Hepatol. 56:167–175. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bao W, Fu HJ, Xie QS, et al: HER2
interacts with CD44 to up-regulate CXCR4 via epigenetic silencing
of microRNA-139 in gastric cancer cells. Gastroenterology.
141:2076–2087. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen K, Liang Q, Xu K, et al: MiR-139
inhibits invasion and metastasis of colorectal cancer by targeting
the type I insulin-like growth factor receptor. Biochem Pharmacol.
84:320–330. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schmitz KJ, Helwig J, Bertram S, et al:
Differential expression of microRNA-675, microRNA-139–3p and
microRNA-335 in benign and malignant adrenocortical tumours. J Clin
Pathol. 64:529–535. 2011.PubMed/NCBI
|
|
27
|
Corbetta S, Vaira V, Guarnieri V, et al:
Differential expression of microRNAs in human parathyroid
carcinomas compared with normal parathyroid tissue. Endocr Relat
Cancer. 17:135–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wong CC, Wong CM, Tung EK, et al: The
microRNA miR-139 suppresses metastasis and progression of
hepatocellular carcinoma by down-regulating Rho-kinase 2.
Gastroenterology. 140:322–331. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Au SL, Wong CC, Lee JM, et al: Enhancer of
zeste homolog 2 epigenetically silences multiple tumor suppressor
microRNAs to promote liver cancer metastasis. Hepatology.
56:622–631. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ker CG, Chen HY, Chen KS, et al: Clinical
significance of cell differentiation in hepatocellular carcinoma.
Hepatogastroenterology. 50:475–479. 2003.PubMed/NCBI
|