Introduction
ADP-ribosylation reactions can be divided into four
major groups: mono-ADP-ribosylation, poly-ADP-ribosylation,
ADP-ribose cyclization and formation of O-acetyl-ADP-ribose
(1). Mono-ADP-ribosylation and
poly-ADP-ribosylation are common in both prokaryotes and eukaryotes
and serve as post-translational modification proteins in which the
ADP-ribose is transferred from NAD+ to a specific amino
acid in a target protein (2,3).
Mono-ADP-ribosylation reactions transfer a single
ADP-ribose to various amino acid (arginine, histidine, diphthamid,
cysteine, asparagine) residues by mono-ADP-ribosyl transferases
(ARTs) (4,5). Arginine-specific ADP-ribosyl
transferase 1 (ART1), a member of the arginine-specific ARTs, was
initially found to be expressed and activated in eukaryons
(6–8), and has been mainly researched in
regards to inflammation (9). Yau
et al reported that MIBG, a specific inhibitor of
arginine-dependent mARTs, could inhibit smooth muscle cell (SMC)
proliferation and migration via Rho-dependent signaling (10). However, it has been proven that the
Rho kinase-associated coil-containing protein kinase (RhoA/ROCK)
pathway plays a central role in tumor progression (11). Several reports indicate evidence
that inhibition of the Rho/ROCK signaling pathway induces
hepatocellular cancer (HCC) cell apoptosis (12,13).
Unlike mono-ADP-ribosylation, poly-ADP-ribosylation
transforms NAD+ to nicotinamide and ADP-ribose to form
long and branched (ADP-ribose) polymers on glutamic acid residues
by poly(ADP-ribose) polymerase (PARP). PARP-1 is the principal
member of the PARP enzymes and has been implicated in both the
prevention and aggravation of various diseases (14). Expression of PARP-1 plays a central
role in the cellular response to DNA damage (6), whereas the diverse physiological
functions of PARP-1 in cell survival, several forms of cell death,
gene transcription, immune responses, inflammation, learning,
memory, synaptic functions, angiogenesis and aging have been widely
appreciated recently (14). Our
previous study demonstrated that downregulation of PARP-1 by RNA
interference of PARG inhibits the migration, proliferation and
metastatic potential of colon carcinoma cells (15–18).
Ye reported that a PARP-1 inhibitor enhances the efficacy of
DNA-damaging anticancer drugs by inducing cancer cell apoptosis,
and inhibits necrosis in normal cells and neurons from the toxic
effects of DNA-damaging anticancer drugs, both through processes
that prevent the significant loss of ATP (19).
Pioneer observations found that mART could mediate
the signal transduction from the cell surface to the nucleus
(11), and our previously study
demonstrated that both the expression of ART1 and PARP-1 was
significantly increased in colon carcinoma tissues when compared
with normal colon tissues, and inhibition of ART1 downregulated the
expression of PARP-1 possibly by regulation of NF-κB (20). However, the interaction of ART1 and
PARP-1 in the process of apoptosis has not yet been elucidated. In
the present study, we aimed to research their effect on apoptosis
induced by cisplatin (CDDP) and the possible mechanism in murine
colon carcinoma CT26 cells (21,22).
Materials and methods
Cell line and reagents
CT26, a murine colon carcinoma cell line, was kindly
provided by Professor Y.Q. Wei, Huaxi Hospital, Sichuan University.
ART1-shRNA and control-shRNA were successfully transfected in CT26
cells by us in early experiments (20). The lentiviral vector was obtained
from GeneChem (Shanghai, China). The Annexin V-PE/7-AAD apoptosis
detection kit was from KeyGen Biotechnology Co., Ltd. (Nanjing,
China). Y-27632 was purchased from Santa Cruz Biotechnology (Santa
Cruz, CA, USA). CDDP was obtained from Sigma (St. Louis, MO, USA).
5-Aminoisoquinoline was kindly supplied by Professor M.D.
Threadgill, Bath University, UK. The BCA protein assay kit was from
Beyotime (Shanghai, China).
Cell culture
The murine CT26 colon carcinoma cell line was
cultured in RPMI-1640 medium supplemented with 10% fetal bovine
serum (FBS), 100 μg/ml streptomycin (Thermo Pierce) at 37°C in a 5%
CO2 incubator. CDDP was prepared in 1640 at a final
concentration of 1 mM and stored at 4°C away from light.
Transfection of CT26 cells and
identification
Mouse ART1-cDNA sequence (accession no. NC_000073)
was obtained from GenBank, and the primers of ART1-cDNA were
designed as follows: lentiviral vector-mediated ART1-cDNA primer 1,
GAGGATCCCCGGGTACCGGTCGCCACCATGAAGATTCCTGCTATGATG; lentiviral
vector-mediated ART1-cDNA primer 2,
TCACCATGGTGGCGACCGGACATCGGGTAAGTTGCTG, and was successfully
constructed by GeneChem. Transfection was carried according to the
manufacturer’s instructions. Cells were cultured in 12-well plates
at a density of 3×104 cells/well when they were in the
logarithmic growth phase. Lentivirus particles (10 μl) were added
to each well while the cells covered 50% of each well. Transfection
efficiency was optimized using green fluorescent protein and
detected under a fluorescence microscope after 72 h. The efficiency
of the ART1-cDNA lentivirus transfected into CT26 cells was
determined by reverse transcriptase (RT)-PCR and western blot
analysis.
RNA was extracted from ART1-cDNA CT26 cells with
TRIzol reagent according to the supplied manual (Takara, Dalian,
China), and RNAs were extracted from ART1-shRNA CT26 cells,
control-shRNA CT26 cells and untransfected CT26 cells used as a
control. ART1 (target gene) and β-actin (internal control gene)
were detected using oligonucleotide primers which were designed and
produced by Sangon Biotech Co. (Shanghai, China): ART1,
5′-ACCTTCTTCGGTATCTGGACCT-3′ (F1) and 5′-TAAGTTGCTGGAGACCTGGATT-3′
(R1); β-actin, 5′-ATATCGCTGCGCTGGTCGTC-3′ (F1) and
5′-AGGATGGCGTGAGGGAGAGC-3′ (R1). The cycling conditions were as
follows: the number of PCR cycles (94°C for 30 sec, 50–58°C for 30
sec, 72°C for 1 min and then 5 min for the last extension) was 30
for the amplification of reverse transcriptase products. Finally,
the PCR amplification products were separated on 2.0% agarose gel
(Genview, Tallahasses, FL, USA). This experiment was performed in
triplicate.
Effect of ART1 and PARP-1 on CT26 cell
apoptosis induced by CDDP
ART1-cDNA CT26 cells were treated with or without
CDDP. ART1-shRNA CT26 cells, control-shRNA CT26 cells and
untransfected CT26 cells were treated in the same manner as were
the control. CT26 cells were cultured in 6-well plates at
1×105 cells/well for 24 h and 30 μM of CDDP for a
further 48 h for inducing apoptosis. To confirm the role of PARP-1
in this apoptosis process, ART1-cDNA CT26 cells were treated with
PARP-1 inhibitor 5-AIQ at 100 μM for 24 h and CDDP at 30 μM for 48
h or ART1-cDNA CT26 cells were treated with CDDP only at 30 μM for
48 h or were untreated as control groups. The apoptosis rate was
tested for each cell group by flow cytometric analysis and Hoechst
33342 staining.
For flow cytometric analysis, cells were
trypsinized, washed twice with phosphate-buffered saline (PBS)
before PE-conjugated Annexin V and 7-ADD were added, incubated for
30 min and analyzed by flow cytometry (Becton-Dickinson). As for
Hoechst 33342 staining, cells were washed twice with PBS, and
stained with 5 μl Hoechst 33342 for 30 min according to the
manufacturer’s instructions (Beyotime). Stained nuclei were
observed under a fluorescence microscope. The percentage of
apoptotic cells, the apoptotic ratio (AR), was calculated using the
formula: AR% = [apoptotic cells (A)/total cell count (T)] × l00.
(Ten images from each sample were acquired and analyzed in three
different experiments). All experiments were performed three
times.
Detection of relevant apoptosis-related
proteins
Western blotting was used to detect the expression
of ART1, RhoA, ROCK1, NF-κB, Cox-2, caspase 3, PARP-1 and PARP-1
cleavaged fragments. Total and nuclear proteins in the above groups
were extracted according to protein extraction protocols (Beyotime
P0013 and P0028). Protein concentrations were determined using the
BCA assay (Beyotime), and respective proteins (30 μg/lane) were
loaded onto 6% polyacrylamide gel (PARP-1, ROCK1) or 10%
polyacrylamide gel (ART1, RhoA, Cox-2, caspase 3 and NF-κB).
Proteins were separated by electrophoresis, transferred to
polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA)
and blocked with 5% non-fat dry milk for 1 h at room temperature.
Primary antibodies against ART1, PARP-1, NF-κB (Bioworld, St. Louis
Park, MN, USA); RhoA, ROCK1, Cox-2, caspase 3 (Proteintech Group,
Chicago, IL, USA); β-actin (Boster, Wuhan, China) were diluted to
1:500 and incubated at 4°C overnight. The next day, the membranes
(37°C 30 min) were washed thoroughly 3 times for 10 min with
Tris-buffered saline and Tween-20 (TBST). Secondary antibodies
(peroxidase-conjugated goat or anti-rabbit IgG) (Boster) diluted to
1:1,000 were added and incubation was carried out for 2 h at 37°C,
and washed thoroughly for 15 min with TBST. The bands were detected
by enhanced chemiluminescence method (Beyotime).
To verify the relationship of ART1, PARP-1 and
ROCK1, untransfected CT26 cells were treated with PARP-1 inhibitor
5-AIQ (100 μM) and ROCK inhibitor Y-27632 (20 μM) (23) for 24 h, respectively. Total protein
was extracted and detected by western blotting. All experiments
were repeated three times.
Data measurement and statistical
analysis
Data are expressed as the means ± standard deviation
(SD) in the quantitative experiments. Statistical analysis was
performed by Student’s t-test or one-way ANOVA. Values of P<0.05
were considered to indicate statistically significant results.
Results
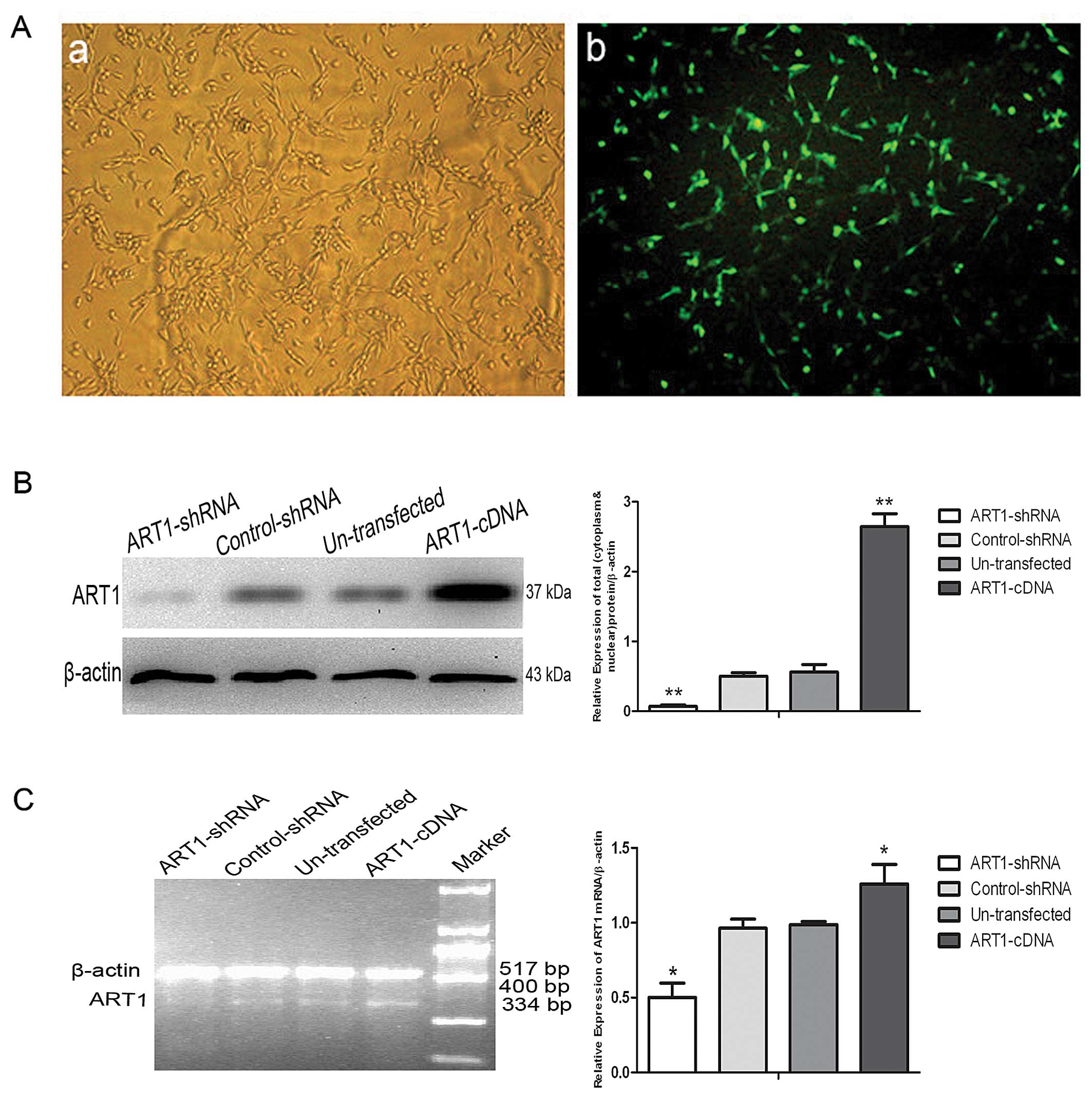
Transfection of lentiviral
vector-mediated ART1-cDNA in CT26 cells
The ratio of lentiviral vector-mediated ART1-cDNA
transfected in the CT26 cells was detected by fluorescence
microscope and indicated that ~80% of CT26 cells were successfully
transfected (Fig. 1A). Western blot
assay and RT-PCR were used to identify the efficiency of the
ART1-cDNA lentivirus transfection in the CT26 cells. The cells
transfected with the lentiviral vector-mediated ART1-cDNA revealed
an increase in ART1 expression when compared to the expression
level in the ART1-shRNA, control-shRNA and untransfected CT26 cells
(Fig. 1B and C; P<0.01 and
P<0.05, respectively).
Effect of ART1 and PARP-1 on CT26 cell
apoptosis induced by CDDP
Based on the flow cytometric analysis, the results
showed that the apoptosis rate increased in each group of CT26
cells induced by CDDP, while the ART1-cDNA CT26 cells induced by
CDDP had a lower apoptosis rate (16.39%), compared with the
ART1-shRNA CT26 cells (80.03%), the untransfected CT26 cells
(50.03%) and the control-shRNA CT26 cells (51.17%) treated in the
same manner (Fig. 2A). The
apoptosis rate of the ART1-cDNA CT26 cells treated with 5-AIQ and
CDDP obviously increased (25.71%), compared to the ART1-cDNA CT26
cells treated with CDDP only (13.23%) and the untreated ART1-cDNA
CT26 cells (4.56%) (P<0.05) (Fig.
2B).
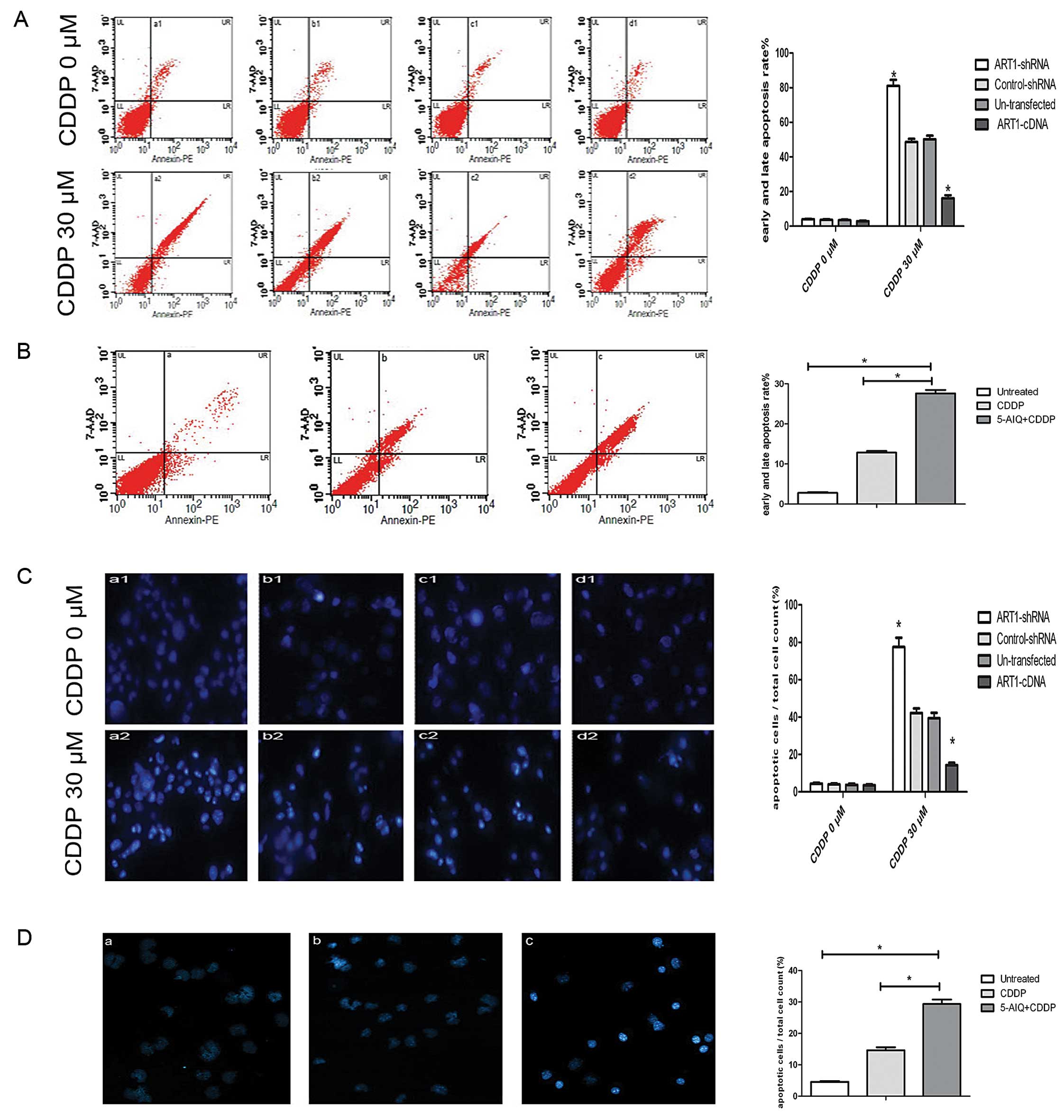 | Figure 2PARP-1 is involved in the
ART1-mediated CT26 cell apoptosis induced by CDDP. (A and C)
ART1-cDNA CT26 cells were untreated (a1) or treated with CDDP (a2),
compared with ART1-shRNA CT26 cells (b1 and b2), control-shRNA CT26
cells (c1 and c2) and untransfected CT26 cells (d1 and d2) treated
in the same manner. a1–d1, untreated cells; a2–d2, cells treated
with CDDP (30 μM) for 48 h. The apoptosis rate was detected by flow
cytometric analysis and Hoechst 33342 staining. (B and D) ART1-cDNA
CT26 cells were treated with 5-AIQ (100 μM) for 24 h and then CDDP
(30 μM) for 48 h (c) or ART1-cDNA CT26 cells were treated with CDDP
(30 μM) for 48 h only (b), or ART1-cDNA CT26 cells were untreated
(c) and apoptosis was detected by flow cytometric analysis and
Hoechst 33342 staining. Diagrams represent the average of three
independent experiments. All experiment were performed three times
(*P<0.05). (In A and B: UL, necrotic cells; LL,
viable cells; LR, early apoptosis; UR, late apoptosis, in
percentages %). PARP-1, poly(ADP-ribose) polymerase-1; ART1,
arginine-specific ADP-ribosyltransferase 1; CDDP, cisplatin; 5-AIQ,
5-aminoisoquinoline. |
The results of Hoechst 33342 staining showed that
apoptotic bodies were significantly increased in each group of CT26
cells treated with CDDP. Moreover, after treatment with CDDP, the
apoptosis rate in the ART1-cDNA CT26 cells declined (15.2%),
compared with the ART1-shRNA CT26 cells (77.8%), the control-shRNA
CT26 cells (37.5%) and the untransfected CT26 cells (35.8%)
(Fig. 2C). ART1-cDNA CT26 cells
treated with 5-AIQ and CDDP had a high apoptosis rate (25.3%),
compared with the ART1-cDNA CT26 cells treated with CDDP only
(14.03%) and the untreated ART1-cDNA CT26 cells (5.06%) (P<0.05)
(Fig. 2D).
Influence of ART1 on the expression of
RhoA, ROCK1, NF-κB, Cox-2, caspase 3, PARP-1 and PARP-1 cleavage
fragments
Among the groups of cells treated with CDDP, the
expression of RhoA, ROCK1, NF-κB, Cox-2 and PARP-1 was increased in
the ART1-cDNA CT26 cells, when compared to these levels in the
ART1-shRNA CT26 cells, the control-shRNA CT26 cells and the
untransfected CT26 cells, while the expression of caspase 3 and
PARP-1 cleavage fragment (p89) had an opposite trend. The
expression of RhoA, ROCK1, NF-κB, Cox2 and PARP-1 had the same
trend in cells not treated with CDDP, except for the PARP-1
cleavage fragment (p89) which was the cleavage substrate for
activated caspase 3 which was noted in the CDDP-treated CT26 cells
only (P<0.05, P<0.01) (Fig
3).
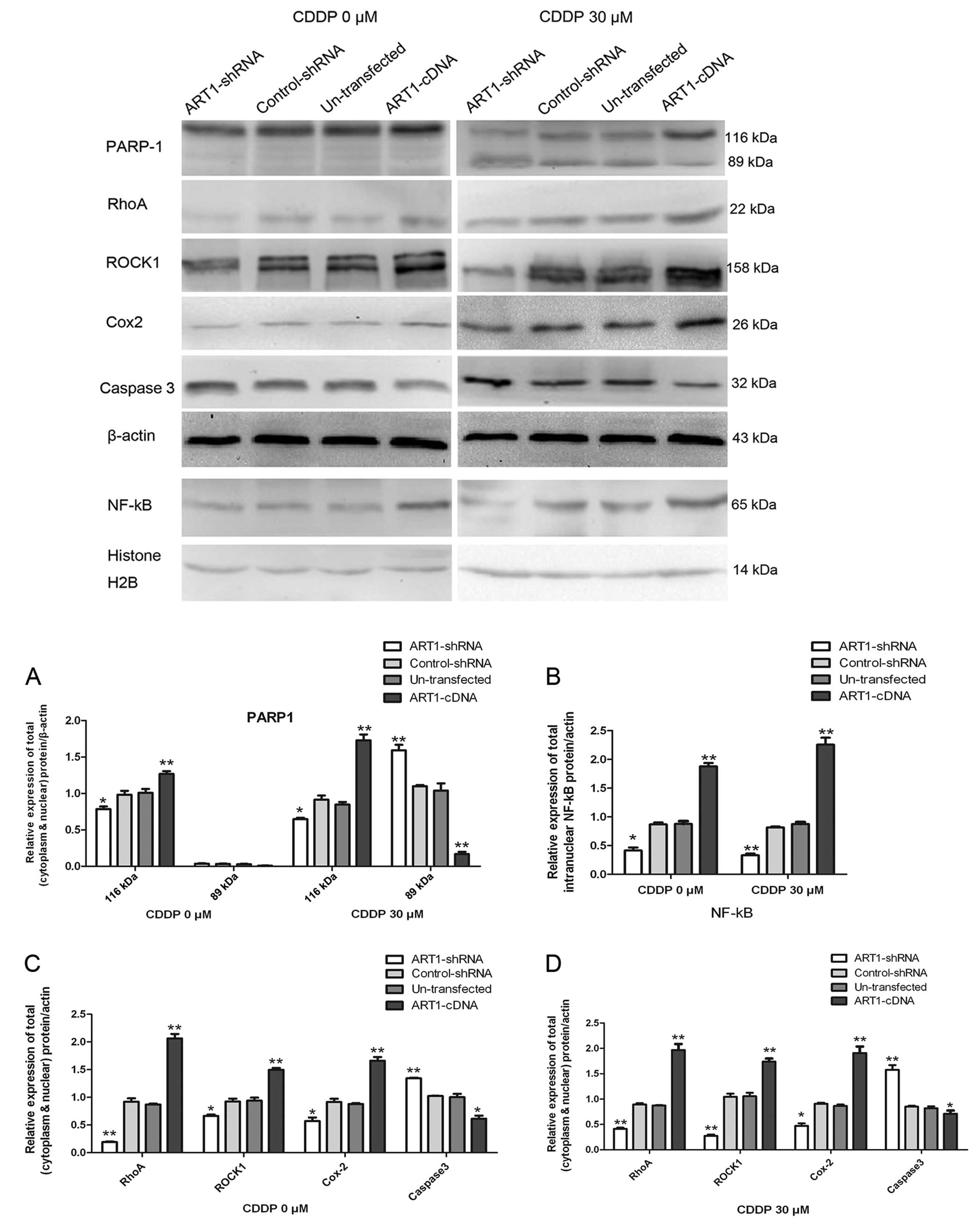 | Figure 3ART1 affects PARP-1, RhoA, ROCK1,
NF-κB, Cox2 and caspase 3 expression in CT26 cells. (A) Expression
of PARP-1 and its cleavage fragment (p89) in ART1-cDNA CT26 cells
treated with or without CDDP (30 μM) for 48 h, compared with the
expression level in the ART1-shRNA CT26 cells, the control-shRNA
CT26 cells and the untransfected CT26 cells treated in the same
manner. (B) The effect of ART1 on the expression of NF-κB in the
ART1-cDNA CT26 cells treated with or without CDDP (30 μM) for 48 h,
compared with the expression level in the ART1-shRNA CT26 cells,
the control-shRNA CT26 cells and the untransfected CT26 cells
treated in the same manner. (C and D) The effect of ART1 on the
expression of RhoA, ROCK1, Cox-2, caspase 3 in the ART1-cDNA CT26
cells treated with or without CDDP (30 μM) for 48 h, compared with
the expression levels in the ART1-shRNA CT26 cells, the
control-shRNA CT26 cells and the untransfected CT26 cells treated
in the same manner. All experiments were repeated three times
(*P<0.05, **P<0.01). ART1,
arginine-specific ADP-ribosyltransferase 1; PARP-1,
poly(ADP-ribose) polymerase-1; ROCK, Rho kinase-associated
coil-containing protein kinase; NF-κB, nuclear factor-κB; CDDP,
cisplatin. |
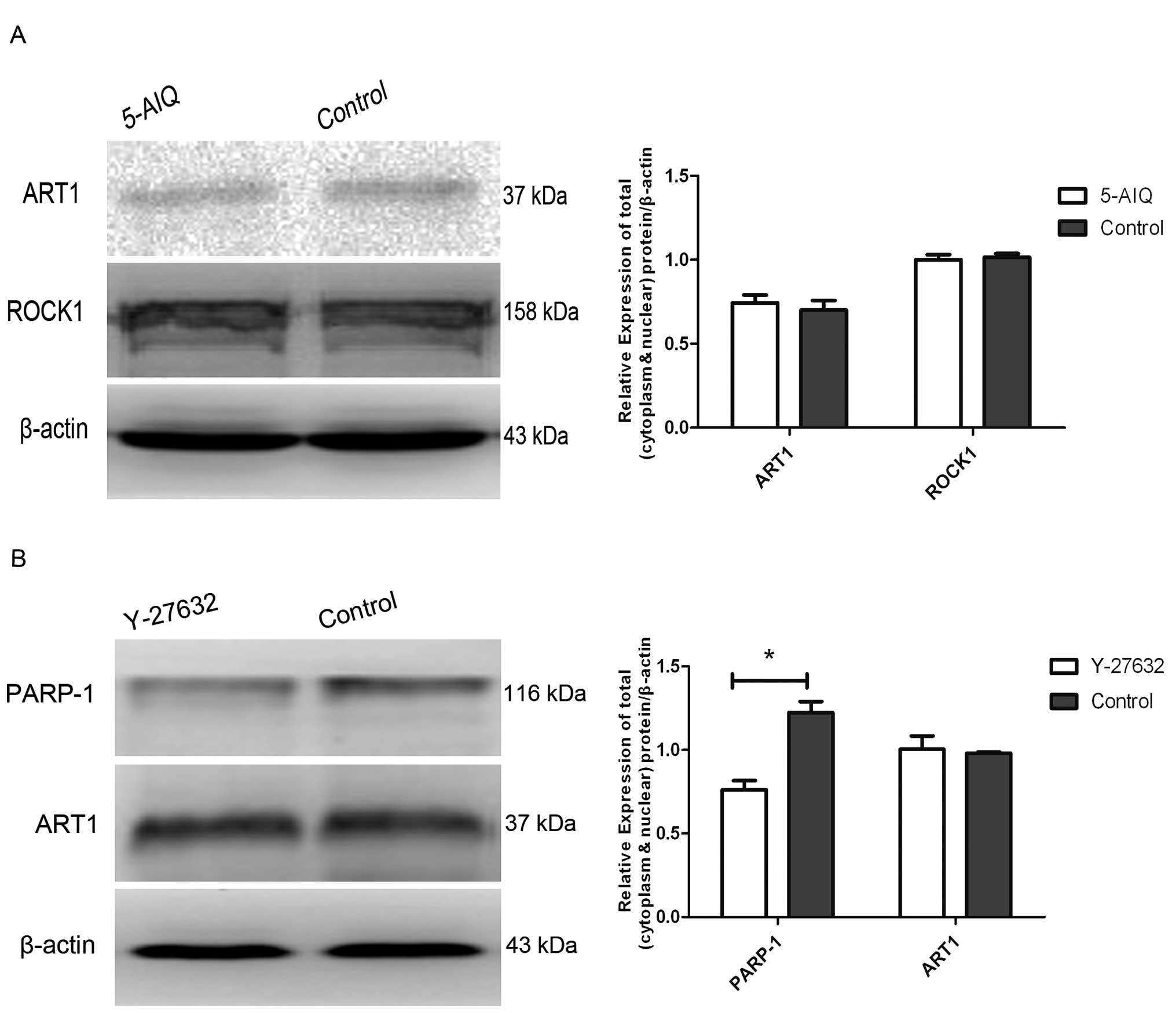
Influence of 5-AIQ and Y-27632 on the
expression of ART1, ROCK1 and PARP-1 in the untransfected CT26
cells
The expression of ROCK1 and ART1 in the 5-AIQ
treated untransfected CT26 cells exhibited no obvious changes when
compared to the untreated untransfected CT26 cells. In the
untransfected CT26 cells treated with Y-27632, the expression of
PARP-1 was significantly decreased while the expression of ART1
exhibited no obvious change (P<0.05) (Fig. 4).
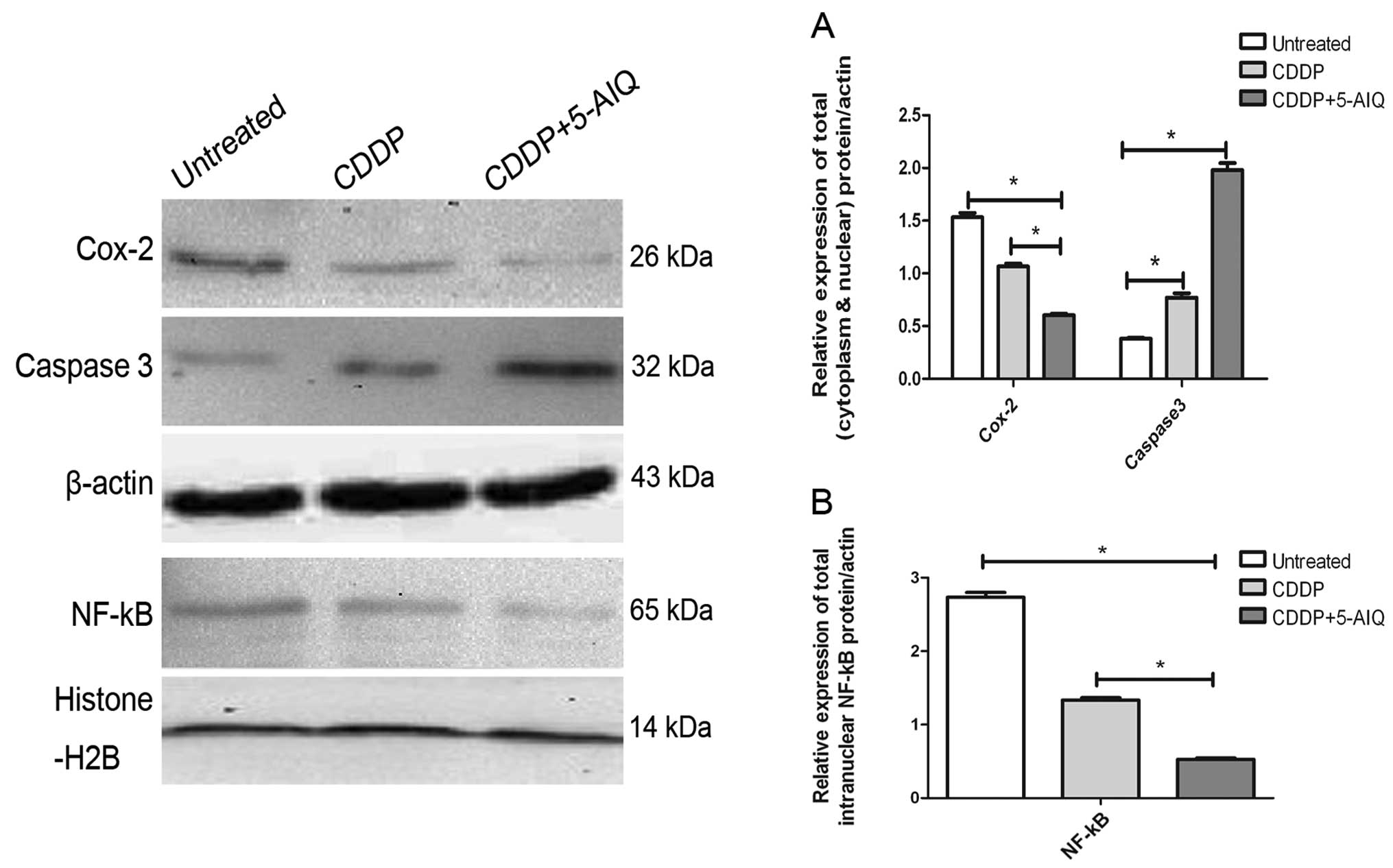
Influence of PARP-1 inhibitor 5-AIQ on
the expression of NF-κB, Cox-2 and caspase 3 in the ART1-cDNA CT26
cells
There was an obvious decreas in NF-κB and Cox-2 and
an increase in caspase 3 in the ART1-cDNA CT26 cells treated with
5-AIQ and CDDP, when compared to the ART1-cDNA CT26 cells treated
with CDDP only and the untreated ART1-cDNA CT26 cells (P<0.05)
(Fig. 5).
Discussion
ADP-ribosylation reactions are currently topics of
intense interest. A tremendous amount of research has been carried
out to decipher the physiological and pathophysiological roles of
ADP-ribosylation reactions at the molecular level. Many research
groups with a wide range of expertise have become involved in
mono-ADP-ribosylation or poly-ADP-ribosylation research (5). It has been determined that both ART1
and PARP-1 play important roles in reticular biological processes,
and ART1 influences the expression of PARP-1 through NF-κB.
However, despite the progress made in recent years in the
biochemistry, molecular biology, physiology and pathophysiology of
ADP-ribosylation, no unified image of the physiological and
pathophysiological correlation of mono- and poly-ADP-ribosylation
in the field of apoptosis has been noted (1). In the present study, the role of ART1
and PARP-1 in CDDP-induced CT26 cell apoptosis was studied. The
results showed that a high level of ART1 expression decreased the
apoptosis rate of CT26 cells induced by CDDP. Moreover, in the
ART1-cDNA CT26 cells treated with 5-AIQ and CDDP an increase in the
apoptosis rate was noted when compared with the ART1-cDNA CT26
cells treated with CDDP only and the untreated ART1-cDNA CT26
cells, which indicated that PARP-1 plays a role in the process of
ART1-mediated apoptosis in CDDP-treated CT26 cells.
Several studies have shown that inhibition of the
RhoA/ROCK pathway increases the apoptosis of human hepatocellular
carcinoma cells (HCC) and endothelial cells through a mitochondrial
apoptosis pathway (12,13). Benitah et al identified that
ROCK is necessary for RhoA-induced expression of Cox-2 at the
transcriptional level through the RhoA/ROCK/NF-κB/COX-2 signaling
pathway (24), and the
Cox-2-specific inhibitor celecoxib induced apoptosis through a
mitochondrial pathway to suppress Akt phosphorylation, to release
cytochrome c to the cytosol and to activate caspase 3
(25,26). Caspase 3 mediates highly specific
proteolytic cleavage events in dying cells, which collectively
manifest the apoptotic phenotype (27). Moreover, inhibition of Cox-2 was
also shown to inhibit NF-κB by directly blocking the activity of
IκB kinase; these feedback loops of the inhibition of Cox-2
mediated inactivation of NF-κB leading to a reconstituted
sensitivity of cancer cells to apoptosis (28). Thus, RhoA/ROCK regulates apoptosis
through an NF-κB/Cox2/caspase 3 pathway. Yau et al reported
that the RhoA/ROCK pathways could be regulated by
ADP-ribosyltransferase (10).
However, the relationship of these factors with ART1 has not been
reported in CDDP-induced colon carcinoma CT26 cell apoptosis. In
the present study, we analyzed these factors, and the results
showed that the expression of RhoA, ROCK1, NF-κB and Cox-2 was
increased in the ART1-cDNA CT26 cells when treated with CDDP, while
caspase 3 had the opposite trend. Thus, we conclude that ART1
probably regulates apoptosis through the
RhoA/ROCK1/NF-κB/Cox2/caspase 3 pathway.
It has been demonstrated that PARP-1 acts as a
co-activator of NF-κB, and the PARP-1 inhibitor decreases the
expression of NF-κB and inhibition of NF-κB could decrease PARP-1
expression through a feedback manner (29–31);
whereas inhibition of PARP-1 is thought to impair DNA repair
function and promote cellular dysfunction and death (32). In models of acute stress, the PARP-1
inhibitor may protect cellular NAD pools and prevent
NF-κB-dependent inflammatory signaling pathway. Research also found
that PARP-1 inhibitor can trigger apoptosis in cancer cells and
inhibit necrosis in normal cells and neurons through a
mitochondrial pathway (33,34). Yau reported that mART could mediate
the signal transduction from the cell surface to the nucleus
(11). Our previous study showed
that ART1 regulated the expression of PARP-1 probably through
NF-κB, while PARP-1 inhibitor 5-AIQ had no effect on ART1 (20). In the present study, we also found
that the expression of PARP-1 was decreased in the ART1-shRNA CT26
cells and was increased in the ART1-cDNA CT26 cells, which is
accordance to the trend of NF-κB. From these findings we can
conclude that ART1 affects the expression of PARP-1 probably
through the regulation of NF-κB.
To clarify the relationship among ART1, ROCK1 and
PARP-1, Y-27632 and 5-AIQ were used to treat the untransfected CT26
cells. Results showed that there was no obvious change in ROCK1 and
ART1 in the 5-AIQ-treated untransfected CT26 cells, and the
expression of PARP-1 in the Y-27632-treated untransfected CT26
cells was significantly decreased while the expression of ART1 had
no obvious change. These results demonstrate that ROCK1 is in the
downstream of ART1 while PARP-1 is regulated by ROCK1. To further
verify the role of PARP-1 in ART1-mediated apoptosis in CT26 cells
treated with CDDP, we used 5-AIQ to treat ART1-cDNA CT26 cells and
determined the expression of NF-κB, Cox-2 and caspase 3. Results
showed that there was an obvious decrease in NF-κB and Cox-2 and an
increase in caspase 3 in the 5-AIQ-treated ART1-cDNA CT26 cells
treated with CDDP, which demonstrated that PARP-1 is involved in
CDDP-induced apoptosis in CT26 cells through the
NF-κB/Cox-2/caspase 3 pathway.
Moreover, Li and Yuan reported that PARP-1 was one
of the first identified substrates of caspase 3 (35). During apoptosis, activated caspase 3
cleaves PARP-1 into two fragments, p89 and p24, resulting in the
inactivation of PARP-1. The cleavage fragments lead to the
suppression of PARP-1 activity by inhibiting the homoassociation
and DNA binding of intact PARP-1, respectively (36–38),
preserving cellular energy for certain ATP-sensitive steps of
apoptosis (39–41). In the present study, the results
also showed that caspase 3 was activated and PARP-1 cleavage
fragments formed when the cells were treated with CDDP. However, in
the untreated cells, caspase 3 was inactivated for there were no
PARP-1 cleavage fragments (p89) observed. Compared to the control
groups, more PARP-1 cleavage fragments were formed in the
ART1-shRNA CT26 cells treated with CDDP and less were formed in the
ART1-cDNA CT26 cells treated with CDDP. These results suggest that
ART1 activates caspase 3 which can cleave PARP-1 into p24 and p89
fragments when induced by CDDP, and this reaction leads to a
decreased level and inactivation of PARP-1, ending in the promotion
of CT26 cell apoptosis.
In brief, this investigation is an initial research
on the synergistic effect of ART1 and PARP-1 on apoptosis induced
by CDDP in murine colon carcinoma CT26 cells. The results showed
that PARP-1 is in the downstream of ART1, and plays a role in
ART1-mediated CT26 cell apoptosis through the ROCK1/NF-κB/PARP-1
pathway when induced by CDDP. In contrast, in CDDP-induced cells,
activated caspase 3 cleaved PARP-1 and the decrease in PARP-1 in
turn decreased the expression of NF-κB, and Cox-2 and increased
caspase 3, resulting in the enhanced ability of ART1-mediated CT26
cell apoptosis. However, despite our findings, further study is
needed to detect the detail mechanism.
Acknowledgements
This study was supported by the Ministry of
Education Specialized Research Fund for the Doctoral Program of
Higher Education (grant no. 20105503110009), and the Science and
Technology Project of the Education Commission of Chongqing (grant
no. KJ110322).
References
|
1
|
Hassa PO, Haenni SS, Elser M and Hottiger
MO: Nuclear ADP-ribosylation reactions in mammalian cells: where
are we today and where are we going? Microbiol Mol Biol Rev.
70:789–829. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Otto H, Reche PA, Bazan F, Dittmar K, Haag
F and Koch-Nolte F: In silico characterization of the family
of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genomics.
6:1392005. View Article : Google Scholar
|
|
3
|
Ueda K and Hayaishi O: ADP-ribosylation.
Annu Rev Biochem. 54:73–100. 1985. View Article : Google Scholar
|
|
4
|
Corda D and Di Girolamo M:
Mono-ADP-ribosylation: a tool for modulating immune response and
cell signaling. Sci STKE. 2002:pe532002.PubMed/NCBI
|
|
5
|
Corda D and Di Girolamo M: Functional
aspects of protein mono-ADP-ribosylation. EMBO J. 22:1953–1958.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao Z, Gruszczynska-Biegala J and
Zolkiewska A: ADP-ribosylation of integrin α7 modulates the binding
of integrin α7β1 to laminin. Biochem J. 385:309–317. 2005.
|
|
7
|
Okazaki IJ, Zolkiewska A, Nightingale MS
and Moss J: Immunological and structural conservation of mammalian
skeletal muscle glycosylphosphatidylinositol-linked
ADP-ribosyltransferases. Biochemistry. 33:12828–12836. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zolkiewska A, Nightingale MS and Moss J:
Molecular characterization of NAD:arginine ADP-ribosyltransferase
from rabbit skeletal muscle. Proc Natl Acad Sci USA.
89:11352–11356. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Paone G, Stevens LA, Levine RL, et al:
ADP-ribosyltransferase-specific modification of human neutrophil
peptide-1. J Biol Chem. 281:17054–17060. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yau L, Litchie B, Thomas S, Storie B,
Yurkova N and Zahradka P: Endogenous mono-ADP-ribosylation mediates
smooth muscle cell proliferation and migration via protein kinase
N-dependent induction of c-fos expression. Eur J Biochem.
270:101–110. 2003. View Article : Google Scholar
|
|
11
|
Gilcrease MZ: Integrin signaling in
epithelial cells. Cancer Lett. 247:1–25. 2007. View Article : Google Scholar
|
|
12
|
Takeba Y, Matsumoto N, Watanabe M, et al:
The Rho kinase inhibitor fasudil is involved in p53-mediated
apoptosis in human hepatocellular carcinoma cells. Cancer Chemother
Pharmacol. 69:1545–1555. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hippenstiel S, Schmeck B, N’Guessan PD, et
al: Rho protein inactivation induced apoptosis of cultured human
endothelial cells. Am J Physiol Lung Cell Mol Physiol.
283:L830–L838. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kirkland JB: Poly ADP-ribose polymerase-1
and health. Exp Biol Med. 235:561–568. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu JX, Wang YL, Tang Y and Xiong W: Effect
of ART1 gene silencing by RNA interference on the proliferation of
mouse colon carcinoma cells and its possible mechanism. TUMOR.
32:949–954. 2012.
|
|
16
|
Li Q, Li M, Wang YL, et al: RNA
interference of PARG could inhibit the metastatic potency of colon
carcinoma cells via PI3-kinase/Akt pathway. Cell Physiol Biochem.
29:361–372. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fauzee NJ, Li Q, Wang YL and Pan J:
Silencing poly (ADP-ribose) glycohydrolase (PARG) expression
inhibits growth of human colon cancer cells in vitro via
PI3K/Akt/NFκ-B pathway. Pathol Oncol Res. 18:191–199.
2012.PubMed/NCBI
|
|
18
|
Fauzee NJ, Pan J and Wang YL: PARP and
PARG inhibitors - new therapeutic targets in cancer treatment.
Pathol Oncol Res. 16:469–478. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ye K: PARP inhibitor tilts cell death from
necrosis to apoptosis in cancer cells. Cancer Biol Ther. 7:942–944.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang Y, Wang YL, Yang L, et al: Inhibition
of arginine ADP-ribosyltransferase 1 reduces the expression of
poly(ADP-ribose) polymerase-1 in colon carcinoma. Int J Mol Med.
32:130–136. 2013.PubMed/NCBI
|
|
21
|
Furuya K, Ozaki T, Hanamoto T, et al:
Stabilization of p73 by nuclear IκB kinase-α mediates
cisplatin-induced apoptosis. J Biol Chem. 282:18365–18378.
2007.
|
|
22
|
Ohnishi K, Ota I, Takahashi A, Yane K,
Matsumoto H and Ohnishi T: Transfection of mutant p53 gene
depresses X-ray- or CDDP-induced apoptosis in a human squamous cell
carcinoma of the head and neck. Apoptosis. 7:367–372. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
van Houdt WJ, Hoogwater FJ, de Bruijn MT,
et al: Oncogenic KRAS desensitizes colorectal tumor cells to
epidermal growth factor receptor inhibition and activation.
Neoplasia. 12:443–452. 2010.PubMed/NCBI
|
|
24
|
Benitah SA, Valerón PF and Lacal JC: ROCK
and nuclear factor-κB-dependent activation of cyclooxygenase-2 by
Rho GTPases: effects on tumor growth and therapeutic consequences.
Mol Biol Cell. 14:3041–3054. 2003.
|
|
25
|
Zhang Z, Lai GH and Sirica AE:
Celecoxib-induced apoptosis in rat cholangiocarcinoma cells
mediated by Akt inactivation and Bax translocation. Hepatology.
39:1028–1037. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lax AJ and Thomas W: How bacteria could
cause cancer: one step at a time. Trends Microbiol. 10:293–299.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nicholson DW and Thornberry NA: Caspases:
killer proteases. Trends Biochem Sci. 22:299–306. 1997. View Article : Google Scholar
|
|
28
|
Yin MJ, Yamamoto Y and Gaynor RB: The
anti-inflammatory agents aspirin and salicylate inhibit the
activity of IκB kinase-β. Nature. 396:77–80. 1998.
|
|
29
|
Cai L, Threadgill MD, Wang Y and Li M:
Effect of poly (ADP-ribose) polymerase-1 inhibition on the
proliferation of murine colon carcinoma CT26 cells. Pathol Oncol
Res. 15:323–328. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chiti F, Stefani M, Taddei N, Ramponi G
and Dobson CM: Rationalization of the effects of mutations on
peptide and protein aggregation rates. Nature. 424:805–808. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hassa PO and Hottiger MO: The functional
role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-κB
in inflammatory disorders. Cell Mol Life Sci. 59:1534–1553.
2002.PubMed/NCBI
|
|
32
|
Ratnam K and Low JA: Current development
of clinical inhibitors of poly(ADP-ribose) polymerase in oncology.
Clin Cancer Res. 13:1383–1388. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jagtap P and Szabó C: Poly(ADP-ribose)
polymerase and the therapeutic effects of its inhibitors. Nat Rev
Drug Discov. 4:421–440. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schreiber V, Dantzer F, Ame JC and De
Murcia G: Poly(ADP-ribose): novel functions for an old molecule.
Nat Rev Mol Cell Biol. 7:517–528. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li J and Yuan J: Caspases in apoptosis and
beyond. Oncogene. 27:6194–6206. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim JW, Kim K, Kang K and Joe CO:
Inhibition of homodimerization of poly(ADP-ribose) polymerase by
its C-terminal cleavage products produced during apoptosis. J Biol
Chem. 275:8121–8125. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim JW, Won J, Sohn S and Joe CO:
DNA-binding activity of the N-terminal cleavage product of
poly(ADP-ribose) polymerase is required for UV mediated apoptosis.
J Cell Sci. 113:955–961. 2000.PubMed/NCBI
|
|
38
|
D’Ambrosio SM, Gibson-D’Ambrosio RE, Brady
T, Oberyszyn AS and Robertson FM: Mechanisms of nitric
oxide-induced cytotoxicity in normal human hepatocytes. Environ Mol
Mutagen. 37:46–54. 2001.PubMed/NCBI
|
|
39
|
Chaitanya GV, Steven AJ and Babu PP:
PARP-1 cleavage fragments: signatures of cell-death proteases in
neurodegeneration. Cell Commun Signal. 8:312010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Soldani C and Scovassi A: Poly(ADP-ribose)
polymerase-1 cleavage during apoptosis: an update. Apoptosis.
7:321–328. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Virág L and Szabó C: The therapeutic
potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev.
54:375–429. 2002.
|