Introduction
Gastric cancer (GC) is the fourth most common
malignancy and the second leading cause of cancer-related mortality
in men worldwide (1). A total of
989,600 new cases of GC were diagnosed and 738,000 GC-related
deaths occurred in 2008, accounting for 8% of the total cases and
10% of total deaths. More than 2/3 of new cases and deaths occur in
developing countries, with the highest frequency in Eastern Asia
(2). In spite of multi-treatments
including surgery, chemotherapy, radiotherapy, and
molecular-targeted therapy, the overall 5-year survival rate of GC
patients remains extremely low. In the United States where fewer
GCs are diagnosed at an early stage, the 5-year survival rate of
stomach cancer is only ~24% (1).
The crude mortality rate of GC in China was 25.2 per 100,000
individuals, which accounted for 23.2% of the total cancer-related
deaths in 1990–1992 (3). Therefore,
there is an urgent need to elucidate the molecular mechanisms of GC
and to develop novel therapeutic strategies to treat GC.
Dysregulation of gene expression is associated with
the neoplastic process, and there is compelling evidence
implicating differential expression of multiple genes in the
development and progression of GC. S100P belongs to the S100 family
of proteins. S100 is a family of calcium-binding proteins
characterized by EF-hand motifs. The name S100 is due to the 100%
solubility of these proteins in a 100% saturated ammonium sulphate
solution. The S100 proteins form homodimers or heterodimers and are
involved in multifunctional cellular activities including protein
phosphorylation, gene transcription, cell proliferation, survival
and apoptosis (4). S100P is one of
the least studied members of this family. S100P is a 95-amino acid
protein, which was first purified and characterized from placenta
by Becker et al (5) in 1992.
Recently, studies indicate that S100P is overexpressed in a variety
of cancers, including pancreatic (6), colon (7), breast (8) and lung carcinomas (9), and its expression has been shown to be
associated with poor clinical outcomes. In pancreatic and colon
cancer, S100P was found to promote proliferation, invasion and
migration of cancer via the receptor for activated glycation end
products (RAGE) (6,7). S100P levels have been correlated with
poor patient survival in breast (8)
and lung cancer (9). In addition,
intracellular S100P binds to the N-terminal domain of dormant
ezrin, resulting in the activation of ezrin and stimulation of the
transendothelial migration of tumor cells (10). Furthermore, the interaction between
S100P and calcyclin-binding protein and Siah-1-interacting protein
(CacyBP/SIP) was found to lead to the degradation of β-catenin,
which is involved in the tumorigenesis for many different types of
cancers (11). GC cells exhibit an
increase in S100P expression following the treatment of
all-trans retinoic acid (12). However, the functional roles of
S100P in cancer progression, including GC, remain to be
elucidated.
In the present study, we applied RNA interference
(RNAi) technology to knock down the expression of S100P in two GC
cell lines MGC-803 and SGC-7901, and we investigated the apoptosis
rate and colony-formation capacity in both cell lines.
Subsequently, changes in the expression of apoptosis-related genes
following S100P downregulation were detected. Our data revealed
that the inhibition of S100P significantly promoted apoptosis and
suppressed colony-formation ability in both cell lines. S100P was
significantly involved in the regulation of apoptosis-related gene
expression. Thus, S100P appears to be a potential target for gene
therapy in GC.
Materials and methods
Patients and tissue microarray
In the present study, the GC tissue microarray (TMA)
was purchased from Fanpu Biotech, Inc. (Guilin China), which
contains 198 GCs (adenocarcinoma, mucinous adenocarcinoma, signet
ring cell carcinoma and undifferentiated carcinoma). All of the
tissues were obtained from archives of paraffin-embedded tissues
between 2005 and 2008. The patient with GCs included 146 males and
52 females. Ages of the 198 patients with gastric carcinoma ranged
from 24 to 83 years (mean age, 58.1 years). Patient consent was
obtained, along with approval from the local medical ethics
committee, for the use of human tissue samples for research
purposes.
Immunohistochemical staining
Immunohistochemistry (IHC) was performed using a
standard streptavidin biotin-peroxidase complex method. Consecutive
sections were deparaffinized with xylene and dehydrated with
alcohol. Endogenous peroxidase was quenched with 3%
H2O2 in 1X PBS. For antigen retrieval, TMA
slides were treated in 10 mM citrate buffer (pH 6.0) and boiled by
electronic wave or microwave for 10 min. Bovine serum albumin 5%
was then applied for 20 min at 37°C to prevent non-specific
binding. The TMA slides were incubated with anti-S100P (1:250
dilution; Abcam, Cambridge, UK) in a moist chamber overnight at
4°C, followed by HRP-conjugated anti-rabbit secondary antibodies
(Shanghai Weiao Ltd., Shanghai, China) for 20 min at 37°C. After
each treatment, the slides were washed with PBS (pH 7.2–7.6) three
times for 2 min. All slides were colored with the DAB substrate
chromogen system (Dako Co.) and counterstained with Mayer’s
hematoxylin. Omission of the primary antibody was used as a
negative control, and appropriate positive controls were used as
recommended by the manufacturers. Slides were analyzed
independently by two pathologists using fluorescence microscopy.
Photographs were captured using a Nikon digital still camera. The
sections were evaluated by Aperio ImageScope, and the number of
positive cells was calculated at ×200 magnification.
Evaluation of immunohistochemistry
One hundred cells were randomly selected and counted
from five representative fields of each section by two independent
pathologists blinded to the clinicopathologic information. The
proportion of positively staining cells was evaluated according to
five levels: <1%, negative; 1–5%, borderline; 6–25%,
intermediate; 26–50%, moderate; >50% of the carcinoma cells
stained, strong (8). For each core,
the intensity of the tumor immunoreactivity was evaluated on a
scale from 0 to 3 based on the method by Allred et al
(13): 0, negative; 1, detectable
but weak; 2, moderate but submaximal; or 3, maximal. Staining
scores for S100P were calculated as the sum of each intensity
multiplied by the percentage of positive cells, giving a range from
0 (0×100%) to 300 (3×100%). For statistical purposes, the IHC
scores were arbitrarily divided into two groups: a staining score
≤18 was considered as negative and a staining score >18 was
considered as positive.
Cell culture and infection
The following human gastric adenocarcinoma cell
lines were employed: adenocarcinoma cell line MGC-803 purchased
from the Shanghai Cell Bank (Shanghai, China) and SGC-7901
preserved at the Medical College of Southeast University (Nanjing,
China). The cell lines were maintained in RPMI-1640 medium
containing 10% fetal bovine serum (FBS; Gibco), 2 mM L-glutamine
and 1% penicillin/streptomycin mixture at 37°C in a humidified
atmosphere of 5% CO2 and 95% air.
For the lentiviral infection, MGC-803 and SGC-7901
cells were cultured in 6-well plates. The S100P short hairpin RNA
(shRNA) expressing the lentivirus (sh-S100P) or non-targeting shRNA
expressing GFP (negative control) were added, with a multiplicity
of infection of 100 in the MGC-803 and 20 in the SGC-7901 cells,
respectively. After 72 h of infection, cells were observed using
fluorescence microscopy (Olympus, Tokyo, Japan).
Lentiviral vector production
Five small interfering RNAs (siRNAs) targeting the
sequence of S100P were transformed into shRNAs with stem-loop-stem
structure and were cloned into GV115-hU6-EGFP-lentiviral vectors
with AgeI and EcoRI sites. The positive recombinant
clone was identified by PCR and DNA sequencing. The effective
targeting sequence of S100P (TCA GTG AGT TCA TCG TGT T) was
selected by western blot analysis. A non-silencing shRNA sequence
(TTC TCC GAA CGT GTC ACG T) was used as a negative control. The
recombined GV115-S100P-lentiviral vector or the negative control
lentiviral vector plasmid and pHelper 1.0 plasmid, the pHelper 2.0
plasmid (Shanghai GeneChem Co., Ltd., Shanghai, China) were
cotransfected into HEK293T cells via Lipofectamine 2000
(Invitrogen, Carlsbad, CA, USA) to generate the lentivirus. After 3
days of incubation, the lentivirus from the culture medium was
collected and concentrated with Centricon Plus-20 (Millipore,
Billerica, MA, USA). The concentrated viral supernatant was
aliquoted and kept at −80°C before use.
Real-time PCR
MGC-803 and SGC-7901 cells were cultured in 6-well
plates and were then infected with the lentivirus for 96 and 120 h,
respectively. Total RNA was isolated from the cultured cells using
TRIzol reagent (Invitrogen). cDNA was generated by reverse
transcription from total mRNA. Two sets of primers were used for
real-time PCR. Primers for S100P and GAPDH were designed by Primer
Premier software as follows: S100P-F, 5′-AGG AAG GTG GGT CTG AAT
CTA-3′ and S100P-R, 5′-TCC ACG GCA TCC TTG TCT-3′; GAPDH-F, 5′-TGA
CTT CAA CAG CGA CAC CCA-3′ and GAPDH-R, 5′-CAC CCT GTT GCT GTA GCC
AAA-3′. The SYBR-Green Real-Time PCR assay kit (Takara, Bio Inc.,
Otsu, Japan) was used, and quantitative real-time PCR (qRT-PCR) was
performed according to the ABI manufacturer’s protocols (Applied
Biosystems, Foster City, CA, USA). All samples were examined in
triplicate.
The 20-μl reaction mixture contained 10 μl
SYBR-Green PCR Master Mix (Takara), 1 μl cDNA template, 0.5 μl PCR
forward primer, 0.5 μl PCR reverse primer and 8.0 μl RNase-free
H2O. PCR running conditions consisted of 30 sec at 95°C
for the initial denaturation, 45 cycles of 5 sec at 95°C and 30 sec
at 60°C. The melting curve was monitored with the following
conditions: 15 sec at 95°C, 30 sec at 55°C, followed by a
temperature range from 55°C to 95°C increased by 0.5°C every 4 sec.
GAPDH was employed as an internal reference under the same
experimental conditions. The threshold cycle (Ct values), which was
the cycle number at which the amount of the amplified gene of
interest reached a fixed threshold, was subsequently determined.
Relative quantification of the S100P mRNA levels was normalized to
human GAPDH levels and calculated with the 2−ΔΔCt method
(14).
Western blot analysis
MGC-803 and SGC-7901 cells were cultured in 6-well
plates and were then infected with the lentivirus for 168 and 96 h,
respectively. Total protein was isolated from whole cells via
ice-cold protein lysis buffer (100 mM Tris-HCl, pH 6.8, 2%
mercaptoethanol, 4% SDS, 20% glycerine 1, 0.01% bromcresol blue),
followed by 15 min of incubation on ice and centrifugation at
12,000 × g for 5 min at 4°C. The protein concentration was
determined by BCA protein assay (Pierce, Rockford, IL, USA).
Protein extracts were separated on a SDS-polyacrylamide gel (12%),
blotted onto a PVDF membrane and incubated with the anti-S100P
antibody (1:1,000; Abcam, Cambridge, UK) or the anti-GAPDH antibody
(1:5,000; Santa Cruz Biotechnology, Santa Cruz, CA, USA). Western
blotting was developed using horseradish peroxidase-conjugated goat
anti-mouse or goat anti-rabbit IgG (1:5,000; Santa Cruz
Biotechnology). The membranes were detected with enhanced
chemiluminescence reagent (Amersham, Braunschweig, Germany) and
recorded on film in the linear detectable range.
Colony-formation assay
Cells were seeded in 12-well plates at a density of
300 cells/well and incubated at 37°C for 14 days to form colonies.
The medium was replaced every 2 days. At the indicated time-point,
cells were washed twice with PBS, fixed with paraform for 1 h,
stained with Giemsa for 20 min, washed with ddH2O three
times and then photographed with a digital camera. The number of
colonies (>50 cells/colony) was counted using fluorescence
microscopy (MicroPublisher 3.3 RTV; Olympus).
Analysis of apoptosis as determined by
fluorescence-activated cell sorter (FACS)
Analysis of apoptosis was performed with the Annexin
V-APC apoptosis detection kit according to the manufacturer’s
instructions (88-8007; eBioscience). Cells were seeded and infected
with the lentivirus. The cells were washed once with PBS and
centrifugated at 1500 × g for 5 min, washed once in 1X binding
buffer and resuspended in 1X staining buffer to
106–107 cells/ml. Furthermore, 100 μl of the
cells (~105–106 cells) was incubated with 5
μl Annexin V-APC at room temperature for 10–15 min in the dark.
Annexin V-stained cells were assessed using FACS flow cytometer
(FACSCalibur; Becton-Dickinson).
Real-time PCR gene microarray assay
Differential expression of apoptosis-related genes
was analyzed using the human apoptosis PCR array (Shanghai
GeneChem). RNA isolation, DNase treatment, and RNA clean-up were
performed according to the manufacturer’s protocol (Qiagen, Hilden,
Germany). The isolated RNA was reverse transcribed into cDNA using
the RT2 First Strand kit (Invitrogen). The template was added to an
instrument-specific, ready-to-use RT2 SYBR-Green qPCR Master Mix
(Invitrogen). PCR was performed on a Takara TP800 instrument
(Takara). Each assay was conducted in triplicate. Data
normalization was based on correcting all Ct values for the average
Ct values of several constantly expressed housekeeping genes
present on the array. The threshold cycle (Ct) values for all the
genes on each PCR array were calculated using the
instrument-specific software, and the fold-changes in gene
expression for pair-wise comparison were calculated using the
2−ΔΔCt method. The analysis was completed by Shanghai
GeneChem.
Statistical analysis
The data shown are presented as the mean ± standard
deviation (SD). Each experiment was repeated at least three times.
All statistical analyses were performed using SPSS v17.0 software
(SPSS, Inc., Chicago, IL, USA). Student’s t-test was used for
comparison of data obtained between the control and test group. The
significance was conventionally set at P-value <0.05.
Results
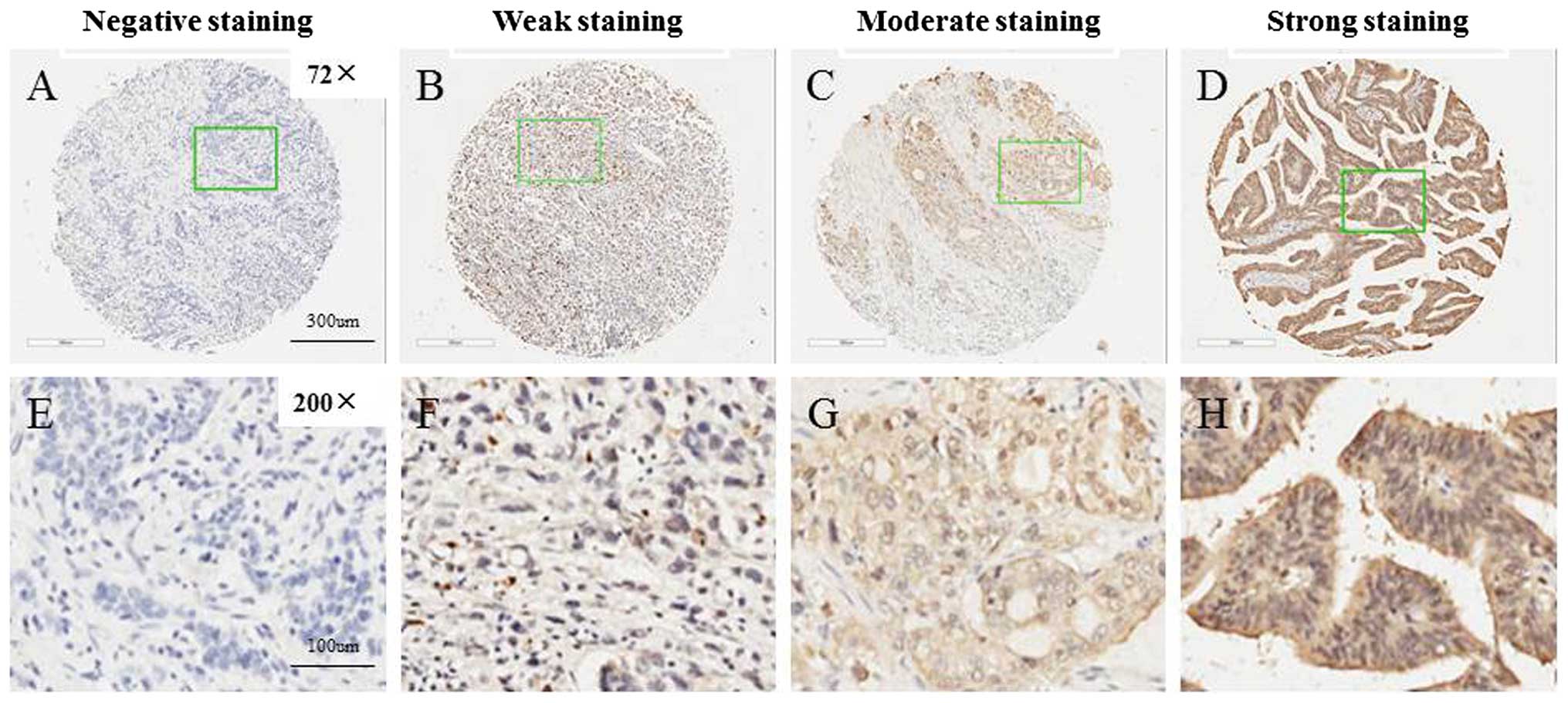
S100P is frequently expressed in GC
tissues
S100P expression was able to be evaluated in the TMA
for 166/198 gastric carcinoma samples. The exclusion criteria
included unrepresentative samples, no tumor tissue or samples with
too few tumor cells (300 cells/case) and lost samples. The
intensity of immunoreactivity ranged from negative to strong
(Fig. 1). Of the 166 GCs in the TMA
evaluated, 38 were negative, and the remaining 128 were classified
into the positive group including moderate and strong expression
subgroups (Table I). In addition,
staining for S100P was found both in the cytoplasm and in the
nucleus in 77.1 (128/166) and 7.7% (13/166) of cases,
respectively.
 | Table IS100P expression in the TMA of GC
tissues. |
Table I
S100P expression in the TMA of GC
tissues.
| S100P |
|---|
|
|
|---|
| Negative | Moderate | Strong |
|---|
| Score interval | (0,18) | (18,90) | (90,300) |
| Mean ± SD | 2.204±4.063 | 57.763±15.670 | 129.639±40.565 |
| No. of cases | 38 | 38 | 90 |
| Percentage | 22.90% | 22.90% | 54.2% |
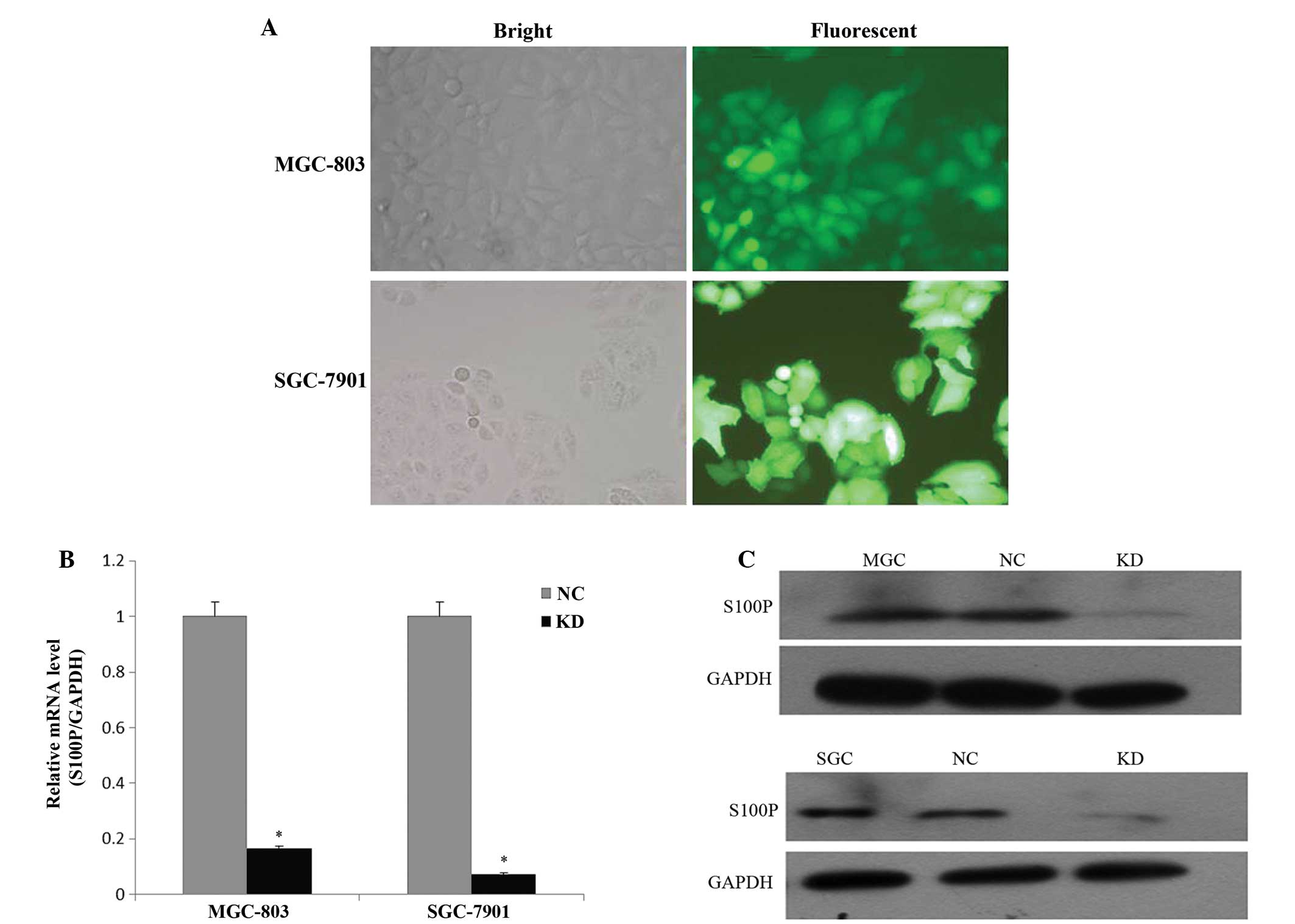
Knockdown of S100P by shRNA lentivirus
system in GC cells
To investigate the role of S100P in GC, shRNA
targeting S100P or non-silencing sequences were cloned into the
GV-115 lentiviral vector. Then, the S100P shRNA lentivirus or
non-silencing shRNA lentivirus expressing GFP were generated and
infected into two GC cell lines with a high multiplicity of
infection of 100 in the MGC-803 and 20 in the SGC-7901 cells,
respectively. As shown in Fig. 2A,
the infection efficiency of the lentivirus was >80% after 72 h
of infection. The qRT-PCR assay revealed that S100P mRNA expression
in the GC cells treated with the S100P shRNA lentivirus was
decreased by 83.4% in the MGC-803 and 92.7% in the SGC-7901 cells
(P<0.05), when compared with the negative control group
(Fig. 2B). We also determined the
level of S100P protein in the MGC-803 and SGC-7901 cells after 168
and 96 h of lentiviral infection by western blot analysis (Fig. 2C). In the MGC-803 and SGC-7901 cell
lines, the protein expression of S100P was significantly reduced by
~80% following S100P shRNA lentivirus treatment (P<0.05).
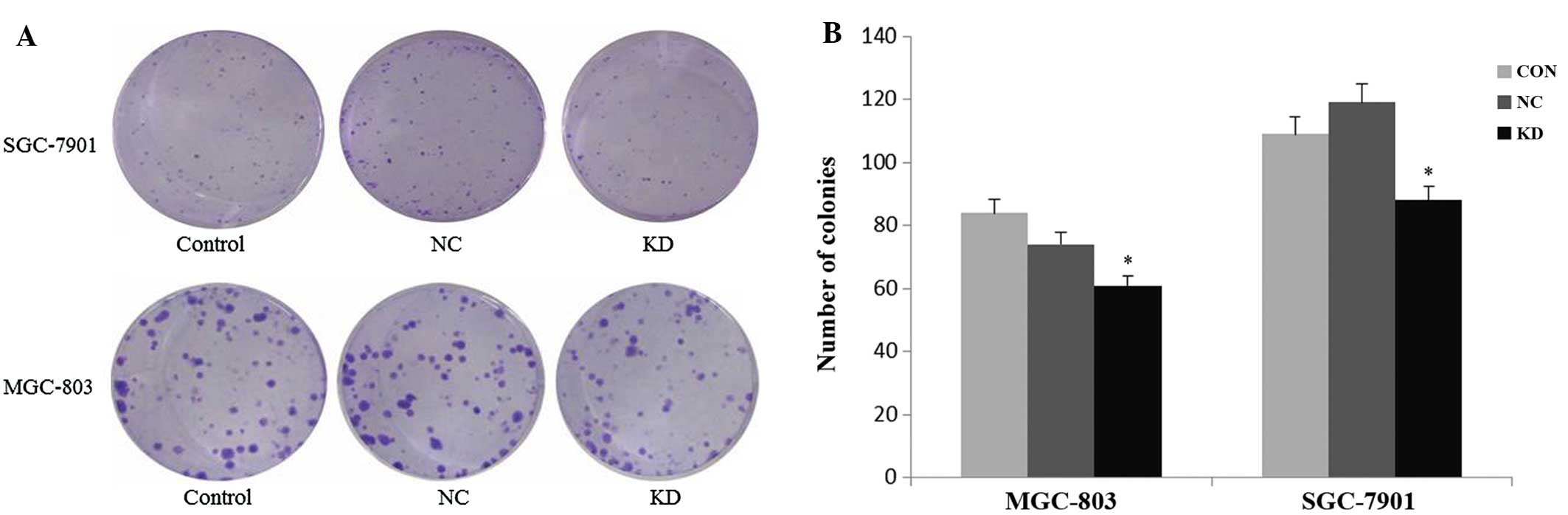
Downregulation of S100P inhibits
cell-colony formation
We then assessed the colony-formation capacity of
MGC-803 and SGC-7901 cells treated with the S100P shRNA lentivirus.
Three groups of MGC-803 and SGC-7901 cells (control, negative
control and knockdown) were allowed to grow for 14 days to form
colonies. As shown in Fig. 3, S100P
knockdown resulted in a notable decrease in the number of colonies,
as compared with the two control groups (P<0.05). The average
colony number in the MGC-803 and SGC-7901 cells was 61±10 and 88±8,
with nearly a 0.27-fold and 0.19-fold decrease, respectively, in
the knockdown group compared with the negative control group.
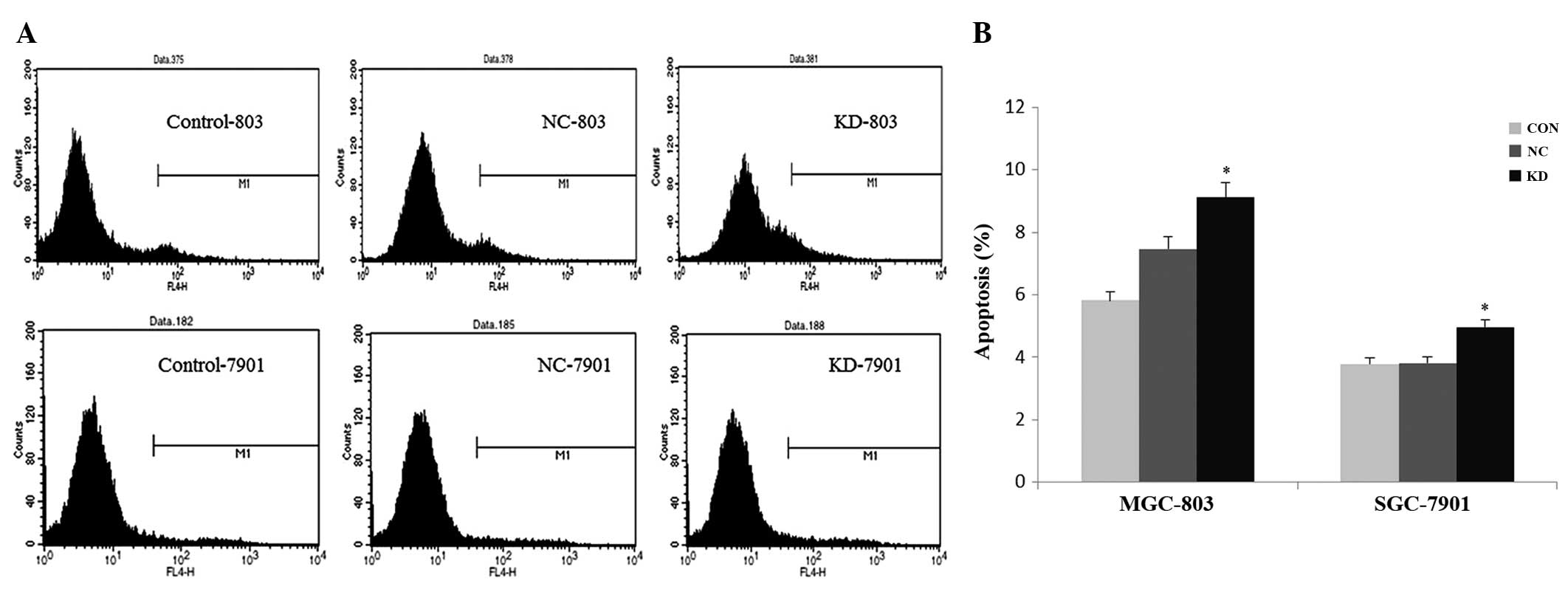
Knockdown of S100P promotes the apoptosis
of gastric cancer cells
To study the function of S100P on gastric cell
apoptosis, MGC-803 and SGC-7901 cells were analyzed with FACS and
Annexin V-APC staining. As shown in Fig. 4, a significant induction in
apoptosis was noted in the S100P shRNA group compared to the
control and negative control groups both in the MGC-803 and
SGC-7901 cells (P<0.05). The percentage of apoptotic cells (M1)
was increased to 9.12 and 4.96% in the MGC-803 and SGC-7901 cells,
respectively. These data verified that knockdown of S100P by the
lentiviral-mediated RNAi specifically promoted apoptosis of the GC
cell lines, and S100P may be involved in the cell survival
pathway.
Differential gene expression analysis by
PCR array
To obtain further insight into the mechanism of
S100P in gastric carcinoma cell apoptosis, the mRNA expression
profile of S100P shRNA-transfected SGC-7901 cells was compared with
that of the negative control using a human apoptosis PCR array
containing 86 cell apoptosis-related genes. Table II shows the fold difference between
the knockdown and negative control. Of the 86 apoptosis-associated
genes in this array, 12 genes demonstrated at least a 1.5-fold
difference in expression between the two groups. Nine genes
exhibited downregulation, while 3 genes were upregulated in the
knockdown group. FOS, DDIT3 and FN1 were significantly elevated,
while FASLG, DAPK1, CTNNB1 and CASP2 were notably downregulated
before and after S100P silencing.
 | Table IICandidate genes whose expression is
altered in the S100P knockdown vs. the control group. |
Table II
Candidate genes whose expression is
altered in the S100P knockdown vs. the control group.
| Gene | Description | Gene ID | Fold-change | P-value |
|---|
| S100P | S100 calcium-binding
protein P | 6286 | −4.02102 | 0.0052 |
| NFAT5 | Nuclear factor of
activated T-cells 5, | 10725 | −2.76273 | 0.0002 |
| FGFR3 | Fibroblast growth
factor receptor 3 | 2261 | −2.13584 | 0.0001 |
| CASP2 | Caspase-2,
apoptosis-related cysteine peptidase | 835 | −1.95844 | 0.0006 |
| STAT2 | Signal transducer and
activator of transcription 2 | 6773 | −1.90022 | 0.0054 |
| FASLG | Fas ligand | 356 | −1.86964 | 0.0401 |
| HSPA1A | Heat shock protein
1A | 3303 | −1.72966 | 0.0021 |
| DAPK1 | Death-associated
protein kinase 1 | 1612 | −1.67947 | 0.0009 |
| CTNNB1 | Catenin
(cadherin-associated protein), β1 | 1499 | −1.65682 | 0.0002 |
| TNFRSF25 | Tumor necrosis factor
receptor superfamily, member 25 | 8718 | −1.61876 | 0.0122 |
| FOS | FBJ murine
osteosarcoma viral oncogene homolog | 2353 | 1.54247 | 0.0024 |
| FN1 | Fibronectin 1 | 2335 | 1.75283 | 0.0000 |
| DDIT3 |
DNA-damage-inducible transcript 3, also
known as CHOP, GADD153 | 1649 | 2.70239 | 0.0001 |
Discussion
Gastric cancer is one of the most common types of
cancer and is the leading cause of death worldwide, with an
incidence which varies greatly across different geographic
locations. The American Cancer Society estimates that ~21,320 cases
of GC were diagnosed and ~10,540 individuals died from GC in 2012
(15). More new cases of GC are
diagnosed in China each year than in any other country. Previous
research indicates that S1100P is involved in the development of
various types of cancers. In the present study, we hypothesized
that S100P serves as an oncogenic factor in GC development.
Firstly, we measured S100P expression in GC tissues. The results
showed that S100P was frequently expressed in GC tissues, with
54.2% of cases demonstrating strong immunoreactivity, consistent
with the findings of Parkkila et al (16). Ge et al (17) demonstrated that S100P was
upregulated in GC in comparison with a normal control and was
correlated with TNM stage and prognosis. Moreover, the 5-year
survival rate was significantly lower in patients with high levels
of S100P expression than in patients with low levels of expression.
In contrast, Jia et al (18)
reported decreased expression of S100P in GC compared to normal
tissues. Patients with S100P-positive cancers showed a statistical
better survival than patients with negative S100P expression. Liu
et al (19) found that the
expression of S100P between normal and cancerous tissues did not
significantly differ. Nevertheless, the expression of S100P in GC
is still in dispute at the present time. Therefore, investigation
of more clinical samples, particularly GC, and their non-cancerous
counterparts is required to conclude whether there is a difference
in S100P expression and whether there is a correlation between
S100P expression and clinicopathological features.
Lentiviral vectors with large coding capacity of
transgene cassettes into dividing and quiescent cells have become
some of the most widely used vectors for fundamental biological
research, function genomics and gene therapy (20). Secondly, we employed a lentiviral
short hairpin delivery system in our trial to infect and
efficiently silence S100P in the GC cell lines. As shown in
Fig. 2, we constructed the S100P
shRNA lentivirus, which sufficiently knocked down the expression
levels of S100P mRNA and protein in the GC cells compared with the
negative control, thus, ensuring the credibility of the subsequent
assays. We also confirmed that knockdown of S100P markedly
inhibited the colony formation capacity of GC cells (Fig. 3). These results indicate that S100P
plays an important role in GC cell growth in the short or relative
long term. We then performed apoptotic analysis by FACS, attempting
to uncover the mechanism by which shRNA of S100P controls gastric
cell growth. Our data revealed that S100P shRNA had a marked
function on GC survival via induction of apoptosis (Fig. 4).
S100P has been found to protect NIH3T3 cells and
pancreatic cancer cells from apoptosis induced by the cytotoxic
agent 5-FU and from detachment from a solid substrate (6,21).
Overexpression of S100P in PC3 cells was found to promote cell
growth, proliferation and reduced basal apoptosis rate.
Furthermore, prostate cancer cells overexpressing S100P are
protected against camptothecin-induced apoptosis (22). In hepatocellular carcinoma, S100P
expression downregulated by RNA interference-mediated gene
silencing increased the basal levels of apoptotic cells, which
suggests that S100P may act as an aggressiveness factor in HCC
cells by conferring resistance to basal apoptosis (23). These findings together with our
results suggest that S100P serves as an important factor that
contributes to the aggressive nature of cancer via inhibition of
apoptosis. To date, however, the specific signaling pathway of
S100P involved in GC cell apoptosis remains unclear.
To clarify the molecular mechanism by which S100P
affects cell apoptosis, we employed the PCR array to profile the
expression of apoptosis-associated genes. The result showed that 12
genes had a >1.5-fold difference in gene expression between the
S100P shRNA-transfected cells and the negative control. Expression
of 3 genes was significantly elevated, while 9 genes appear to be
downregulated in the S100P-knockdown group.
Celecoxib inhibits cancer cell growth and stimulates
apoptosis. Upregulation of S100P expression by celecoxib in GC
cells was found to negatively affect the drug antitumorigenic
activity through inhibition of apoptosis. Moreover, apoptosis
induced by celecoxib was stimulated by a small interfering RNA
targeting S100P. Yet, there was no significant difference in
apoptotic cells between the siRNA S100P and non-silenced group
(24). DDIT3 (also known as CHOP,
GADD153), upregulated in the S100P-knockdown cells, is considered a
key event in endoplasmic reticulum stress-mediated apoptosis
(25). Expression of the ER
stress-responsive transcription factor CHOP was found to be
correlated with patient survival and remained an independent
prognostic variable in pairwise comparisons with all clinical
variables tested (26). CHOP
expression had no statistical difference between the mock and
S100P-overexpressing cells. Celecoxib-induced expression of CHOP
mRNA was suppressed in S100P-overexpressing cells, which suggests
that S100P induction inhibits celecoxib-induced apoptosis through
suppression of CHOP expression (24). This finding is similar to our
presumption that S100P affects apoptosis through dysregulation of
the expression of CHOP. Consequently, our future research aims to
confirm the protein level changes in genes with differential
expression following S100P depletion and to collect sufficient GC
samples with controls to detect S100P and validate gene expression
and find a correlation between S100P and the validated genes in
GC.
In conclusion, to the best of our knowledge, the
present study proved for the first time that lentiviral-mediated
RNAi targeting S100P promotes cell apoptosis and inhibits
colony-formation ability of GC cells. Our data indicate that S100P
serves as an oncogene in GC development. Therefore, knockdown of
S100P expression by a lentiviral-delivered shRNA may be a potential
and valuable strategy for the treatment of GC.
Abbreviations:
|
GC
|
gastric cancer, RNAi, RNA
interference
|
|
TMA
|
tissue microarray
|
|
shRNA
|
short hairpin RNA
|
|
FACS
|
fluorescence-activated cell sorter
|
References
|
1
|
Garcia M, Jemal A, Ward EM, et al: Global
Cancer facts & Figures 2007. American Cancer Society; Atlanta:
2007
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
3
|
Sun XD, Mu R, Zhou YS, et al: Analysis of
mortality rate of stomach cancer and its trend in twenty years in
China. Zhonghua Zhong Liu Za Zhi. 26:4–9. 2004.(In Chinese).
|
|
4
|
Donato R: S100: a multigenic family of
calcium-modulated proteins of the EF-hand type with intracellular
and extracellular functional roles. Int J Biochem Cell Biol.
33:637–668. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Becker T, Gerke V, Kube E and Weber K:
S100P, a novel Ca2+-binding protein from human placenta.
cDNA cloning, recombinant protein expression and
Ca2+-binding properties. Eur J Biochem. 207:541–547.
1992.
|
|
6
|
Arumugam T, Simeone DM, Golen KV and
Logsdon CD: S100P promotes pancreatic cancer growth, survival, and
invasion. Clin Cancer Res. 11:5356–5364. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fuentes MK, Nigavekar SS, Arumugam T,
Logsdon CD, Schmidt AM, Park JC and Huang EH: RAGE activation by
S100P in colon cancer stimulates growth, migration, and cell
signaling pathways. Dis Colon Rectum. 50:1230–1240. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang G, Platt-Higgins A, Carroll J, de
Silva Rudland S, Winstanley J, Barraclough R and Rudland PS:
Induction of metastasis by S100P in a rat mammary model and its
association with poor survival of breast cancer patients. Cancer
Res. 66:1199–1207. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartling B, Rehbein G, Schmitt WD, Hofmann
HS, Silber RE and Simm A: S100A2-S100P expression profile and
diagnosis of non-small cell lung carcinoma: impairment by advanced
tumour stages and neoadjuvant chemotherapy. Eur J Cancer.
43:1935–1943. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Austermann J, Nazmi AR, Muller-Tidow C and
Gerke V: Characterization of the Ca2+-regulated
ezrin-S100P interaction and its role in tumor cell migration. J
Biol Chem. 283:29331–29340. 2008.
|
|
11
|
Filipek A, Jastrzebska B, Nowotny M and
Kuznicki J: CacyBP/SIP, a calcyclin and Siah-1-interacting p
rotein, binds EF-hand proteins of the S100 family. J Biol Chem.
277:28848–28852. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shyu RY, Huang SL and Jiang SY: Retinoic
acid increases expression of the calcium-binding protein S100P in
human gastric cancer cells. J Biomed Sci. 10:313–319. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Allred DC, Harvey JM, Berardo M and Clark
GM: Prognostic and predictive factors in breast cancer by
immunohistochemical analysis. Mod Pathol. 11:155–168.
1998.PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
|
|
15
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
16
|
Parkkila S, Pan PW, Ward A, et al: The
calcium-binding protein S100P in normal and malignant human
tissues. BMC Clin Pathol. 8:22008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ge F, Wang C, Wang W and Wu B: S100P
predicts prognosis and drug resistance in gastric cancer. Int J
Biol Markers. 28:e387–e392. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jia SQ, Niu ZJ, Zhang LH, et al:
Identifcation of prognosis-related proteins in advanced GC by mass
spectrometry-based comparative proteomics. J Cancer Res Clin Oncol.
135:403–411. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu J, Li X, Dong GL, et al: In
silico analysis and verification of S100 gene expression in
gastric cancer. BMC Cancer. 8:2612008. View Article : Google Scholar
|
|
20
|
Mátrai J, Chuah MK and VandenDriessche T:
Recent advances in lentiviral vector development and applications.
Mol Ther. 18:477–490. 2010.
|
|
21
|
Arumugam T, Simeone DM, Schmidt AM and
Logsdon CD: S100P stimulates cell proliferation and survival via
receptor for advanced glycation end products (RAGE). J Biol Chem.
279:5059–5065. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Basu GD, Azorsa DO, Kiefer JA, et al:
Functional evidence implicating S100P in prostate cancer
progression. Int J Cancer. 123:330–339. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim JK, Jung KH, Noh JH, et al: Targeted
disruption of S100P suppresses tumor cell growth by downregulation
of cyclin D1 and CDK2 in human hepatocellular carcinoma. Int J
Oncol. 35:1257–1264. 2009.PubMed/NCBI
|
|
24
|
Namba T, Homan T, Nishimura T, Mima S,
Hoshino T and Mizushima T: Up-regulation of S100P expression by
non-steroidal anti-inflammatory drugs and its role in
anti-tumorigenic effects. J Biol Chem. 284:4158–4167. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Oyadomari S and Mori M: Roles of
CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ.
11:381–389. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dalton LE, Clarke HJ, Knight J, et al: The
endoplasmic reticulum stress marker CHOP predicts survival in
malignant mesothelioma. Br J Cancer. 108:1340–1347. 2013.
View Article : Google Scholar
|