Introduction
Ovarian carcinoma is the most lethal gynecologic
malignancy in the United States (1). Approximately 23% of gynecologic
cancers originate in the ovary, but 47% of all deaths from cancer
of the female genital tract occur in women with ovarian cancer
(2). The high rate of lethality
from ovarian carcinoma is mainly due to the advanced stage (stage
III and IV) of disease at the time of diagnosis and lack of
effective therapies for advanced disease (2). A better understanding of molecular
changes in ovarian carcinoma is required to identify better
therapeutic strategies for this fatal disease.
The epithelial-to-mesenchymal transition (EMT) has
been recognized as an important physiological process that is
associated with cancer progression and metastasis in multiple
epithelial cancer types, including ovarian carcinoma (3). A hallmark of EMT is the loss of
epithelial markers (such as E-cadherin) and gain of mesenchymal
markers (such as vimentin, N-cadherin). The change of cellular
lineage has been extensively studied and we have begun to
appreciate the signaling pathways involved. Signaling molecules
that affect EMT of ovarian carcinoma include MAPK (4), Notch3 (5), SFRP4 (6) and ALX1 (7). A major area for EMT investigations has
been the key cell membrane protein E-cadherin, which is responsible
for holding neighboring epithelial cells together in a classic
cobblestone structure. During EMT, E-cadherin is invariably lost
from the membrane, allowing cells to assume a loosely connected
structure that is associated with enhanced cell migration/invasion,
thus contributing to metastasis. A number of transcription factors
have been shown to be key regulators of E-cadherin expression,
including SNAI1, SNAI2, ZEB1 and ZEB2.
The regulatory circuit of E-cadherin has been
further illustrated by the discoveries that these key
transcriptional factors are post-transcriptionally regulated by a
group of microRNAs (miRNAs) (8–14).
miRNAs are short 20–22-nucleotide RNA molecules that are negative
regulators of gene expression in a variety of eukaryotic organisms.
Single-stranded miRNAs bind to specific target mRNAs through partly
complementary sequences that are predominantly in the
3′-untranslated region (3′-UTR) (15,16).
The best-characterized miRNAs that suppress EMT belong to the
miR-200 family (miR-200a, miR-200b, miR-200c, miR-141, miR-429),
which have been shown to directly target the 3′-UTRs of ZEB1
and ZEB2 (13,17). Recent studies identified miR-506 as
one of a new class of robust EMT repressors; miR-506 directly
targets the 3′-UTRs of SNAI2 (18,19),
CD151 (19) and vimentin
(VIM) (19). Our
investigations of the miRNA network regulating the EMT of ovarian
carcinoma have identified miR-101 as another key regulator
(18). miR-101 has been shown by
recent studies to regulate EMT through EZH2 (20,21)
and the Wnt signaling pathway (22). However, how miR-101 suppresses EMT
in ovarian carcinoma has not been reported; this is the focus of
the investigation reported here.
In the present study, we comprehensively examined
the mechanisms through which miR-101 regulates EMT in ovarian
carcinoma. We provide evidence that miR-101 directly targets ZEB1
and ZEB2 through an miR-200-independent mechanism.
Materials and methods
Materials and cell culture
The ovarian cancer cell line SKOV3 was obtained from
American Type Culture Collection (Manassas, VA, USA) and maintained
in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS).
The cells were incubated at 37°C in an atmosphere containing 5%
CO2. Human miR-101 mimics hsa-miR-101 and hsa-miR-Ctrl
were obtained from Dharmacon (Chicago, IL, USA). Small-interfering
RNA (siRNA) targeting ZEB1 or ZEB2 and scrambled
negative siRNA control were purchased from Sigma (St. Louis, MO,
USA).
miRNA/siRNA transfection
Cells (2×105/well) were seeded in 6-well
plates and allowed to attach for at least 16 h. miR-101 mimic
(miR-101), scrambled negative miR-101 control (miR-Ctrl), or siRNA
was transfected into cells using Lipofectamine RNAiMAX (Invitrogen,
Grand Island, NY, USA) at a final concentration of 50 nM.
Luciferase reporter assay
The 3′-UTRs of the ZEB1 and ZEB2
genes, each of which contains one or two putative miR-101-binding
sites, were amplified by polymerase chain reaction (PCR) from cDNA
derived from HeyA8 cells and inserted into the multiple cloning
site of the pmirGLO vector (Promega, Fitchburg, WI, USA). The
primers used for the ZEB1 gene 3′-UTR were:
5′-AAACTCGAGTACTTCAATTC CTCGGTATTG-3′, and 5′-AAATCTAGACACACTGTTCTA
CAGTCCAAGGC-3′; the primers used for the ZEB2 gene 3′-UTR
were: 5′-AAACTCGAGTACACCCATGTC AGTATTAGAAG-3′, and
5′-AAATCTAGACACAGATCAA CGTCATGTTCC-3′. Two mutant ZEB1 and
ZEB2 3′-UTR reporter vectors that lacked the binding sites
for miR-101 were created through site-directed mutagenesis using a
QuikChange kit (Stratagene, La Jolla, CA, USA). The primers used
for site-directed mutagenesis of the ZEB1 gene 3′-UTR were:
ZEB1 3′-UTR-M1, 5′-CTGTGCAACATTTTTTGTA
CAAATGTCTTCAAACCTGG-3′, and 5′-CCAGGTTTGAA
GACATTTGTACAAAAAATGTTGCACAG-3′; ZEB1 3′-UTR-M2,
5′-CACAGTGTAGTGTATAAGTGCACAGTTT GTATTAATACAATAATAT-3′, and
5′-ATATTATTGTATTA ATACAAACTGTGCACTTATACACTACACTGT G-3′. The primers
used for site-directed mutagenesis of the ZEB2 gene 3′-UTR
were as follows: ZEB2 3′-UTR-M, 5′-CCTAATTTTA
TTTATTTCAGAGCTCAGTGTACAGTATTATAGTTCTT C-3′, and
5′-GAAGAACTATAATACTGTACACTGAGCTCT GAAATAAATAAAATTAGG-3′. All clones
were verified by DNA sequencing.
For the luciferase reporter assay, 0.5 μg of
pmirGLO, pmirGLO-3′-UTR-WT, or pmirGLO-3′-UTR-MT was transfected
into HeLa cells that were cultured in 24-well plates, together with
50 nM miR-101 or miR-Ctrl, using Lipofectamine 2000 (Invitrogen).
Twenty-four hours after transfection, cells were subjected to lysis
and firefly luciferase and Renilla luciferase activities
were determined using a dual-luciferase reporter assay system
(Promega) as previously described (23). Relative firefly luciferase activity
(firefly luciferase activity/Renilla luciferase activity)
for each construct was compared to that of the control mimics. For
each transfection, luciferase activity was averaged from
triplicates.
Real-time reverse transcription PCR
analysis (RT-PCR)
RT-PCR was performed as previously described
(18). In brief, total RNA was
isolated with the mirVana miRNA isolation kit (Ambion, Grand
Island, NY, USA). Reverse transcription was performed using
SuperScript II reverse transcriptase (Invitrogen) according to the
manufacturer’s protocol. TaqMan real-time PCR assays for
ZEB1 and ZEB2 were purchased from Applied Biosciences
(Grand Island, NY, USA). RNU6B was used as a normalization
control.
Western blot analysis
Primary antibodies for β-actin, fibronectin and
vimentin were obtained from Santa Cruz Biotechnology, Inc. (Santa
Cruz, CA, USA). The N-cadherin antibody was obtained from
Invitrogen [N-Cadherin Monoclonal Antibody, Mouse (3B9)]. The
E-cadherin antibody was purchased from BD Biosciences (San Jose,
CA, USA). Western blotting was performed as previously described
(24). In brief, equal amounts of
protein from whole cell lysates of each sample were loaded on a 10%
polyacrylamide gel for electrophoresis; the membrane was blocked in
5% non-fat milk in 1X Tris-buffered saline (pH 7.4) containing
0.05% Tween-20 and probed with primary antibodies at concentrations
of 1:500 (for E-cadherin), 1:1,000 (for β-actin, fibronectin,
N-cadherin), or 1:2,500 (for vimentin). The secondary antibodies
were used at a concentration of 1:4,000 to 1:5,000. The proteins
were visualized using the SuperSignal West Pico or SuperSignal
Femto chemiluminescent substrate from Pierce Biotechnology, Inc.
(Rockford, IL, USA).
Immunofluorescence staining
Immunofluorescence staining was performed as
previously described (18). In
brief, SKOV3 cells were seeded onto an uncoated glass slide
coverslip and cultured in complete medium under standard cell
culture conditions. Cells were fixed in 4% paraformaldehyde at
ambient temperature for 15 min, followed by permeabilization in 1X
phosphate-buffered saline solution (PBS) containing 0.05% NP-40 for
30 min at ambient temperature. The cells were blocked in blocking
solution (1X PBS containing 10% normal goat serum and 0.05% NP-40)
for at least 4 h. After washing briefly with 1X PBS, the cells were
incubated with a mouse monoclonal anti-human E-cadherin antibody
(1:100 dilution in the blocking solution) at 4°C overnight. After
washing, a goat anti-mouse IgG conjugated with Alexa Fluor 488
(Invitrogen, #A11029; 1:1,000 in the blocking solution) was
incubated with the cells at ambient temperature for 1 h. Phalloidin
staining was performed at ambient temperature for 45 min using
phalloidin-TRITC (Molecular Probes, Invitrogen) at a concentration
of 2.5 μg/ml. Phase images were captured by a ZEISS Axiovert 200
microscope at a magnification of ×200. The fluorescence images were
captured using a ZEISS Axioplan 2 imaging microscope at a
magnification of ×630 or ×400.
Cell invasion assays
Cell invasion assays were performed in triplicate
using Matrigel-coated Transwell chambers (8-μm pore size; BD
Pharmingen, Franklin Lakes, NJ, USA). Briefly, cells
(5×104) transfected with miR-101 or miR-Ctrl were plated
48 h after transfection in 500 μl of serum-free medium in the upper
chamber of the wells and allowed to migrate toward 10% FBS medium
(750 μl) in the lower chamber for 22 h. Cells that remained on top
of the filter were mechanically removed, and those that migrated to
the underside of the filter were fixed and stained with Hema-Diff
Solution (Fisher Scientific, Pittsburgh, PA, USA). Invaded cells
were counted under a microscope in six randomly chosen fields and
representative images were captured. Data are expressed as number
of invaded cells (means ± standard deviation) normalized to the
number of control cells that migrated. Each result represents an
average of triplicates.
Wound healing assay
SKOV3 cells (~2×105) were plated in each
well of a 6-well plate. Following overnight incubation, the cells
were transfected with 50 nM miR-Ctrl or 50 nM of miR-101 for 48 h,
to allow the cells reached full confluence. The cell monolayers
were wounded by scraping with a micropipette tip, washed several
times with medium to remove dislodged cells and placed back in
growth medium. Images were captured using a phase-contrast
microscope (Olympus, Japan) immediately and 16 h after
wounding.
Generation of SKOV3-ZEB1/ZEB2 stable cell
lines
The pcDNA3.1(+)-ZEB1 and
pcDNA3.1(+)-ZEB2 vectors were generated by digesting a
ZEB1 fragment from MGC human ZEB1 cDNA
(MHS4426-211690344) or ZEB2 from MGC human ZEB2 cDNA
(MHS4426-211690995) (both from Thermo Scientific, Waltham, MA, USA)
and subcloning it into the pcDNA3.1(+) vector using BamHI
and NotI restriction enzymes and KpnI and XbaI
restriction enzymes, respectively. The correct sequence and
orientation were confirmed by DNA sequencing. The pcDNA3.1-ZEB1 or
pcDNA3.1-ZEB2 vector or empty vector alone was transfected into
SKOV3 cells using Lipofectamine 2000. At 48 h after transfection,
the cells were placed in culture in complete medium with 1,000
μg/ml G418 for 4 weeks.
Results
miR-101 directly targets ZEB1 and
ZEB2
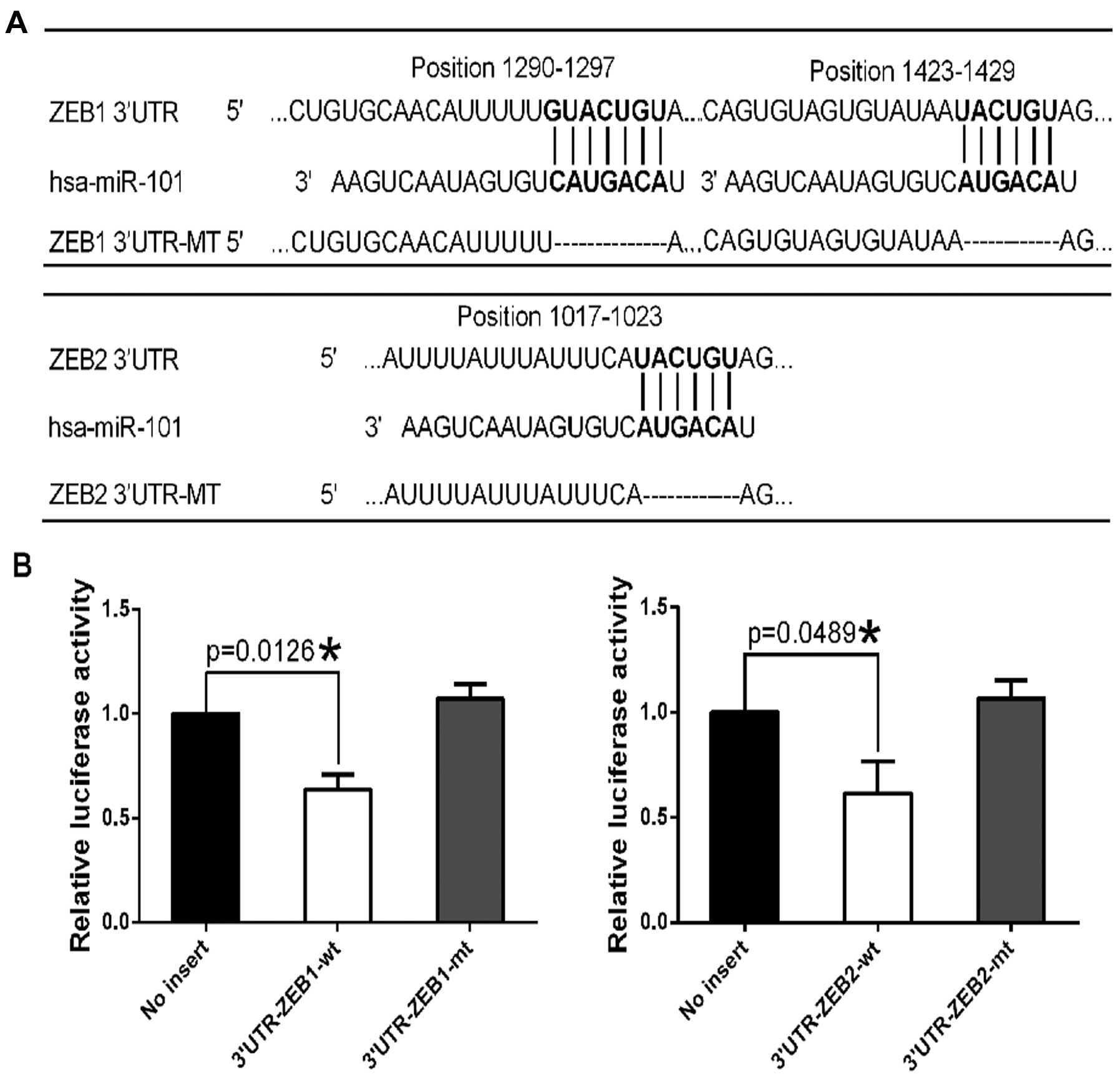
TargetScan analysis predicted two miR-101-binding
sites in the 3′-UTR of the ZEB1 gene and one site in the
3′-UTR of the ZEB2 gene (Fig.
1A), suggesting that miR-101 may directly target ZEB1
and ZEB2. We performed luciferase reporter assays to examine
whether miR-101 mimic (miR-101) directly targets ZEB1 and
ZEB2. We cloned the 3′-UTR of ZEB1 or the 3′-UTR of
ZEB2 into the pmirGLO-ctrl vector to generate pmirGLO-ZEB1
and pmirGLO-ZEB2 constructs. Co-transfection of pmirGLO-ZEB1 and
miR-101 into HeLa cells resulted in 36.3% less luciferase activity
than co-transfection with miR-Ctrl, suggesting that miR-101
directly targets ZEB1 (Fig.
1B). Similarly, co-transfection of pmirGLO-ZEB2 and miR-101
resulted in 38.6% less luciferase activity than co-transfection
with miR-Ctrl, suggesting that miR-101 directly targets ZEB2
(Fig. 1B).
To confirm that miR-101 specifically regulates
ZEB1 and ZEB2 through the predicted binding sites, we
generated the constructs pmirGLO-ZEB1-mt and pmirGLO-ZEB2-mt, from
which the miR-101-binding site sequences on the 3′-UTR of ZEB1 or
ZEB2 were deleted. We then co-transfected the mutant constructs
with miR-101 or miR-Ctrl into HeLa cells. Deletion of the
miR-101-binding sites from the 3′-UTR of ZEB1 or ZEB2
abolished the effect of miR-101 on luciferase activity (Fig. 1B). These results indicate that the
ZEB1 and ZEB2 genes are direct targets of
miR-101.
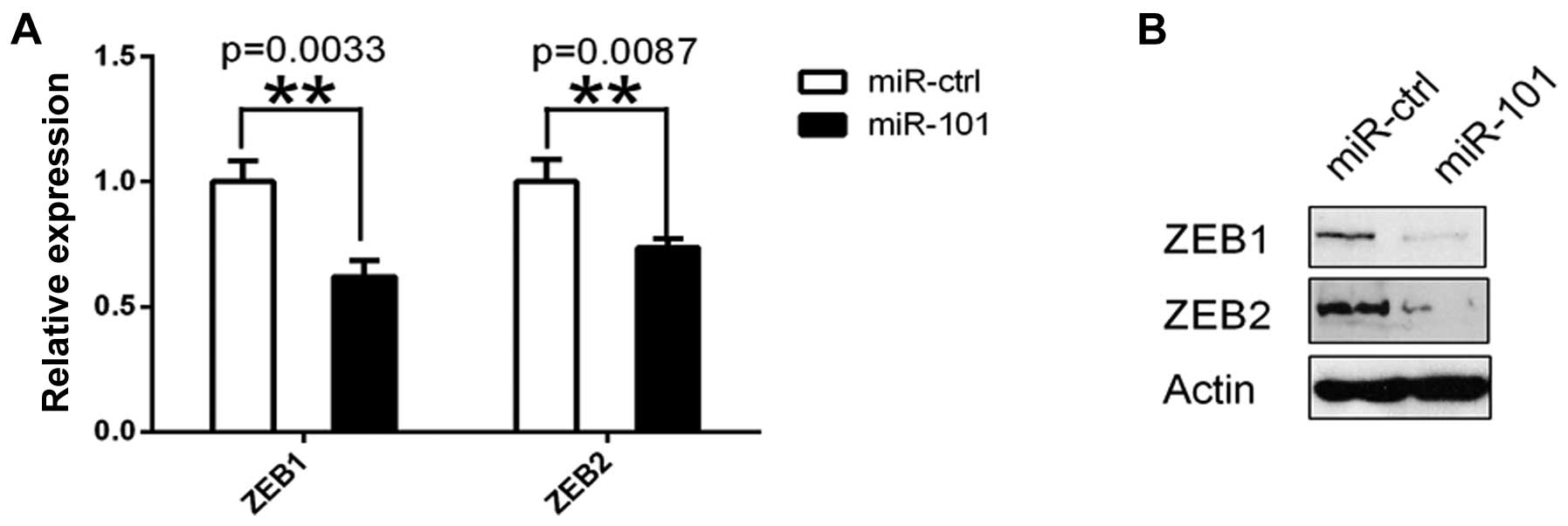
To demonstrate that miR-101 is an endogenous
regulator of ZEB1 and ZEB2 in ovarian carcinoma
cells, we transfected SKOV3 ovarian cancer cells with miR-101 or
miR-Ctrl followed by measurement of ZEB1 and ZEB2
mRNA and protein at 48 h after transfection. The results showed
that ZEB1 and ZEB2 mRNA and proteins in SKOV3 cells
were significantly downregulated after miR-101 transfection
(Fig. 2).
miR-101 inhibits EMT, migration and
invasion of ovarian cancer cells
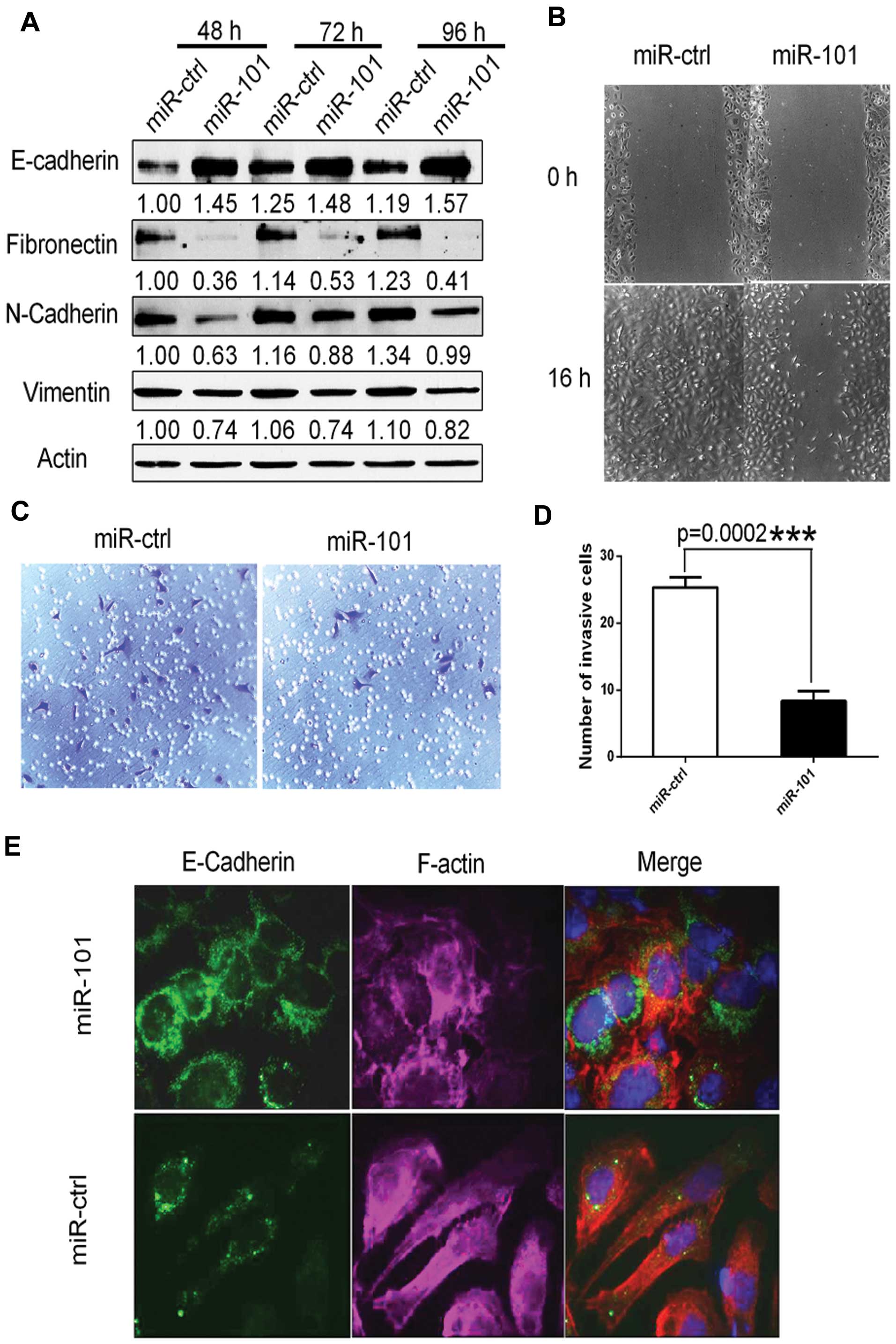
To determine whether forced expression of miR-101
promotes the epithelial phenotype, we transfected SKOV3 cells with
either miR-101 or miR-Ctrl. miR-101 overexpression significantly
increased E-cadherin protein levels, while it downregulated
mesenchymal markers fibronectin, N-cadherin and vimentin (Fig. 3A).
It is well recognized that the EMT is involved in
cell migratory and invasive capacity in ovarian carcinoma (6,25). To
this end, we performed a wound-healing assay and found that cell
migration was markedly reduced in cells expressing ectopic miR-101
when compared with cells transfected with miR-Ctrl (Fig. 3B). Similarly, invasion assays
revealed that ectopic miR-101 expression significantly decreased
cell invasion by ~3-fold when compared with miR-Ctrl-transfected
cells (Fig. 3C and D). We also
performed immunofluorescence staining to directly visualize the
effect of miR-101 on E-cadherin expression and localization and
cell morphology (Fig. 3E). In this
experiment, the E-cadherin protein was localized on the membrane at
cell-cell junctions of miR-101-transfected SKOV3 cells, which is
indicative of epithelial cells. In addition, F-actin distribution
was rearranged to a cortical pattern, which is another hallmark of
the epithelial phenotype (Fig. 3E).
In contrast, the cells transfected with miR-Ctrl showed the
elongated mesenchymal cell phenotype indicated by an absence of
E-cadherin on the cell membrane and rearrangement of F-actin from a
cortical to a stress-fiber pattern (Fig. 3E).
Inhibition of EMT by miR-101 is mediated
by ZEB1 and ZEB2
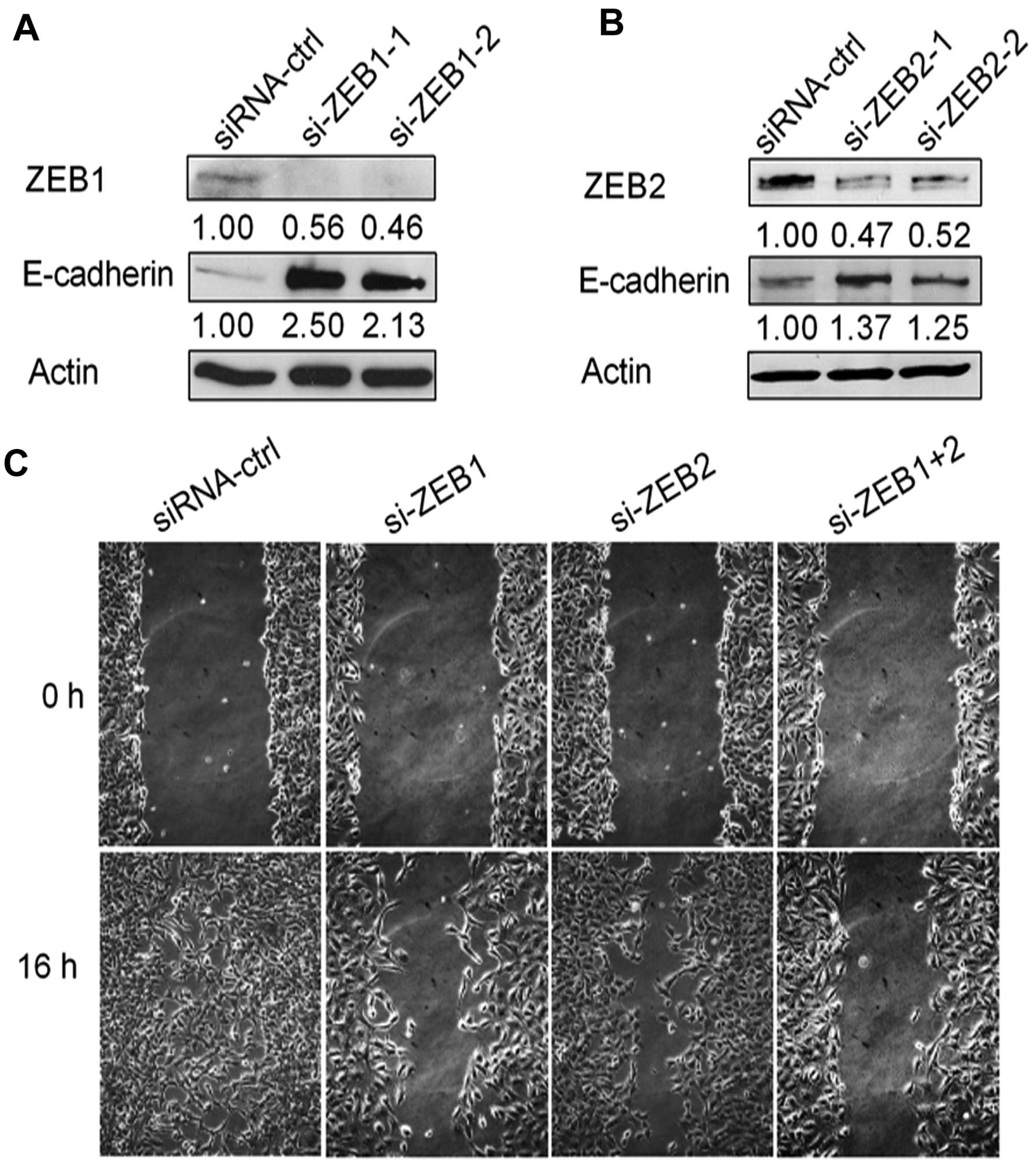
To determine whether the inhibition of EMT by
miR-101 was mediated by ZEB1 or ZEB2, we knocked down
ZEB1 by two different siRNAs and found that both siRNAs led
to increased E-cadherin protein levels in SKOV3 cells (Fig. 4A). Similarly, knockdown of
ZEB2 in SKOV3 cells by two different siRNAs led to the same
result (Fig. 4B). We then performed
the wound-healing assay in SKOV3 cells transfected with si-ZEB1 or
si-ZEB2 or both. As expected, knockdown of ZEB1 or ZEB2 markedly
decreased cell migration when compared with siRNA-ctrl, especially
with dual silencing of both genes (Fig.
4C).
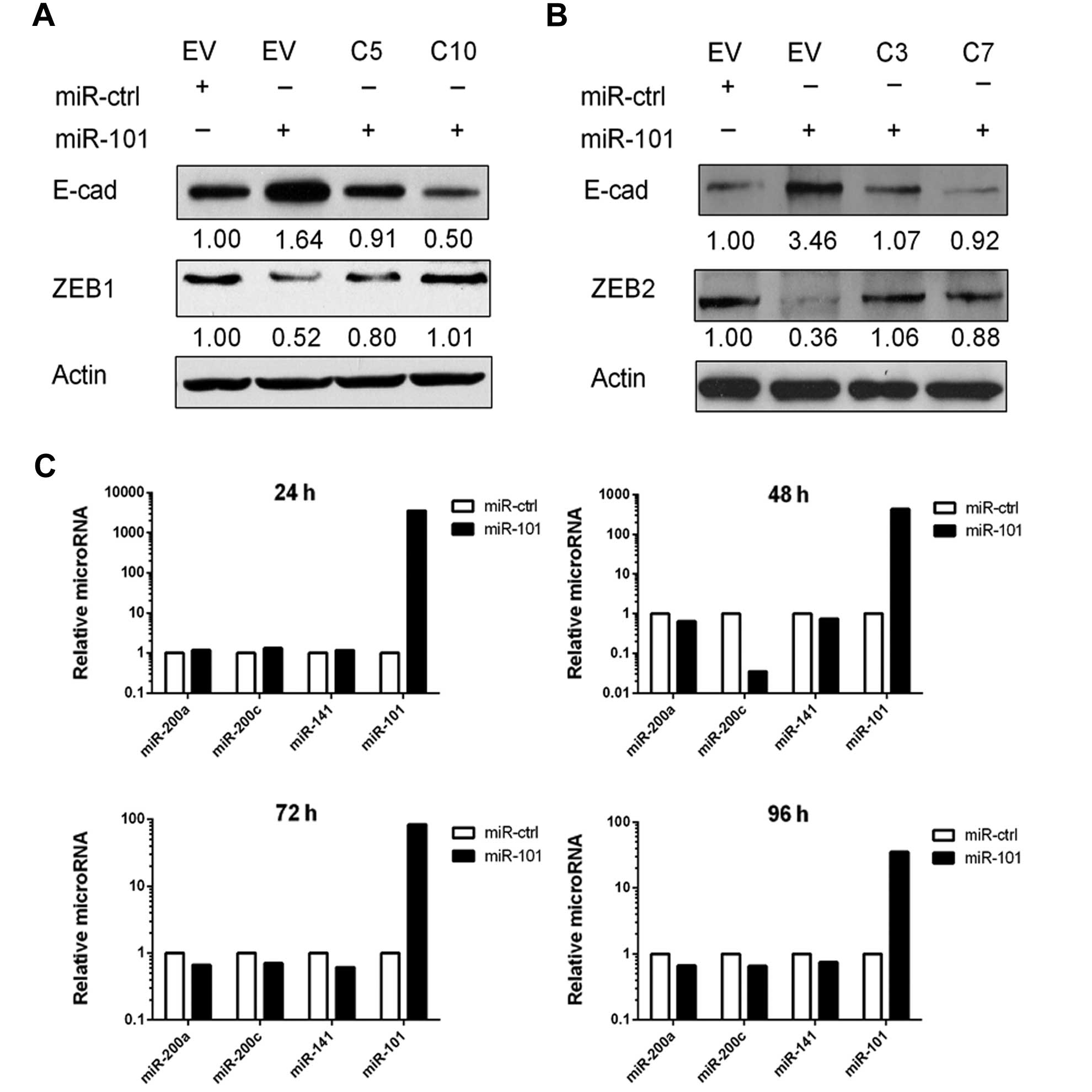
We next performed ZEB1 and ZEB2 rescue experiments.
To obtain stable clones, we first transfected SKOV3 cells with
pcDNA3.1(+)-ZEB1 or pcDNA3.1(+)-ZEB2 that did not contain a 3′-UTR.
When miR-101 was transfected into these cells, the expression of
ZEB1 and ZEB2 was not downregulated and E-cadherin was not
increased (Fig. 5A and B). These
results showed that miR-101 upregulated E-cadherin through the
miR-101-binding sites on the 3′-UTRs of ZEB1 and
ZEB2. We further examined the possibility that miR-101 may
somehow increase expression of miR-200, which has been shown to
also target the 3′-UTRs of ZEB1 and ZEB2. We isolated
RNA from miR-101- or miR-Ctrl-transfected SKOV3 cells and measured
the level of miR-200 family members. We detected no significant
changes in the levels of miR-200 family members in
miR-101-transfected cells (Fig.
5C).
Discussion
Ovarian cancer is the leading cause of mortality in
gynecologic malignancies, as the high death rates from ovarian
cancer remain largely unchanged over the past 30 years, with a
5-year overall survival rate of only 30–39% (26). A better understanding of the
mechanisms involved in progression of ovarian carcinoma is urgently
required. EMT has emerged as an important mechanism that promotes
ovarian carcinoma progression. Through analysis of the Cancer
Genome Atlas data, our previous study identified a master miRNA
regulatory network that regulated EMT in serous ovarian carcinoma,
and this network includes miR-101 (18). In the present study, we provided
evidence for the first time that miR-101 suppresses EMT by directly
targeting E-cadherin-suppressor genes ZEB1 and ZEB2
through specific binding sites on their 3′-UTRs.
There are two separate copies of the miR-101 gene,
located on 1p31.3 and 9p24. Both regions have been identified as
fragile regions of the genome that are associated with abnormal
deletion or amplification in cancer (27). Downregulation of miR-101 has been
observed in bladder cancer (28),
intraductal papillary mucinous neoplasms of the pancreas (29) and ovarian carcinoma (30), suggesting that miR-101 plays a role
in tumor suppression.
ZEB1 and ZEB2, members of the ZEB family, have been
shown to induce EMT through repression of E-cadherin and to promote
tumor progression and metastatic spread (31–33).
Higher expression of ZEB1 and ZEB2 were found in diverse types of
cancer (34–36) including ovarian carcinoma (37,38),
suggesting that the two factors may play an essential role in EMT
of ovarian carcinoma. The discovery that there are putative binding
sites on the 3′-UTRs of both ZEB1 and ZEB2 genes for
miR-101 suggests a potential mechanism through which miR-101
regulates EMT. Using reporter gene assays and rescue experiments,
we validated this regulatory mechanism. Furthermore, ectopic
miR-101 expression significantly upregulated E-cadherin and
decreased mesenchymal markers and cell motility, indicating that
miR-101 acted as a strong EMT suppressor in ovarian cancer
cells.
Transcriptional regulators of E-cadherin expression
include SNAI1, SNAI2, TWIST1, TWIST2, ZEB1 and ZEB2. These key
regulators of E-cadherin expression can in turn be regulated by
multiple miRNAs, revealing an impressive redundancy in regulation
of this physiological process at multiple levels. Notably, both
miR-200 (39) and miR-101 (30), both regulators of ZEB1 and ZEB2, are
downregulated in ovarian carcinoma. Furthermore, miR-506, which
regulates another E-cadherin repressor, SNAI2, is also
downregulated in ovarian carcinoma (18). EMT in cancer is generally a late
event associated with metastasis. This is consistent with the
requirement that multiple redundant regulatory mechanisms be lost
for EMT to occur.
With regard to the causes of the simultaneous loss
of multiple miRNAs, results of our previous study, which showed
that miR-506 expression is partially lost through methylation,
suggested that deletion of miRNA genes may not explain this
(18). Of note, recent reports
showed that miR-200 family members as well as miR-101 are
methylated in several types of cancer, explaining their decreased
expression (40–42). It has been reported consistently
that EZH2, which regulates the methylation program, may regulate
E-cadherin expression in ovarian carcinoma (43). EZH2 has been proposed as an oncogene
in many types of cancer including ovarian carcinoma (43,44).
Conceivably, EZH2 upregulation may downregulate the EMT-suppressing
miRNAs, removing a highly redundant mechanism in controlling
E-cadherin expression. However, the redundant mechanism appears to
be restricted to downregulation, as each of the suppressing miRNAs
can be sufficient to inhibit EMT without requiring other miRNAs.
This provides an opportunity for using one of these miRNAs as a
therapeutic tool.
In conclusion, we demonstrated for the first time
that miR-101 can directly target ZEB1 and ZEB2, resulting in
suppression of the EMT in ovarian carcinoma. miR-101 may have
therapeutic value in the treatment of ovarian carcinoma.
Acknowledgements
The present study was partially supported by a grant
from the Blanton-Davis Ovarian Cancer Research Program to W. Zhang
and by the University of Texas M.D. Anderson Cancer Center core
grant CA016672 from the National Institutes of Health. The authors
thank Kathryn Hale from the Department of Scientific Publications
at M.D. Anderson Cancer Center for editing this manuscript. F.G. is
supported by a fellowship from the China Education Council. D.Y. is
an Odyssey Fellow at M.D. Anderson Cancer Center and is supported
by the Diane Denson Tobola Fellowship in Ovarian Cancer Research
and the Harold C. and Mary L. Daily Endowment Fund.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar
|
|
2
|
Berek JS, Crum C and Friedlander M: Cancer
of the ovary, fallopian tube, and peritoneum. Int J Gynaecol
Obstet. 119(Suppl 2): S118–S129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013.PubMed/NCBI
|
|
4
|
Arvizo RR, Saha S, Wang E, Robertson JD,
Bhattacharya R and Mukherjee P: Inhibition of tumor growth and
metastasis by a self-therapeutic nanoparticle. Proc Natl Acad Sci
USA. 110:6700–6705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gupta N, Xu Z, El-Sehemy A, Steed H and Fu
Y: Notch3 induces epithelial-mesenchymal transition and attenuates
carboplatin-induced apoptosis in ovarian cancer cells. Gynecol
Oncol. 130:200–206. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ford CE, Jary E, Ma SS, Nixdorf S,
Heinzelmann-Schwarz VA and Ward RL: The Wnt gatekeeper SFRP4
modulates EMT, cell migration and downstream Wnt signalling in
serous ovarian cancer cells. PLoS One. 8:e543622013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yuan H, Kajiyama H, Ito S, et al: ALX1
induces snail expression to promote epithelial-to-mesenchymal
transition and invasion of ovarian cancer cells. Cancer Res.
73:1581–1590. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yoshino H, Enokida H, Itesako T, et al:
Epithelial-mesenchymal transition-related microRNA-200s regulate
molecular targets and pathways in renal cell carcinoma. J Hum
Genet. 58:508–516. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen J, Wang L, Matyunina LV, Hill CG and
McDonald JF: Overexpression of miR-429 induces
mesenchymal-to-epithelial transition (MET) in metastatic ovarian
cancer cells. Gynecol Oncol. 121:200–205. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ke Y, Zhao W, Xiong J and Cao R: miR-149
inhibits non-small-cell lung cancer cells EMT by targeting FOXM1.
Biochem Res Int. 2013:5067312013.PubMed/NCBI
|
|
11
|
Zhou Y, Li Y, Ye J, et al: MicroRNA-491 is
involved in metastasis of hepatocellular carcinoma by inhibitions
of matrix metalloproteinase and epithelial to mesenchymal
transition. Liver Int. 33:1271–1280. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Harazono Y, Muramatsu T, Endo H, et al:
miR-655 is an EMT-suppressive microRNA targeting ZEB1
and TGFBR2. PLoS One. 8:e627572013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gregory PA, Bert AG, Paterson EL, et al:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Paterson EL, Kazenwadel J, Bert AG,
Khew-Goodall Y, Ruszkiewicz A and Goodall GJ: Down-regulation of
the miRNA-200 family at the invasive front of colorectal cancers
with degraded basement membrane indicates EMT is involved in cancer
progression. Neoplasia. 15:180–191. 2013.PubMed/NCBI
|
|
15
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang D, Sun Y, Hu L, et al: Integrated
analyses identify a master microRNA regulatory network for the
mesenchymal subtype in serous ovarian cancer. Cancer Cell.
23:186–199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Arora H, Qureshi R and Park WY: miR-506
regulates epithelial mesenchymal transition in breast cancer cell
lines. PLoS One. 8:e642732013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Carvalho J, van Grieken NC, Pereira PM, et
al: Lack of microRNA-101 causes E-cadherin functional deregulation
through EZH2 up-regulation in intestinal gastric cancer. J Pathol.
228:31–44. 2012.PubMed/NCBI
|
|
21
|
Varambally S, Cao Q, Mani RS, et al:
Genomic loss of microRNA-101 leads to overexpression of histone
methyltransferase EZH2 in cancer. Science. 322:1695–1699. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Strillacci A, Valerii MC, Sansone P, et
al: Loss of miR-101 expression promotes Wnt/β-catenin signalling
pathway activation and malignancy in colon cancer cells. J Pathol.
229:379–389. 2013.PubMed/NCBI
|
|
23
|
Parker BC, Annala MJ, Cogdell DE, et al:
The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a
regulation in glioblastoma. J Clin Invest. 123:855–865.
2013.PubMed/NCBI
|
|
24
|
Holmes KM, Annala M, Chua CY, et al:
Insulin-like growth factor-binding protein 2-driven glioma
progression is prevented by blocking a clinically significant
integrin, integrin-linked kinase, and NF-κB network. Proc Natl Acad
Sci USA. 109:3475–3480. 2012.PubMed/NCBI
|
|
25
|
Zhang L, Yang M, Gan L, et al: DLX4
upregulates TWIST and enhances tumor migration, invasion and
metastasis. Int J Biol Sci. 8:1178–1187. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li X, Lu Y, Chen Y, Lu W and Xie X:
MicroRNA profile of paclitaxel-resistant serous ovarian carcinoma
based on formalin-fixed paraffin-embedded samples. BMC Cancer.
13:2162013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cui J, Eldredge JB, Xu Y and Puett D:
MicroRNA expression and regulation in human ovarian carcinoma cells
by luteinizing hormone. PLoS One. 6:e217302011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu Z, Lin Y, Chen H, et al: MicroRNA-101
suppresses motility of bladder cancer cells by targeting
c-Met. Biochem Biophys Res Commun. 435:82–87. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Caponi S, Funel N, Frampton AE, et al: The
good, the bad and the ugly: a tale of miR-101, miR-21 and miR-155
in pancreatic intraductal papillary mucinous neoplasms. Ann Oncol.
24:734–741. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Semaan A, Qazi AM, Seward S, et al:
MicroRNA-101 inhibits growth of epithelial ovarian cancer by
relieving chromatin-mediated transcriptional repression of
p21waf1/cip1. Pharm Res. 28:3079–3090. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Browne G, Sayan AE and Tulchinsky E: ZEB
proteins link cell motility with cell cycle control and cell
survival in cancer. Cell Cycle. 9:886–891. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vandewalle C, Van Roy F and Berx G: The
role of the ZEB family of transcription factors in development and
disease. Cell Mol Life Sci. 66:773–787. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: an alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Spoelstra NS, Manning NG, Higashi Y, et
al: The transcription factor ZEB1 is aberrantly expressed in
aggressive uterine cancers. Cancer Res. 66:3893–3902. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pena C, García JM, García V, et al: The
expression levels of the transcriptional regulators p300 and CtBP
modulate the correlations between SNAIL, ZEB1, E-cadherin and
vitamin D receptor in human colon carcinomas. Int J Cancer.
119:2098–2104. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Imamichi Y, König A, Gress T and Menke A:
Collagen type I- induced Smad-interacting protein 1 expression
downregulates E-cadherin in pancreatic cancer. Oncogene.
26:2381–2385. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Elloul S, Elstrand MB, Nesland JM, et al:
Snail, Slug, and Smad-interacting protein 1 as novel parameters of
disease aggressiveness in metastatic ovarian and breast carcinoma.
Cancer. 103:1631–1643. 2005. View Article : Google Scholar
|
|
38
|
Elloul S, Silins I, Tropé CG, Benshushan
A, Davidson B and Reich R: Expression of E-cadherin transcriptional
regulators in ovarian carcinoma. Virchows Arch. 449:520–528. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dahiya N, Sherman-Baust CA, Wang TL, et
al: MicroRNA expression and identification of putative miRNA
targets in ovarian cancer. PLoS One. 3:e24362008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Castilla MÁ, Díaz-Martín J, Sarrió D, et
al: MicroRNA-200 family modulation in distinct breast cancer
phenotypes. PLoS One. 7:e477092012.PubMed/NCBI
|
|
41
|
Hur K, Toiyama Y, Takahashi M, et al:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. 62:1315–1326. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wei X, Xiang T, Ren G, et al: miR-101 is
down-regulated by the hepatitis B virus × protein and induces
aberrant DNA methylation by targeting DNA methyltransferase 3A.
Cell Signal. 25:439–446. 2013.
|
|
43
|
Rao ZY, Cai MY, Yang GF, et al: EZH2
supports ovarian carcinoma cell invasion and/or metastasis via
regulation of TGF-β1 and is a predictor of outcome in ovarian
carcinoma patients. Carcinogenesis. 31:1576–1583. 2010.
|
|
44
|
Lu C, Bonome T, Li Y, et al: Gene
alterations identified by expression profiling in tumor-associated
endothelial cells from invasive ovarian carcinoma. Cancer Res.
67:1757–1768. 2007. View Article : Google Scholar : PubMed/NCBI
|