Introduction
Human hepatocellular carcinoma (HCC) is highly
malignant and is the second cause of cancer-related death in China.
The incidence rates of HCCs are high in Western countries, central
Africa and eastern and southeastern Asia. However, the molecular
mechanisms underlying HCC pathogenesis remain poorly understood.
The clinical efficacy of current chemotherapies and available
targeted therapies for HCC is also limited (1). Thus, basic, translational and clinical
studies are urgently needed to identify new therapeutic targets for
the treatment of HCCs.
Vigilin, a multi-KH domain protein, is highly
conserved from yeast to the human. As an RNA-binding protein,
vigilin is localized in the nuclear envelope (2), nucleus (3,4) and
rough endoplasmic reticulum (ER) where it associates with the
polysome (5,6), translation elongation factor 1A and
WD-repeat protein Asc1p (2,5,7,8).
Vigilin is involved in translational control (9), nuclear export of tRNA (10), cytoplasmic transport of RNA
(4,11), metabolism of specific mRNAs
(12) and RNAi-mediated vigilin
silencing (13). Human vigilin was
initially characterized as a high density lipoprotein binding
protein (HDLBP; HBP) (14), and was
subsequently shown to play an important role in the cellular sterol
metabolism in human atherogenesis (15). However, the role of vigilin in human
types of cancers remains controversial. Studies showing vigilin
overexpression in the TOV112D human ovarian cancer cells and human
prolactinomas and gastric cancer suggest that vigilin may play a
promotive role in carcinogenesis (16–19).
In contrast, vigilin can bind to the 3′ untranslated mRNA region of
the c-fms proto-oncogene encoding the receptor of macrophage colony
stimulating factor in breast cancer cells to destabilize this mRNA
and inhibit its translation, suggesting vigilin may have a
tumor-suppressor role in these cells (20). Similarly, the role of vigilin in
human HCC cell growth is also poorly understood.
The present study was designed to examine the
effects of vigilin on HCC cells in culture and in mouse models. We
showed that vigilin expression was upregulated in a subset of human
HCCs and vigilin was required for HCC cell proliferation, survival,
migration and tumor growth. Knockdown of vigilin sensitized cells
to cisplatin, a widely used drug for cancer chemotherapy, to
inhibit HCC cell growth.
Materials and methods
Clinical specimens and cell lines
Thirty-three human primary hepatocellular carcinomas
(HCCs) and paired adjacent non-tumor tissues were collected at the
Department of Surgery, West China Hospital of Sichuan University.
Demographic patient data and clinicopathological characteristics
were also obtained from the same hospital. The criteria for grading
HCCs were based on the classification of the World Health
Organization. Seven normal liver tissues were obtained as controls
from subjects affected by hepatic hemangioma, intrahepatic stones,
liver cysts, other non-cancerous or non-cirrhosis liver diseases.
Eight cirrhosis liver tissues were obtained from subjects with
liver cirrhosis other than HCC. Informed consent was obtained from
each patient, and the study was approved by the appropriate
institutional review committees. Cold cup biopsies of tumor tissues
were snap frozen and stored in liquid nitrogen. The remaining
tissues were fixed in 10% formalin in saline for 24 h, followed by
dehydration in ethanol and embedding in paraffin for diagnostic
assessment.
The L-02 human embryonic liver cell line and the
three human HCC cell lines including HepG2, SMMC7721 and BEL-7402
were obtained from the Chinese Type Culture Collection (CTCC).
Cells were cultured in standard media at 37°C in 5%
CO2.
Immunohistochemistry (IHC)
For detection of vigilin protein by IHC, clinically
collected human HCC specimens and BEL7402 xenograft tumor tissues
collected from nude mice were fixed in 10% formalin in saline for
24 h, dehydrated and embedded in paraffin. Tissue sections were
prepared, deparaffinized, rehydrated and treated with 3% hydrogen
peroxide in a citric acid buffer (pH 6.0) at 95°C for 40 min. The
vigilin antibody was made in our laboratory and its specificity was
validated using lymph cells, cancer cells and tumor tissues with
known high and low vigilin protein expression as described
previously (21). Primary
antibodies against vigilin (1:200), Ki67 (1:50) and activated
caspase 3 (1:50) (both from Abcam, UK) were diluted with
phosphate-buffered saline (PBS) containing 10% normal goat serum.
Pretreated tissue sections were incubated with the diluted primary
antibodies in a humidified chamber at 37°C for 45 min. Tissue
sections were subsequently inoculated with the Dako Rapid EnVision
secondary antibody system (Dako, USA) and developed using the
3,3′-diaminobenzidine (DAB) substrate. Sections were further
counterstained with hematoxylin. A blank control was obtained by
excluding the primary antibody in the staining. Stained sections
were examined under an Olympus BX41 microscope and imaged using a
CCD camera. The intensity of vigilin immunoreactivity was scored
using a four-scale system (22): 0,
no expression; 1, weak expression; 2, medium expression and 3, high
expression. Cells positive for Ki67 or activated caspase-3
immunostaining were counted against the total number of cells in
the viewing fields.
Western blotting
Cultured cells or xenograft tumors were lysed in an
ice-cold lysis buffer (KaiJi, China). The lysates were cleared by
centrifugation at 16,000 × g at 4°C for 10 min. Supernatants were
collected, and their protein concentrations were determined using
the Bradford protein assay reagent (KaiJi). Sixty micrograms of
total protein for each sample was analyzed by western blotting.
Antibodies used for western blotting were against vigilin (1:100)
and β-actin (1:1,000 dilution; Santa Cruz, USA). The bound primary
antibodies were visualized using the enhanced chemiluminescence
detection system (Pierce Chemical, USA).
Lentiviral-mediated delivery of shRNA and
generation of vigilin stable knockdown cell lines
Human vigilin shRNAs were constructed into the
pLKO.1-puro vector from Sigma-Aldrich (23). The targeting sequence of vigilin
mRNA was 5′-UCCCAACACAAGUAUGUCAUU-3′ (24). A non- targeting shRNA,
5′-CGCUGAGUACUUCGAAAUGUC, from Sigma-Aldrich was used as a negative
control. The shRNA vectors were co-transfected with the lentiviral
packaging plasmids psPAX2 and pMD2.G (Addgene, Cambridge, MA, USA)
into HEK293T cells using Lipofectamine 2000 (Invitrogen, USA). The
media containing lentivirus particles were collected, filtered and
overlaid onto BEL7402 cells in the presence of 8 μg/ml polybrene
for 24 h. The lentivirus expressing the luciferase-specific shRNA
was used to generate the control BEL7402 cell line. Subsequently,
the infected cells were selected with 3 μg/ml of puromycin (Acros,
Belgium). The resulting stable control and vigilin knockdown cell
lines were termed BEL7402-Ctrl and BEL7402-KD cells,
respectively.
Cell growth assay
The 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl
tetrazolium bromide (MTT) assay was employed to measure cell growth
in culture. BEL7402-Ctrl and BEL7402-KD cells (2×103 in
150 μl of medium) were plated in 96-well plates and cultured at
37°C under 5% CO2. MTT assays were performed at 24, 48,
72 and 96 h after cells were seeded. For the MTT assay, 20 μl of
MTT reagent (5 mg/ml in stock solution) was added to each well, and
the cells were incubated for another 4 h under normal culture
condition. After removing the culture medium, DMSO was added to
each well and the plate was shaken horizontally for 10 min at room
temperature to dissolve the formazan crystals formed in live cells.
Absorbance at 570 nm was read with a micro-ELISA reader (Bio-Rad,
USA). Each assay was carried out in 6 replicates.
Colony formation assay
BEL7402-Ctrl and BEL7402-KD cells were seeded in
6-well plates (200 cells/well) and cultured in RPMI-1640 medium
with 3 μg/ml puromycin for 2 weeks to allow colony formation. The
cell colonies were fixed with methanol, stained with Giemsa,
photographed under a microscope and counted manually at high-power
magnification. Only colonies containing ≥50 number of cells were
counted. Assays were repeated 3 times.
Wound-healing assay
BEL7402-Ctrl and BEL7402-KD cells were cultured in
RPMI-1640 medium with 3 μg/ml puromycin in a 6-well plate. The
monolayer of cell culture at 60% confluency was carefully scratched
with a 200-μl pipette tip as described previously (25). Non-adherent cells and cellular
debris were washed away with PBS. Cells migrated into the scratched
area were monitored and photographed at 24, 48 and 72 h after cells
were scratched. The scratch width was measured at 5 random points
using ImageJ 1.43 software. Cell migration distance (in μm) was
calculated by the formula (original width - current width)
(25). Experiments were carried out
in triplicates and repeated 3 times.
Xenograft tumor growth assay
All animal procedures were approved by the Sichuan
University Animal Care and Use Committee. Nude mice were maintained
in a special animal facility for immune-defective mice. Male
BALB/c-nu/nu mice (4–6 weeks of age, 16–18 g) were obtained from
the Laboratory Animal Center of Sichuan University. The
BEL7402-Ctrl and BEL7402-KD cells were collected in PBS and
inoculated subcutaneously into the left and right dorsal flanks of
the nude mice. A total of 2×106 cells in 200 μl of PBS
were injected to each site. The tumor size was measured weekly with
a caliper, and tumor volume was calculated by the formula: Tumor
volume (mm3) = [width (mm)]2 × [length
(mm)]/2, as described previously (26). Five weeks later, the mice were
sacrificed and tumors dissected from the mice were weighed. Part of
every tumor was snap-frozen in liquid nitrogen, and used for
western blotting and semi-quantitative RT-PCR assays. The rest of
the tumor was fixed for 24 h in 10% phosphate-buffered formalin and
paraffin-embedded. Then, 5-μm sections were prepared for IHC
staining.
RT-PCR
Total RNA was extracted from BEL7402 cells using the
TRIzol reagent (Invitrogen). One microgram of RNA was reversely
transcribed using the reverse transcriptase kit (Promega, USA)
according to the manufacturer’s instructions. The reaction without
adding the transcriptase was used as a negative control for RT-PCR.
PCR analysis was performed using the β-actin as an internal
control. Vigilin cDNA levels were analyzed using the following
primers: vigilin-F, 5′-CGTTATTGGGCAGAAAGGAA and vigilin-R,
5′-CTCTGTGGGAAGCGAATGTC; β-actin-F, 5′-TCATCACCATTGGCAATGAG and
β-actin-R, 5′-CACTGTGTTGGCGTACAGGT. PCR products were separated on
agarose gel and visualized under UV light after being stained with
ethidium bromide.
Cisplatin treatment
BEL7402-Ctrl and BEL7402-KD cells were seeded in a
96-well plate at a density of 2×103 cells/well and were
cultured for 24 h. After removing the initial medium, the cells
were treated with culture medium containing different
concentrations of cisplatin for 72 h. Six-wells were used for each
concentration of cisplatin treatment. Relative cell number was
measured by MTT assay. Experiments were independently repeated 3
times.
Statistical analysis
The data are expressed as means ± SEM of independent
measurements. Statistical analysis was performed based on the 3
repeated tests by using the Student’s t-test with SPSS 13.0
software. Statistical significance was set at p<0.05 (95%
confidence level).
Results
Vigilin is overexpressed in human
HCCs
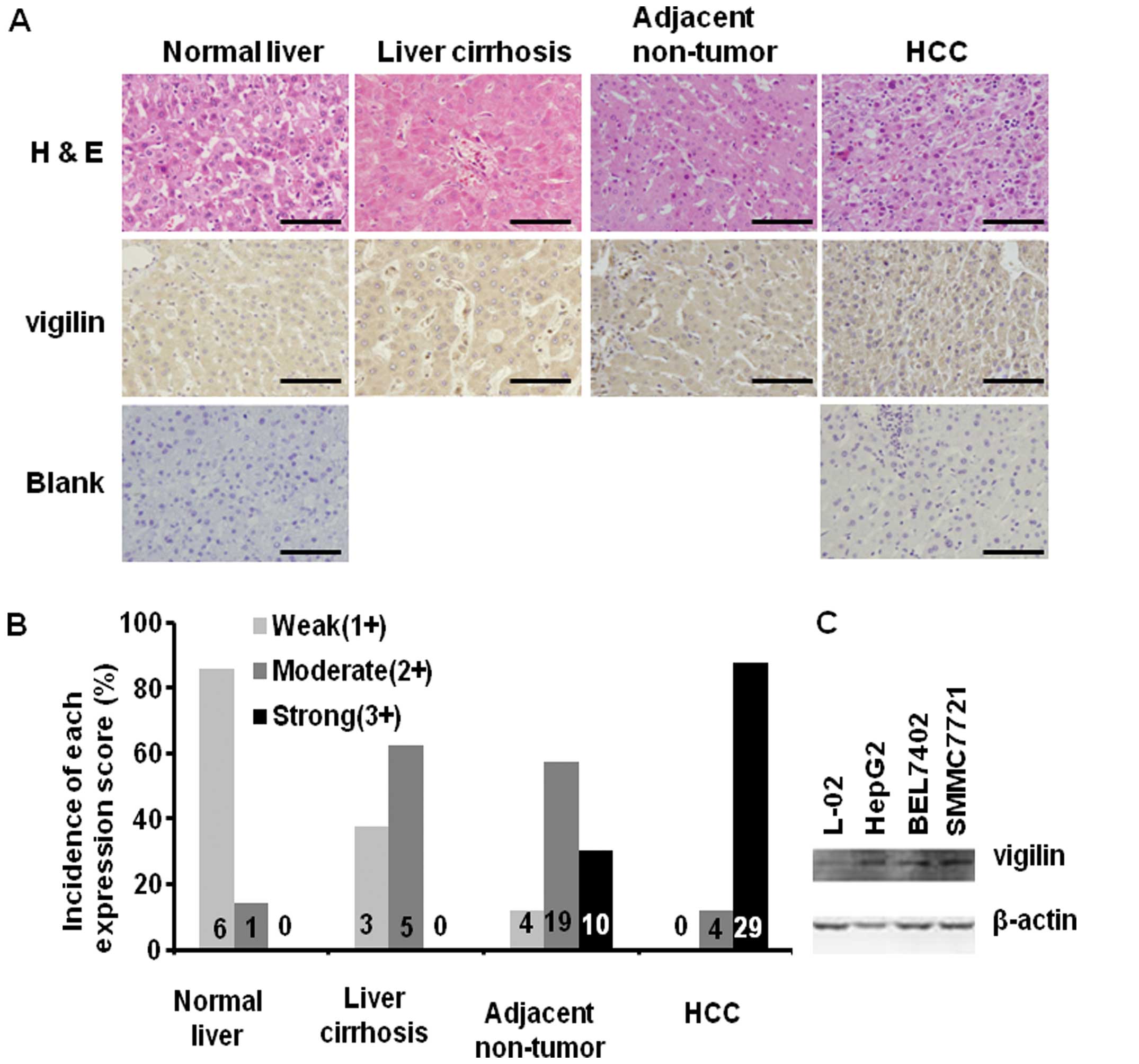
To explore the link between vigilin and HCC, we
performed immunohistochemistry to compare vigilin expression in
normal human liver, liver cirrhosis, adjacent non-tumor liver and
HCC tissues (Fig. 1A). Six out of 7
normal livers exhibited low levels (score=1) of vigilin staining
and the other one had a medium (score=2) level of vigilin staining.
Among the 8 liver cirrhosis samples examined, 3 showed low vigilin
staining and 5 exhibited medium vigilin staining (Fig. 1B). Among the 33 samples containing
HCC and adjacent non-tumor liver tissues, 19 and 10 adjacent
non-tumor hepatocytes exhibited medium and high (score=3) levels of
vigilin expression, respectively, and only one showed low level
vigilin expression. Notalby, as many as 29 of these 33 (88%) HCCs
had high levels of vigilin expression and the other 4 exhibited
medium level of vigilin expression (Fig. 1B). Statistical analysis revealed
that vigilin expression in normal liver and liver cirrhosis had no
significant difference (p>0.05). The level of vigilin expression
in the adjacent non-tumor liver hepatocytes was significantly
higher than that in the normal liver (p<0.001), but was not
significantly different from that in liver cirrhosis (p>0.05).
The level of vigilin expression in HCCs was significantly higher
than the level in all other 3 types of liver tissues (p<0.01).
These results suggest that vigilin is expressed in an increasing
gradient from normal liver to cirrhosis, to adjacent non-tumor
hepatocytes and to HCC. Vigilin is overexpressed in most human
HCCs.
We also compared the levels of vigilin protein in an
embryonic hepatocyte cell line (L-02) and 3 HCC cell lines (HepG2,
BEL7402 and SMMC7721). We found that vigilin was expressed at much
higher levels in the 3 HCC cell lines vs. the L-02 non-tumor
hepatocellular cell line (Fig. 1C).
These results were consistent with the elevated vigilin expression
found in the human HCC specimens. Taken together, our results
suggest that increased vigilin expression may play an important
role in HCC progression.
Knockdown of vigilin decreases HCC cell
proliferation and clonogenicity
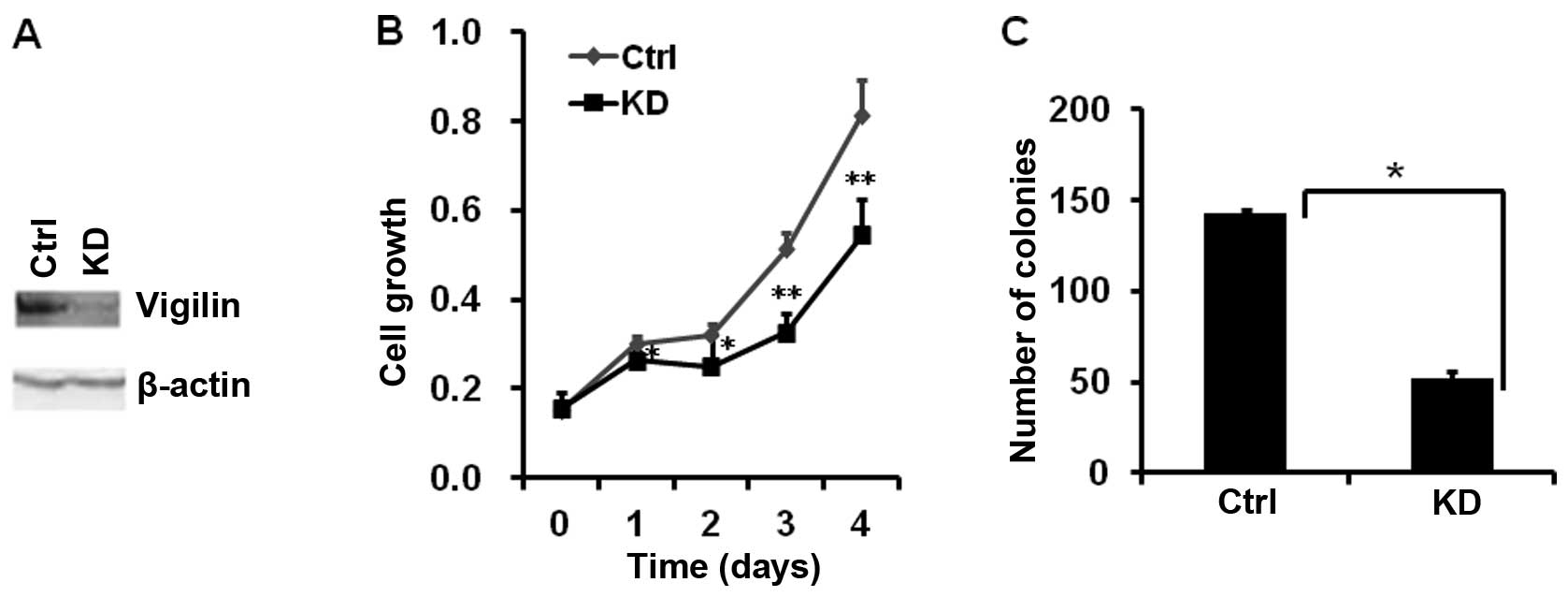
In order to assess the biological role of vigilin in
HCC cells, we generated a stable BEL7402-KD cell line with vigilin
knockdown. The effective knockdown of vigilin in this cell line was
confirmed by RT-PCR and western blotting (Fig. 2A). Knockdown of vigilin in
BEL7402-KD cells significantly decreased their proliferation vs.
BEL7402-Ctrl cells bearing an empty vector. After 4 days of
culture, the number of BEL7402-KD cells was 33% less than the
number of BEL7402-Ctrl cells (Fig.
2B). Furthermore, knockdown of vigilin also drastically reduced
the number of colonies formed from the culture with a low density
of BEL7402-KD cells vs. control cells. The mean numbers of colonies
developed from 200 cells/well in a 6-well plate were 143 from
BEL7402-KD cells and 52 from the control cells (Fig. 2C). These data indicate that vigilin
is required for HCC cell proliferation and survival.
Knockdown of vigilin decreases HCC cell
migration
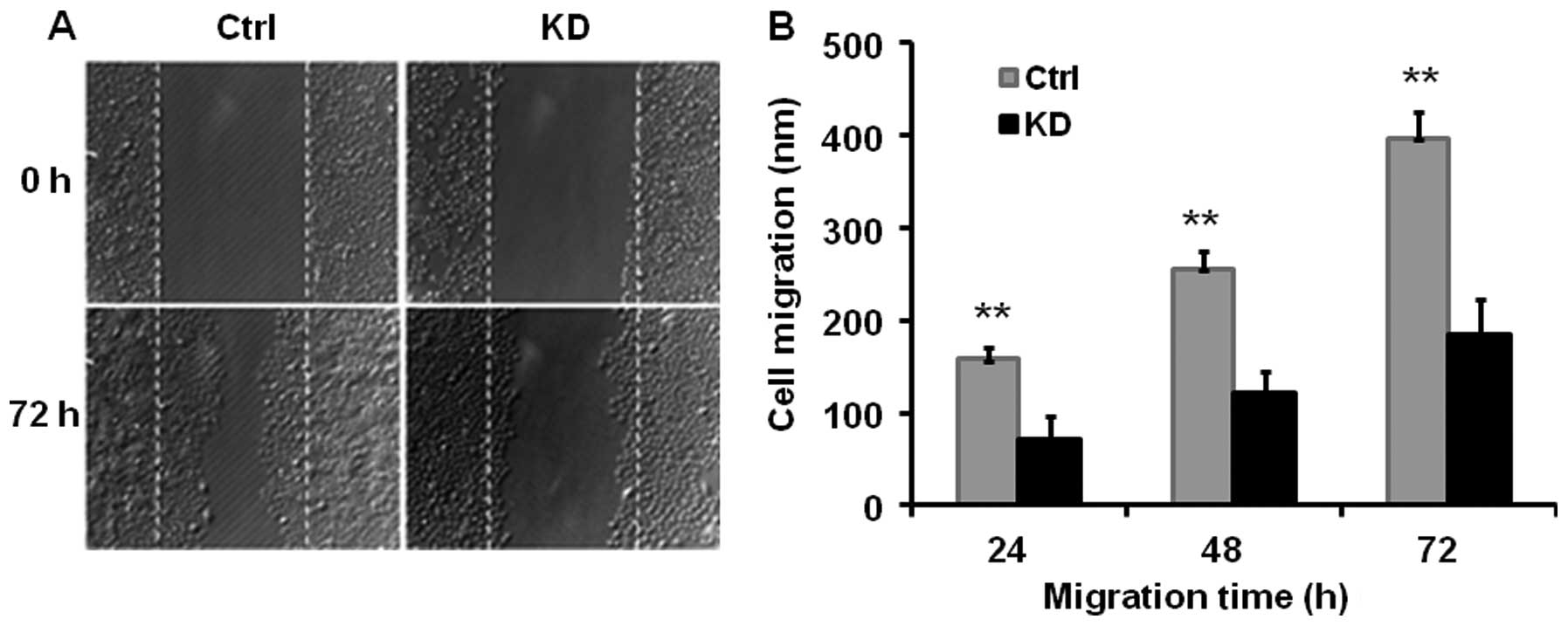
We performed a wound-healing assay to estimate the
effect of vigilin knockdown on HCC cell migration. We found that
knockdown of vigilin in BEL7402-KD cells reduced their migration
capability by ~52% vs. BEL7402-Ctrl cells as measured at 24, 48 and
72 h after performing the scratch wound (Fig. 3). These data indicate that vigilin
plays a promotive role in HCC cell migration.
Knockdown of vigilin inhibits HCC
xenograft tumor growth in nude mice
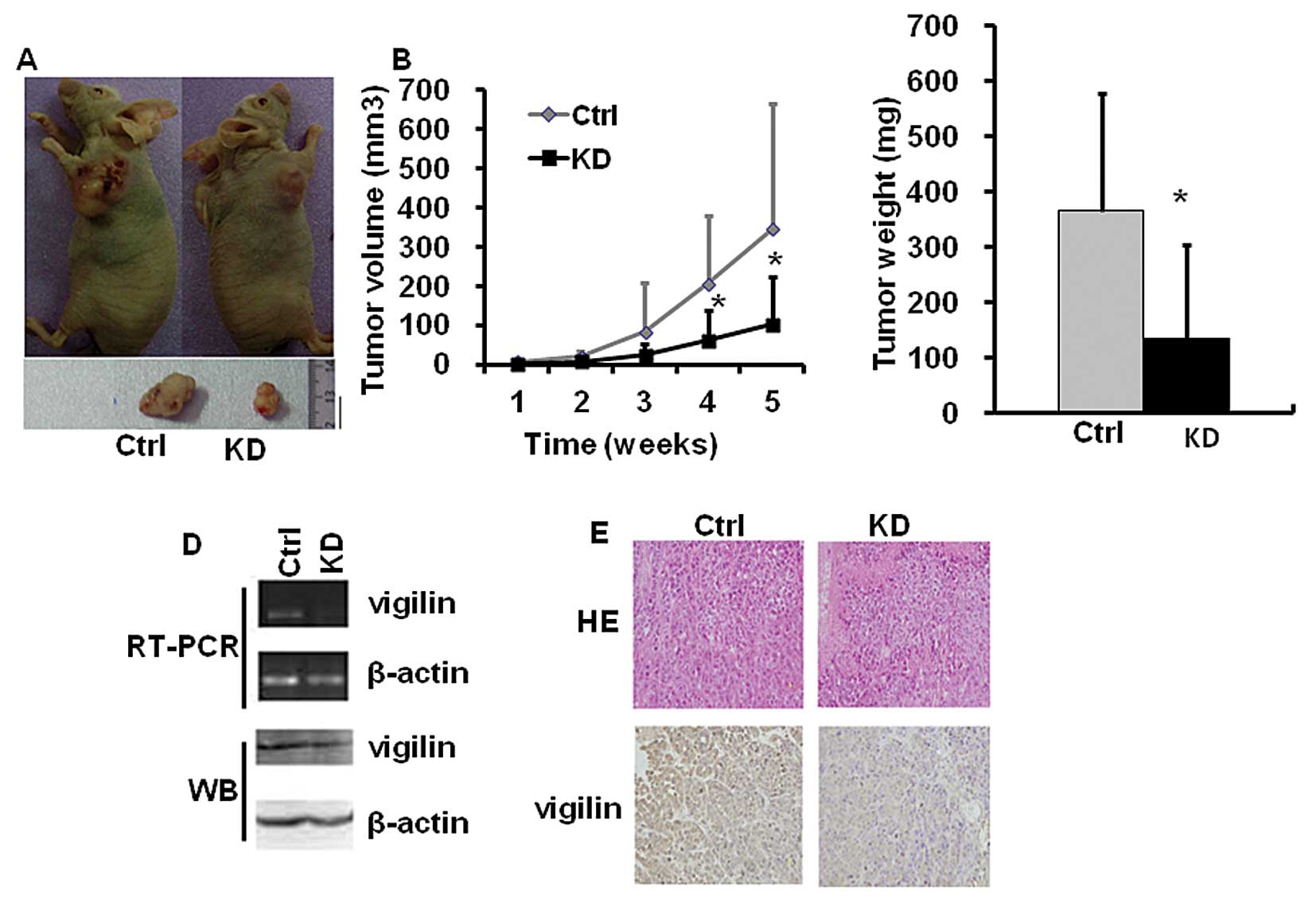
To determine the role of vigilin in HCC tumor
growth, we subcutaneously injected BEL7402-KD cells with vigilin
knockdown and BEL7402-Ctrl cells with vigilin expression into nude
mice, and monitored their tumorigenesis. Vigilin knockdown
significantly attenuated the growth of BEL7402-KD cell-derived
tumors as compared to the growth of BEL7402-Ctrl cell-derived
tumors in nude mice (Fig. 4A). The
average size of BEL7402-Ctrl tumors reached ~400 mm3
within 35 days, while the average size of BEL7402-KD tumors was
only ~110 mm3 within the same period (Fig. 4B). At the experimental end point,
the average weight of BEL7402-KD tumors was only 37% of the weight
of BEL7402-Ctrl tumors (Fig. 4C).
RT-PCR, western blotting and IHC analyses revealed that the levels
of vigilin expression remained low in the BEL7402-KD tumors vs.
BEL7402-Ctrl tumors (Fig. 4D and
E). These results demonstrate that vigilin is required for the
growth of BEL7402 HCC cell-derived tumors in mice.
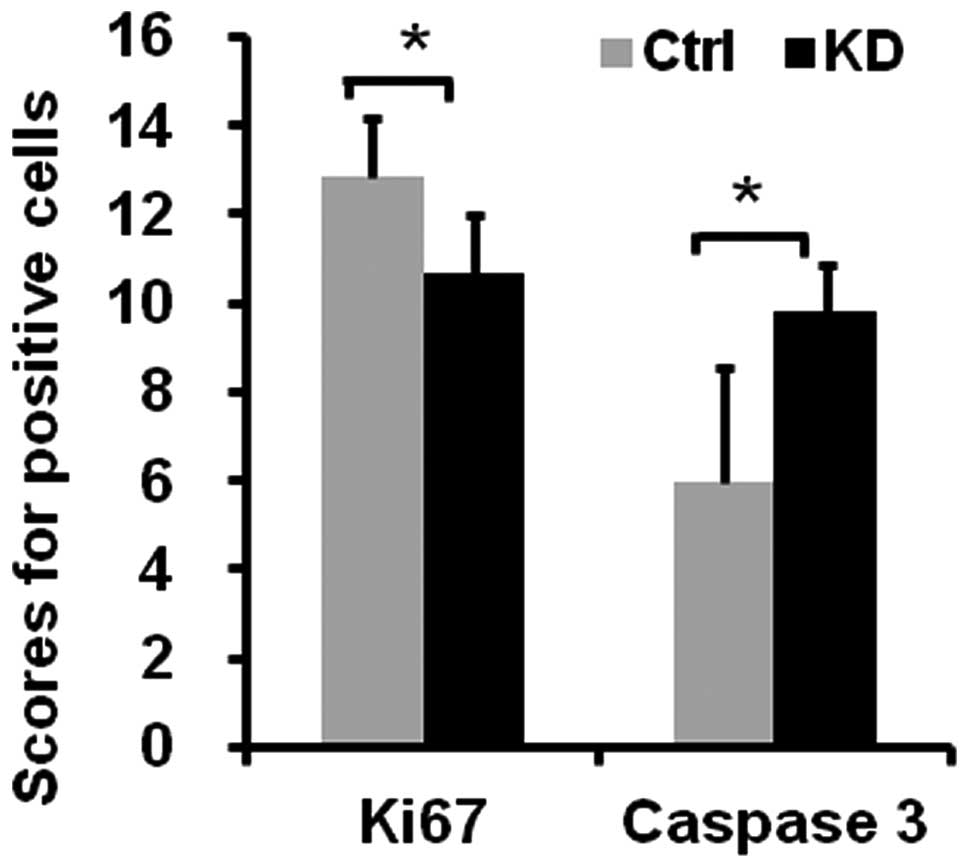
To identify the cellular mechanisms responsible for
vigilin knockdown-suppressed tumor growth in mice, we performed IHC
for Ki67 to detect proliferative cells and IHC for cleaved caspase
3 to detect apoptotic cells. The number of Ki67-positive cells was
decreased 16.8%, while the number of apoptotic cells was increased
63.8% in the BEL7402-KD tumors vs. the BEL7402-Ctrl tumors
(Fig. 5). These results suggest
that knockdown of vigilin suppresses the growth of BEL7402-KD
tumors in mice by inhibiting cell proliferation and promoting cell
apoptosis.
Knockdown of vigilin enhances
cisplatin-mediated inhibition of HCC cell growth
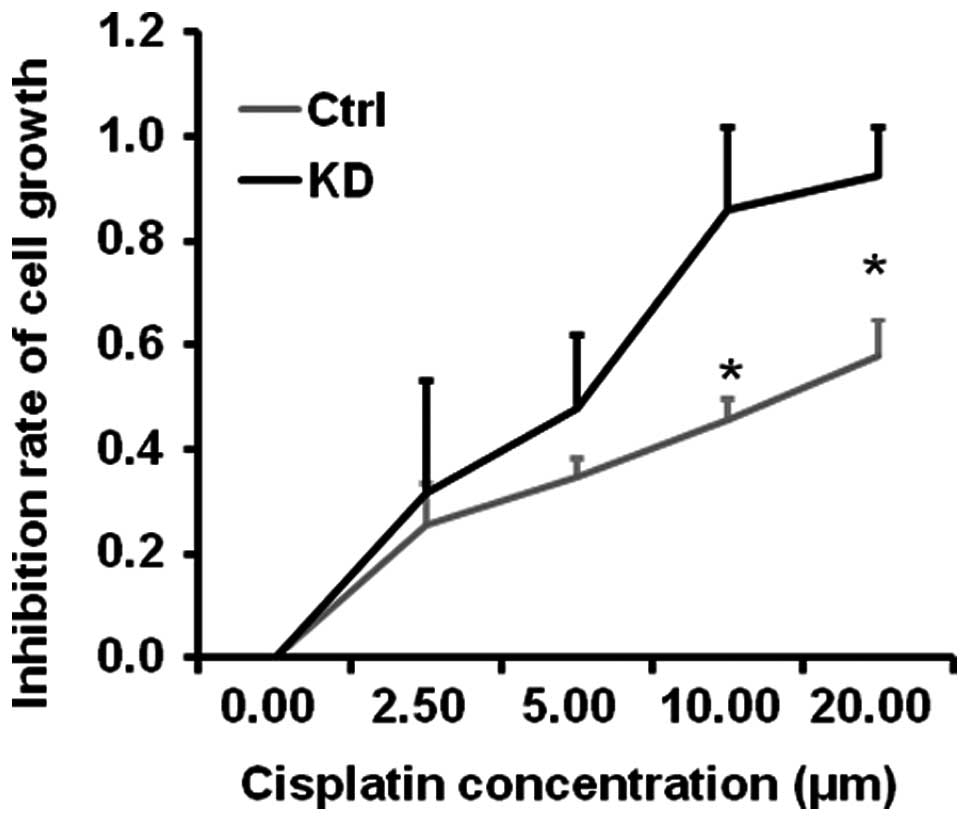
Cisplatin is a commonly used drug for cancer
chemotherapy. To examine whether vigilin knockdown enhances
cisplatin-inhibited HCC cell growth, we treated BEL7402-Ctrl and
BEL7402-KD cells with different concentrations of cisplatin for 72
h. We found that BEL7402-KD cells exhibited much higher sensitivity
than BEL7402-Ctrl cells in response to cisplatin treatment. The
EC50 of cisplatin was ~6 μM for BEL7402-Ctrl cells and
~15 μM for BEL7402-KD cells (Fig.
6). These results suggest that targeting vigilin may sensitize
HCC cells to cisplatin to promote cell killing.
Discussion
In the present study, we demonstrated that the
expression of vigilin is increased in human HCC tissues. The
frequency and degree of vigilin overexpression were also increased
in a gradient from benign lesions, liver cirrhosis to HCC, and each
step showed a significant elevation in vigilin expression. This
suggests that vigilin may promote HCC progression and its
overexpression may also serve as a molecular marker for HCC
progression. Knockdown of vigilin in HCC cells suppressed their
proliferation, clonogenic ability and mobility, suggesting that
vigilin is an important factor that promotes HCC cell proliferation
and tumorigenesis.
Our results demonstrating that vigilin is
overexpression in human HCC specimens and cell lines and that
vigilin promotes HCC cell proliferation and tumorigenesis are
consistent with previous studies showing vigilin overexpression in
other types of cancers. For example, it has been reported that
vigilin is upregulated in TOV112D ovarian cancer cells (16,17),
prolactinomas, gastric cancer (18,19),
Hep-2 larynx carcinoma, HeLa cervix carcinoma, MG63 osteosarcoma,
U937 and HL60 leukemia (27),
pancreatic carcinoma (28) and
LNCaP prostate cancer cells (29).
Together, these results suggest a detrimental role of vigilin in
the development and progression of multiple human types of cancers,
including human HCC. Targeting vigilin in these cancer cells may
have therapeutic value.
Intriguingly, other studies have suggested that
vigilin may be a tumor suppressor in other types of human cancers.
For example, vigilin was recently shown to accelerate the
degradation and inhibit the translation of the c-fms proto-oncogene
mRNA in breast cancer cells (20).
These observations suggest that vigilin may either inhibit or
promote carcinogenesis, depending on different cancer types and
cellular and tissue contexts.
Carcinogenesis, including HCC development, consists
of multiple steps. Most HCCs developed in Chinese patients undergo
a progression from HBV hepatitis infection, followed by liver
cirrhosis and finally to carcinoma (30). However, there are few biomarkers to
screen patients at risk for liver cirrhosis. Notably, the present
study revealed that vigilin expression is frequently and
progressively increased from benign lesions to liver cirrhosis, and
then to HCC, suggesting that vigilin may serve as a potential
molecular marker for evaluating the risk of HCC development. This
assumption is coincident with evidence indicating that vigilin is a
cancer-related antigen in the sera of breast cancer patients
(31) and the existence of vigilin
antibody in the sera of melanoma patients (32).
In conclusion, our findings indicate that vigilin is
frequently overexpressed in human HCCs and may play a crucial role
in HCC cell proliferation, survival, migration and tumor growth.
Since knockdown of vigilin inhibits HCC cell growth, survival and
tumorigenesis, vigilin may be a potential therapeutic target for
HCC treatment, alone or combined with chemotherapeutic agents such
as cisplatin.
Acknowledgements
This study was supported by a grant from the
National Natural Science Foundation of China (no. 30870957). We
thank Mr. Sheng-Fu Li of the Key Laboratory of Transplant
Engineering and Immunology, Sichuan University for experimental
assistance and Dr Jun-Hui Zhang of the West China School of Public
Health of Sichuan University for statistical analysis.
References
|
1
|
Leong TY and Leong AS: Epidemiology and
carcinogenesis of hepatocellular carcinoma. HPB. 7:5–15. 2005.
View Article : Google Scholar
|
|
2
|
Wintersberger U, Kühne C and Karwan A:
Scp160p, a new yeast protein associated with the nuclear membrane
and the endoplasmic reticulum, is necessary for maintenance of
exact ploidy. Yeast. 11:929–944. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kügler S, Grünweller A, Probst C, et al:
Vigilin contains a functional nuclear localisation sequence and is
present in both the cytoplasm and the nucleus. FEBS Lett.
382:330–344. 1996.PubMed/NCBI
|
|
4
|
Vollbrandt T, Willkomm D, Stossberg H, et
al: Vigilin is co-localized with 80S ribosomes and binds to the
ribosomal complex through its C-terminal domain. Int J Biochem Cell
Biol. 36:1306–1318. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Frey S, Pool M and Seedorf M: Scp160p, an
RNA-binding, polysome-associated protein, localizes to the
endoplasmic reticulum of Saccharomyces cerevisiae in a
microtubule-dependent manner. J Biol Chem. 276:15905–15912. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lang BD and Fridovich-Keil JL: Scp160p, a
multiple KH-domain protein, is a component of mRNP complexes in
yeast. Nucleic Acids Res. 28:1576–1584. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baum S, Bittins M, Frey S and Seedorf M:
Asc1p, a WD40-domain containing adaptor protein, is required for
the interaction of the RNA-binding protein Scp160p with polysomes.
Biochem J. 380:823–830. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Klinger MH and Kruse C: Immunocytochemical
localization of vigilin, a tRNA-binding protein, after cell
fractionation and within the exocrine pancreatic cell of the rat.
Ann Anat. 178:331–335. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hilgendorf I, Gellersen O, Emmrich J, et
al: Estradiol has a direct impact on the exocrine pancreas as
demonstrated by enzyme and vigilin expression. Pancreatology.
1:24–29. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kruse C, Grünweller A, Willkomm DK, et al:
tRNA is entrapped in similar, but distinct, nuclear and cytoplasmic
ribonucleoprotein complexes, both of which contain vigilin and
elongation factor 1 α. Biochem J. 329:615–621. 1998.PubMed/NCBI
|
|
11
|
Kruse C, Willkomm D, Gebken J, et al: The
multi-KH protein vigilin associates with free and membrane-bound
ribosomes. Cell Mol Life Sci. 60:2219–2227. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cunningham KS, Dodson RE, Nagel MA, et al:
Vigilin binding selectively inhibits cleavage of the vitellogenin
mRNA 3′-untranslated region by the mRNA endonuclease polysomal
ribonuclease 1. Proc Natl Acad Sci USA. 97:12498–12502.
2000.PubMed/NCBI
|
|
13
|
Zhou J, Wang Q, Chen LL and Carmichael GG:
On the mechanism of induction of heterochromatin by the RNA-binding
protein vigilin. RNA. 14:1773–1781. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Plenz G, Kügler S, Schnittger S, et al:
The human vigilin gene: identification, chromosomal localization
and expression pattern. Hum Genet. 93:575–582. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chiu DS, Oram JF, LeBoeuf RC, et al:
High-density lipoprotein-binding protein (HBP)/vigilin is expressed
in human atherosclerotic lesions and colocalizes with
apolipoprotein E. Arterioscler Thromb Vasc Biol. 17:2350–2358.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gagné JP, Gagné P, Hunter JM, et al:
Proteome profiling of human epithelial ovarian cancer cell line
TOV-112D. Mol Cell Biochem. 275:25–55. 2005.PubMed/NCBI
|
|
17
|
Kim H and Lubman DM: Micro-proteome
analysis using micro-chromatofocusing in intact protein
separations. J Chromatogr A. 1194:3–10. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Evans CO, Moreno CS, Zhan X, et al:
Molecular pathogenesis of human prolactinomas identified by gene
expression profiling, RT-qPCR, and proteomic analyses. Pituitary.
11:231–245. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim NS, Hahn Y, Oh JH, et al: Gene
cataloging and expression profiling in human gastric cancer cells
by expressed sequence tags. Genomics. 83:1024–1045. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Woo HH, Yi X, Lamb T, et al:
Posttranscriptional suppression of proto-oncogene c-fms
expression by vigilin in breast cancer. Mol Cell Biol. 31:215–225.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei L, Zhang CL, Jiang L, et al:
Expression of human VIGILIN N terminus and observation of
subcellular localization. Sichuan Da Xue Xue Bao Yi Xue Ban.
40:185–189. 2009.(In Chinese).
|
|
22
|
Reiner A, Neumeister B, Spona J, et al:
Immunocytochemical localization of estrogen and progesterone
receptor and prognosis in human primary breast cancer. Cancer Res.
50:7057–7061. 1990.PubMed/NCBI
|
|
23
|
Stewart SA, Dykxhoorn DM, Palliser D, et
al: Lentivirus-delivered stable gene silencing by RNAi in primary
cells. RNA. 9:493–501. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goolsby KM and Shapiro DJ: RNAi-mediated
depletion of the 15 KH domain protein, vigilin, induces death of
dividing and non-dividing human cells but does not initially
inhibit protein synthesis. Nucleic Acids Res. 31:5644–5653. 2003.
View Article : Google Scholar
|
|
25
|
Liang CC, Park AY and Guan JL: In vitro
scratch assay: a convenient and inexpensive method for analysis of
cell migration in vitro. Nat Protoc. 2:329–333. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Galvez AF, Chen N, Macasieb J and de Lumen
BO: Chemopreventive property of a soybean peptide (lunasin) that
binds to deacetylated histones and inhibits acetylation. Cancer
Res. 61:7473–7478. 2001.PubMed/NCBI
|
|
27
|
Robinson WS: The role of hepatitis B virus
in the development of primary hepatocellular carcinoma: part I. J
Gastroenterol Hepatol. 7:622–638. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wadle A, Mischo A, Imig J, et al:
Serological identification of breast cancer-related antigens from a
Saccharomyces cerevisiae surface display library. Int J
Cancer. 117:104–113. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kalnin̦a Z, Silin̦a K, Meistere I, et al:
Evaluation of T7 and lambda phage display systems for survey of
autoantibody profiles in cancer patients. J Immunol Methods.
334:37–50. 2008.PubMed/NCBI
|
|
30
|
Neu-Yilik G, Zorbas H, Gloe TR, et al:
Vigilin is a cytoplasmic protein. A study on its expression in
primary cells and in established cell lines of different species.
Eur J Biochem. 213:727–736. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schuh A, Assmuth K, Hilgendorf I, et al:
Protein synthesis of eucaryotic cells could be decreased by
antisense-DNA of the multi KH domain protein vigilin. Int J Mol
Med. 12:35–43. 2003.PubMed/NCBI
|
|
32
|
Oh-Ishi M and Maeda T: Disease proteomics
of high-molecular-mass proteins by two-dimensional gel
electrophoresis with agarose gels in the first dimension (Agarose
2-DE). J Chromatogr B Analyt Technol Biomed Life Sci. 849:211–222.
2007. View Article : Google Scholar : PubMed/NCBI
|