1. Introduction
LIM homeobox genes are one of the most important
subfamilies of homeobox genes. They encode a series of
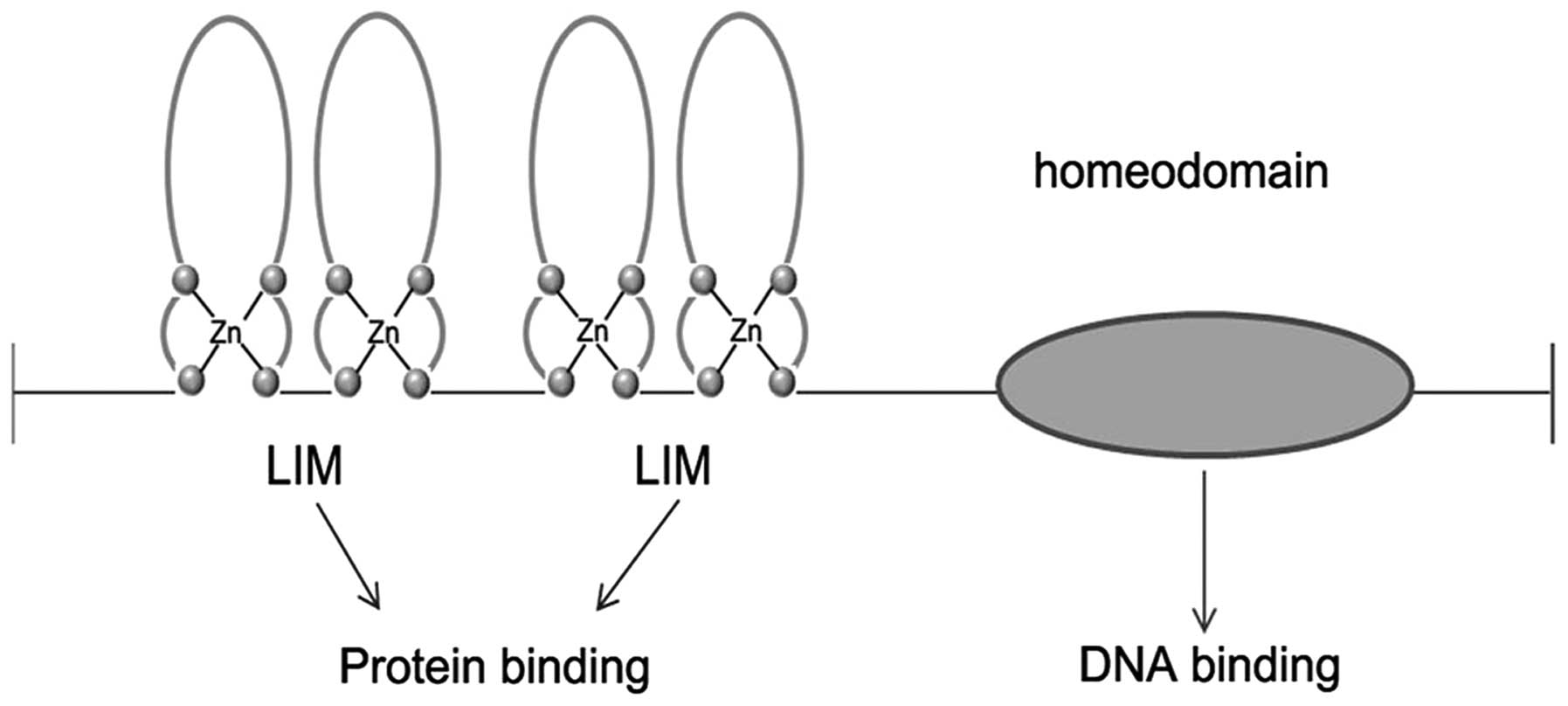
LIM-homeodomain (LIM-HD) proteins featuring two LIM domains in
their amino termini and a centrally located HD that is used to
interact with specific DNA elements in target genes (Fig. 1). This special structure can set
LIM-HD proteins apart from other transcription factors of
homeodomain superfamilies such as paired homeobox (PAX) proteins,
Pit-Oct-Unc (POU)-HD and HOX (homeobox) cluster proteins (1,2). Their
LIM domain contains two tandemly-repeated, cysteine-rich,
double-zinc finger motifs that can be recognized by a number of
co-factors which can mediate LIM-HD functions. These LIM
domain-interacting proteins include NLI/LDB/CLIM/CHIP, MRG1, SLB
and RLIM, a ubiquitin protein ligase. What is more, by means of the
LIM domain, LIM-HD proteins can interact with other transcription
regulators; thus, they can participate in a wide array of
developmental events (3–6).
Mammalian genomes such as those of the mouse, rat
and human contain at least 12 LIM homeobox genes (Table I) that encode key regulators of
developmental pathways. Studies with mouse models and human
patients have shown that these genes are critical for the
development of specialized cells in multiple tissue types including
the nervous system, skeletal muscle, the heart, the kidneys and
endocrine organs such as the pituitary gland and the pancreas
(1).
 | Table IHuman LIM homeobox genes in the HUGO
Gene Nomenclature Committee (HGNC) database. |
Table I
Human LIM homeobox genes in the HUGO
Gene Nomenclature Committee (HGNC) database.
| HGNC name | Symbol | Synonym
symbol(s) | Chromosomal
location |
|---|
| LIM homeobox 1 | LHX1 | LIMK4, LIM-1, LIM1,
MGC126723, MGC138141 | 17q12 |
| LIM homeobox 2 | LHX2 | LH2, MGC138390 | 9q33.3 |
| LIM homeobox 3 | LHX3 | DKFZp762A2013,
LIM3, M2-LHX3, CPHD3 | 9q34.3 |
| LIM homeobox 4 | LHX4 | Gsh-4, Gsh4 | 1q25.2 |
| LIM homeobox
protein 5 | LHX5 | MGC129689 |
12q24.31–q24.32 |
| LIM homeobox
protein 6 | LHX6 | LHX6.1, MGC119542,
MGC119544, MGC119545 | 9q33.2 |
| LIM homeobox 8 | LHX8 | Lhx7 | 1p31.1 |
| LIM homeobox 9 | LHX9 | / | 1q31.3 |
| ISL LIM homeobox
1 | ISL1 | ISL-1, ISLET1 | 5q11.1 |
| ISL LIM homeobox
2 | ISL2 | FLJ10160,
ISL-2 | 15q24.3 |
| LIM homeobox
transcription factor 1, α | LMX1A | LMX1-1, LMX-1,
LMX1 | 1q22–q23 |
| LIM homeobox
transcription factor 1, β | LMX1B | LMX1-2, NPS1,
MGC142051, MGC138325 | 9q33.3 |
Although many researchers have reported their
fundamental roles in the development of various organisms, little
is known concerning the functions of these LIM-HD proteins in
cancer. Recently, studies have shown that LIM homeobox genes also
play an important role in cancer. Among 12 human LIM homeobox
genes, 10 LIM-HD proteins have been reported to be associated with
various types of cancer (Table
II). In this review, we mainly summarize the functions of human
LIM-HD proteins in cancer development and their potential as
biomarkers and the related challenges. In addition, we discuss the
role of these LIM homeobox genes in cancer from a signaling pathway
perspective.
 | Table IISummary of studies on the role of LIM
homeobox transcription factors in cancer. |
Table II
Summary of studies on the role of LIM
homeobox transcription factors in cancer.
| LIM-HD | Main developmental
function | Possible roles in
cancer | Gene expression
control mechanism | Related cancer | Refs. |
|---|
| LHX1 | Essential for
development of human renal and urogenital systems | Oncogene | Unknown | Nephroblastoma | (11) |
| | Clear cell renal
cell carcinoma (CCC) | (12) |
| LHX2 | Functions as a
transcriptional regulatory protein in the control of lymphoid and
neural cell differentiation | Oncogene | Overexpression due
to chromosome translocation or hypomethylation | Chronic myeloid
leukemia (CML) | (15–17) |
| Unknown | Pancreatic
cancer | (18) |
| Methylation
biomarker | Downregulated by
hypermethylation | Non-Hodgkin’s
lymphoma (NHL) | (21) |
| | Breast cancer | (22,23) |
| | Lung cancer | (24) |
| LHX3 | Required for
pituitary development and motor neuron specification | Methylation
biomarker | Downregulated by
hypermethylation | Breast cancer | (29) |
| LHX4 | Plays a role in
nervous system development | Oncogene | Overexpression due
to chromosome translocation | Acute lymphoblastic
leukemia (ALL) | (34,35) |
| Deregulated by
protein - protein interaction | Synovial
sarcoma | (36) |
| Unknown | Prostate
cancer | (37) |
| Methylation
biomarker |
Hypermethylation | Lung cancer | (23) |
| | Hepatocellular
carcinoma | (38) |
| LHX5 | Essential role in
brain development or compensatory actions of LHX1 | Methylation
biomarker |
Hypermethylation | Breast cancer | (23) |
| LHX6 | Involved in the
control of differentiation and development of neural and lymphoid
cells | Methylation
biomarker |
Hypermethylation | Head and neck
squamous cell carcinoma (HNSCC) | (43) |
| | Cervical
cancer | (44,45) |
| LHX7/8 | No report | | | | |
| LHX9 | Plays a role in
gonadal development and may be involved in the control of cell
differentiation of several neural cell types | Tumor-suppressor
gene | Downregulated by
hypermethylation | Pediatric malignant
astrocytomas | (47) |
| Hypermethylation
accompanied by transcriptional repression | Follicular
lymphoma | (48) |
| SL1 | Central to the
development of pancreatic cell lineages and may also be required
for motor neuron generation | Cancer
biomarker | Unknown | Pancreatic
neuroendocrine tumors | (50,54–57) |
| Potential minimal
residual disease (MRD) marker | Unknown | Neuroblastoma
(NB) | (58) |
| ISL2 | No report | | | | |
| LMX1A | Acts as a positive
regulator of insulin gene transcription and it also plays a role in
the development of dopamine producing neurons during
embryogenesis | Oncogene | Unknown | Mucinous
cystadenocarcinoma | (61) |
| | Glioma | (62) |
| | Pancreatic ductal
adenocarcinomas | (63) |
| Tumor suppression
gene and methylation biomarker |
Hypermethylation | Cervical
cancer | (64–66) |
| | Ovarian cancer | (67,68) |
| | Bladder cancer | (69) |
| | Gastric cancer | (70) |
| | Colon cancer | (71) |
| LMX1B | Essential for the
normal development of dorsal limb structures, the glomerular
basement membrane, the anterior segment of the eye, and
dopaminergic and serotonergic neurons | Regulate other
oncogene overexpression | Through Wnt7a-LMX1b
signaling pathway | Breast cancer | (74) |
2. Relationship of LIM homeobox genes and
cancer
LHX1 and cancer
LHX1, also called LIM1, is required for head, kidney
and female reproductive tract development in the murine embryo
(7–9). Most LIM1 null mutants die with a
headless phenotype around E10.5; the few surviving newborns had no
kidney (7). Conditional ablation of
LIM1 in the metanephric mesenchyme blocks the formation of nephrons
at the nephric vesicle stage, leading to the production of small,
non-functional kidneys that lack nephrons (10).
Guertl et al (11) investigated LIM1 expression during
human renal development, in dysplastic kidneys and in renal
neoplasms and found that LIM1 was detected in pretubular
aggregates, S-shaped and comma-shaped bodies as well as immature
glomeruli between 10 and 30 weeks of gestation. Eleven dysplastic
kidneys showed no expression of LIM1. In contrast, 12 of 32
nephroblastomas showed nuclear positivity. One regressive
nephroblastoma had diffuse expression of LIM1 in tubular
structures, all others showed focal positivity in mesenchymal,
blastemal and epithelial structures. Renal cell carcinomas revealed
no expression of LIM1. Their study supports the concept of a
causative role of LIM1 deficiency in the development of multicystic
kidney. In a small subset of nephroblastomas with a more diffuse
expression pattern, LIM1 might also contribute to the pathogenesis
of these lesions.
Dormoy et al (12) further assessed whether LIM1 may be
associated with tumorigenesis. They found that LIM1 is
constitutively and exclusively reexpressed in tumors as a growth
and survival factor in human clear cell renal cell carcinoma (CCC).
More importantly, in nude mice bearing human CCC, LIM1 silencing
abolished tumor growth through the same mechanism as in
vitro. In LIM1-depleted cells and tumors, cell motility was
substantially impaired due to the inhibition of expression of
various proteins involved in metastatic spread, such as paxillin or
tenascin-C.
LHX2 and cancer
The LHX2 gene functions as a transcriptional
regulatory protein in the control of lymphoid and neural cell
differentiation. LHX2 was initially identified as an early marker
in B-lymphocyte differentiation (13). Overexpression of LHX2 in murine
hematopoietic precursors leads to the development of chronic
myeloproliferative disorders (14).
LHX2 was once found to be deregulated by
juxtaposition with the IGH locus in a pediatric case of chronic
myeloid leukemia (CML) in B-cell lymphoid blast crisis. The strong
overexpression of LHX2 induced by t(9;14)(q33;q32) may play a
recurrent role in leukemogenesis, specifically when it is in
association with BCR-ABL chimera of the Philadelphia chromosome
(Ph′), the hallmark of CML (15).
High levels of LHX2 expression were also observed in
all cases of CML tested, regardless of disease status and may prove
useful as a marker of CML for monitoring residual disease (16). Since LHX2 was mapped to chromosome
9q33–34.1, in the same region as the reciprocal translocation that
creates the BCR-ABL. they also confirmed that the transcriptional
activation of LHX2 in CML is likely due to a cis-acting
effect, but not a trans-acting effect, of the Bcr-Abl fusion
protein. The high level of LHX2 mRNA in CML cells probably can also
be a consequence of the low level of methylation of the gene in
leukemic cells (17).
Gorantla et al (18) reported the presence of uPA in the
nucleus as well as the interaction of uPA with LHX2. LHX2 was
overexpressed at the invasive front of tumors while completely
absent in tumors downregulated for both uPAR and uPA. Suppression
of the uPAR-uPA system retards angiogenesis, invasion, and in
vivo tumor development in pancreatic cancer cells. Recently,
LHX2 was also identified as a transcription factor functionally
positioned downstream of p63 and NF-κB, but upstream of signals
such as Wnt/β-catenin, BMP, and SHH that are required to drive
activated stem cells toward terminal differentiation (19). This would indicate that LHX2 may
behave like a gatekeeper molecule mediating the activation of
cancer stem cells.
Notably, besides being overexpressed as an oncogene
in cancer, LHX2 may be downregulated due to hypermethylation in
cancer. Homeobox genes that are upregulated in cancer may be
normally expressed during development and/or in undifferentiated
cells, whereas others that are downregulated in cancer may be
normally expressed in adulthood and/or in differentiated tissues
(20).
Rahmatpanah et al (21) used an array-based technique called
differential methylation hybridization (DMH) to study small B-cell
lymphoma (SBCL) subtypes and found that hypermethylation of only
one gene, LHX2, was present in all Non-Hodgkin’s lymphoma (NHL)
cell lines and a high proportion of patient samples.
Kim et al (22) utilized a genome-wide technique,
methylated DNA isolation assay (MeDIA), in combination with
high-resolution CpG microarray analysis to identify hypermethylated
genes in breast cancer. LHX2 showed significantly higher
frequencies of aberrant hypermethylation in primary tumors (43.6%;
P<0.05) while frequencies were intermediate in paired adjacent
normal tissues and absent in normal tissues. Moreover, a
significant number of genes (2853; P<0.05) exhibited an
expression-methylation correlation in breast cancer. Among these
genes, LHX2 and LHX5 had prognostic value independent of subtypes
and other clinical factors (23).
In lung cancer, Rauch et al (24) identified frequent methylation of
homeodomain-containing genes including LHX2 and LHX4 using
methylated-CpG island recovery assay (MIRA)-assisted microarray
analysis. Although low levels of methylation were detected in some
normal tissues removed by tumor surgery, methylation of LHX2 and
LHX4 was more pronounced in tumor samples.
LHX3 and cancer
LHX3 null mice were found to have defective
development of spinal cord motor neurons (25) and also to exhibit incomplete
pituitary development (26,27). LHX3 functions to specify interneuron
and motor neuron fates during development (28). To date, LHX3 has only been shown to
be correlated with breast cancer. Dietrich et al (29) reported that elevated methylation of
LHX3 was detected in invasive ductal carcinoma (IDC) and ductal
carcinoma in situ (DCIS) compared with normal ducts,
adenosis tissue, stroma and tumor infiltrating lymphocytes
(TILs).
LHX4 and cancer
During embryonic and postnatal development, the LHX4
gene is expressed in hindbrain, cerebral cortex, pituitary gland
and spinal cord, suggesting that LHX4 plays a role in nervous
system development (30,31). Mutations in LHX4 genes are
associated with combined hormone deficiency diseases in human and
animal models (32,33).
Similar to LHX2, LHX4 can be upregulated by
chromosomal translocations and downregulated by hypermethylation in
cancer. Firstly, LHX4 mRNA was found to be expressed at high levels
in the leukemic cells of patients and in an acute lymphoblastic
leukemia (ALL) cell line due to the t(1;14)(q25;q32) suggesting a
rare but recurrent genetic abnormality in leukemogenesis (34,35).
Moreover, de Bruijn et al (36) reported that the SS18 gene on
chromosome 18 is fused to either one of the three closely related
SSX genes on the X chromosome as a result of the synovial
sarcoma-associated t(X;18) translocation. They found that
endogenous LHX4 binds to the CGA promoter and that LHX4-mediated
CGA activation is enhanced by the SS18-SSX protein, but not by the
SSX protein and suggested that this novel protein-protein
interaction may have direct consequences for the deregulation of
SS18-SSX target gene LHX4 in the development of human synovial
sarcomas.
Meanwhile, Goc et al (37) provided an initial report that
simvastatin simultaneously modulated intrinsic and extrinsic
pathways in the regulation of prostate cancer cell apoptosis in
vitro and in vivo and also significantly reduced protein
levels of pro-survival gene LHX4, but the exact function is yet to
be determined.
In contrast, low expression of LHX4 was found in
primary lung tumor samples (23)
and hepatocellular carcinoma (HCC) (38) when compared with the expression
level in paired non-cancerous tissues. Moreover, low expression of
LHX4 was associated with tumor undifferentiation state and a high
AFP level in HCC. Functional studies revealed that ectopic
expression of LHX4 reduced AFP expression, which leads to the
suppression of HCC growth. The results indicate that LHX4 functions
as a potential tumor suppressor in hepatocarcinogenesis.
LHX5 and cancer
Human LHX5 is expressed in the adult central nervous
system, including the spinal cord, thalamus and the cerebellum
(39). The mouse LHX5 protein is
closely related to the LHX1 LIM-HD factor, and complementary or
overlapping roles of these two regulatory proteins have been
suggested (40). Mice that are
heterozygous for a LHX5 gene disruption appear normal, while most
homozygous mice die a few days after birth (41).
Together with LHX2, LHX5 was found to have
prognostic value independent of subtypes and other clinical factors
in breast cancer (22).
LHX6 and cancer
LHX6 encodes an LIM-homeodomain protein involved in
embryogenesis, more specifically in mammalian head development
(42). Estécio et al
(43) identified LHX6 as a new
frequent cancer-associated hypermethylated CpG island in human head
and neck squamous cell carcinoma (HNSCC). The hypermethylation of
this fragment was detected in 13 of 14 (92.8%) HNSCC cell lines
studied and in 21 of 32 (65.6%) primary tumors, whereas little or
no methylation was noted in 10 normal oral mucosa samples. They
extended this investigation to other cancer cell lines and
methylation was found in those derived from colon, breast,
leukaemia, lung and in 12/14 primary colon tumors.
LHX6 methylation was also found in cervical cancer
cell lines and cancer tissues (44). This epigenetic alteration in the
LHX6 promoter begins at a relatively early stage as CIN I,
suggesting its potential as a biomarker for early diagnosis
(45). Moreover, overexpression of
the LHX6 gene in cervical cancer cells was found to suppress the
tumorigenic phenotype, as shown by soft agar colony formation and
migration assays, suggesting that LHX6 could be a novel
tumor-suppressor gene in the cervix.
LHX9 and cancer
The expression pattern and structural
characteristics of LHX9 suggest that it encodes a transcription
factor that might be involved in the control of cell
differentiation of several neural cell types (46). The LHX9 gene is frequently silenced
in pediatric malignant astrocytomas by hypermethylation and this
epigenetic alteration is involved in glioma cell migration and
invasiveness (47). LHX9
hypermethylation accompanied by transcriptional repression was
found in follicular lymphoma (48).
ISL1 and cancer
The ISL1 transcription factor was initially cloned
from pancreatic insulin-producing cells where it is able to bind
the insulin gene enhancer (49).
Mice deficient in ISL1 fail to form the dorsal exocrine pancreas,
and islet cells fail to differentiate (50). In addition to its roles in the
pancreas and heart, ISL1 is one of the earliest markers for motor
neuron differentiation (51).
Thus, it is not surprising that ISL1 has been shown
to be a sensitive marker of pancreatic islet cells and their
neoplasms (50,52–56).
ISL1 has been reported as a sensitive lineage-specific marker for
pancreatic neuroendocrine neoplasms (NENs) and their metastases.
Graham et al (54) also
studied its specificity with large numbers of NENs from other parts
of the gut or other organs. They found that ISL1 does not
distinguish pancreatic NENs from duodenal and colorectal NENs, even
when used in association with CDX2. On the other hand, in order to
better understand the expression of the four transcription factors
(TFs): ISL1, pancreatico-duodenal homeobox 1 gene product (PDX1),
neurogenin 3 gene product (NGN3), and CDX-2 homeobox gene product
(CDX2), that mainly govern the development and differentiation of
the pancreas and duodenum, Hermann et al (57) studied their expression in hormonally
defined pancreatic neuroendocrine tumors (P-NETs) and duodenal
neuroendocrine tumors (D-NETs). They found a correlation between TF
expression patterns and certain hormonally defined P-NET and D-NET
types, suggesting that most of the tumor types originate from
embryologically determined precursor cells. However, the observed
TF signatures cannot distinguish P-NETs from D-NETs.
In addition, Cheung et al (58) explored potential minimal residual
disease (MRD) markers differentially expressed in neuroblastoma
(NB) tumors over normal marrow/blood with genome-wide expression
profiling and identified 8 top-ranking markers: CCND1, CRMP1, DDC,
GABRB3, ISL1, KIF1A, PHOX2B and TACC2. They were abundantly
expressed in stage 4 NB tumors (n=20) and had low to no detection
in normal marrow/blood samples (n=20). Moreover, expression of
CCND1, DDC, GABRB3, ISL1, KIF1A and PHOX2B in 116 marrows sampled
after 2 treatment cycles was highly prognostic of progression-free
and overall survival (P<0.001).
LMX1A and cancer
The LMX1A gene maps to 1q22–q23 and was proven to be
a critical regulator of cell-fate decisions using genetic fate
mapping in wild-type and LMX1A−/− mice (59). LMX1A was also found to play a
pivotal role in the differentiation of human embryonic stem cells
into midbrain dopaminergic neurons (60).
Recent evidence has shown an important role of LMX1A
in cancer. For example, Lin et al (61) reported that higher immunostaining
scores and the percentage of cells stained for LMX1A in mucinous
cystadenocarcinomas correlated with T stage, American Joint
Committee on Cancer clinical stage, poorer tumor differentiation,
and poorer survival rate. In addition, a higher intensity of
immunoreactivity for LMX1A correlated with more advanced grade in
WHO grade I–III gliomas, but not in WHO grade IV tumors (62). In addition, higher expression of
LMX1A and OPN was found to be highly correlated with histologic
grade and pathologic stage of pancreatic ductal adenocarcinomas
(63).
In contrast, LMX1A was identified as a metastasis
suppressor in cervical cancer (64–66).
It was once reported that the methylation of LMX1A correlated with
the recurrence and overall survival due to the mechanisms affecting
epithelial-mesenchymal transition (EMT) and stem-like properties in
ovarian cancer (67,68). The methylation of LMX1A was also
found to be associated with bladder cancer recurrence (69). Dong et al (70) found that the expression of LMX1A was
significantly decreased due to the hypermethylation in gastric
cancer tissues compared with normal tissues. Restoration of LMX1A
induced cell apoptosis and suppressed anchorage-independent growth,
suggesting that LMX1A is a potential biomarker for gastric cancer.
Moreover, LMX1A hypermethylation was reported in a colon cancer
cell line (HCT-116), but was demethylated in DKO cells in which two
major DNA methyltransferases, DNMT1 and DNMT3b, were genetically
disrupted (71). However, its role
in transformation has not yet been characterized.
LMX1B and cancer
Together with LMX1A, LMX1B is part of a related
subfamily of LIM-HD genes. Using targeted disruption of the mouse
LMX1B gene, multiple functions of this factor have been uncovered.
Mice lacking functional LMX1B exhibit numerous abnormalities,
including a lack of ciliary body, iris stroma and corneal dysplasia
(72,73).
To date, LMX1B has only been suggested to be
associated with breast cancer. Rieger et al (74) found that LBH was overexpressed in
highly invasive ER-negative, basal subtype human breast cancers and
suppressed the differentiation of HC11 mammary epithelial cells. In
further support of a clinical association of LBH with Wnt/β-catenin
signaling, additional meta-analysis showed that LBH overexpression
also correlates with Wnt pathway gene expression in colon cancer,
which is primarily driven by Wnt activating mutations. Wnt7a-LMX1B
signaling may be an important repressive mechanism that blocks
Wnt/β-catenin target gene expression, and consequently ventral
differentiation, in dorsal limb ectoderm (75). Thus, LBH may act as a downstream
effector of this signaling in both normal and neoplastic epithelial
development, which is under the tight control of antagonistic
non-canonical Wnt7a signaling (76).
3. Targeting the developmental pathways to
explore the role of LIM homeobox genes in cancer
Although numerous LIM homeobox genes are known to be
involved in cancer, the roles of these genes in cancer remain
unclear. Few LIM homeobox gene signaling pathway studies in cancer
are available. Among these, LMX1A and LIM1 have been the subjects
of a certain degree of intensive study.
Lai et al (65) demonstrated that LMX1A may have a
critical role in preventing cervical cancer invasion and metastasis
by inhibiting different aspects of EMT. They found that besides
inhibiting cervical cancer invasion, the restoration of LMX1A
altered expression of epithelial and mesenchymal markers in two
cervical cancer cell lines (HeLa-3rd and CaSki). Furthermore, by
analyzing TGF-β signaling, they found that BMP4 and BMP6 were
downregulated by LMX1A. BMPs have been confirmed to be important
components of LMX1A-dependent roof plate signaling, in which BMP6
is dependent on LMX1A expression during spinal cord development in
mice (77). Moreover, LMX1A was
found to be a mediator of early BMP signaling, and its activation
of early roof plate development is dependent on BMP4 signaling in
chicks (78). These complex
regulatory networks of different BMPs and LMX1A in diverse
microenvironments and tissues during embryonic development may also
hint to its roles in cancer biology.
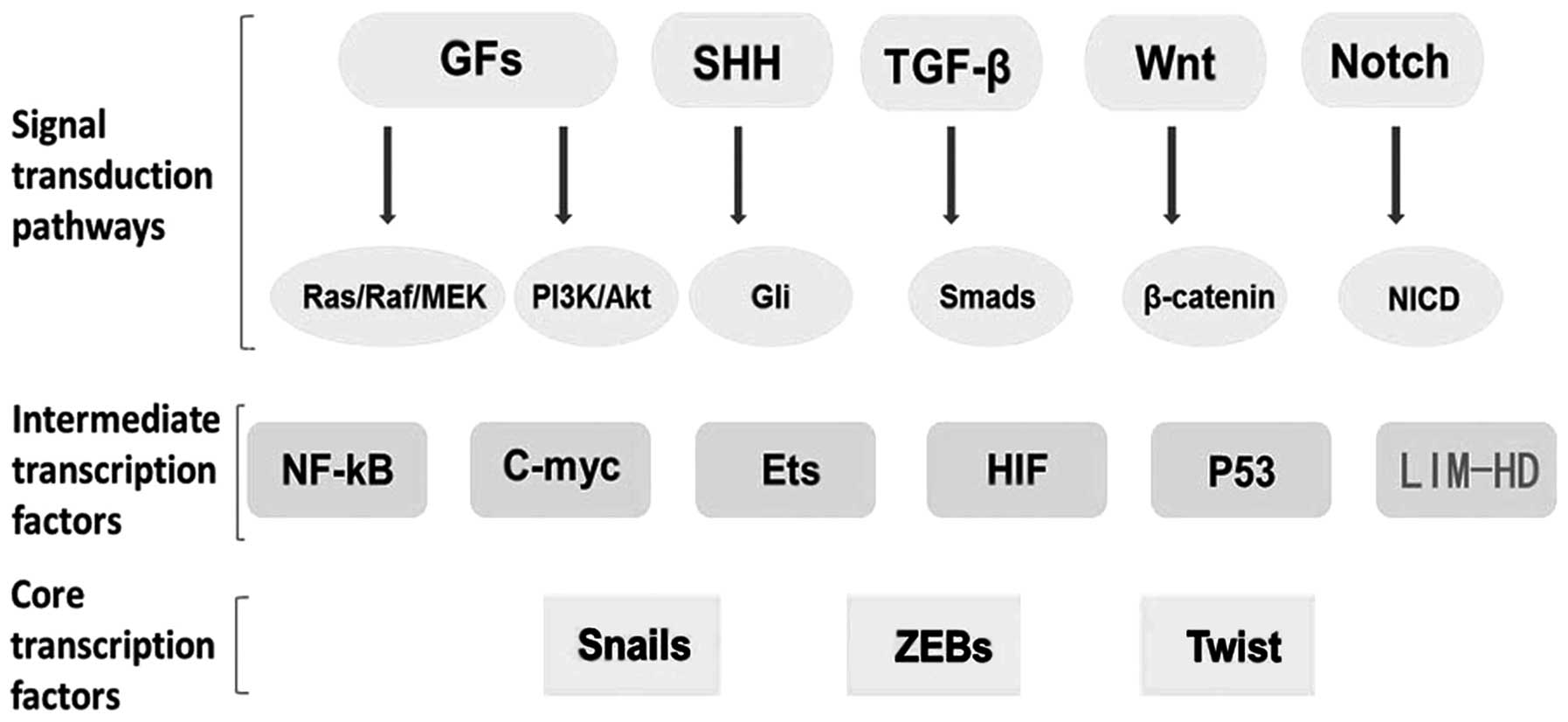
Transcription factors such as SNAIL, SLUG, TWIST,
ZEB and FOXC2 are important markers of EMT. There is much evidence
to confirm that several oncogenic pathways, such as growth factors
(GFs), Ras, SHH, TGF-β, Wnt and Notch, may induce EMT. LIM homeobox
genes such as LMX1A may also be important intermediate
transcription factors which can induce or regulate core
transcription factors and co-operate with them in target
regulation, eventually leading to downregulation of epithelial
genes and upregulation of mesenchymal genes (Fig. 2).
Regarding LIM1, it was found to be a downstream
effector of SHH-Gli signaling functions and its expression is
regulated by Pax2, FGFs and Wnt factors, which were confirmed to be
involved in human tumorigenesis (79). Recently, LIM1 was found to be a
growth and survival factor in human clear cell renal cell carcinoma
through the activation of multiple oncogenic pathways including
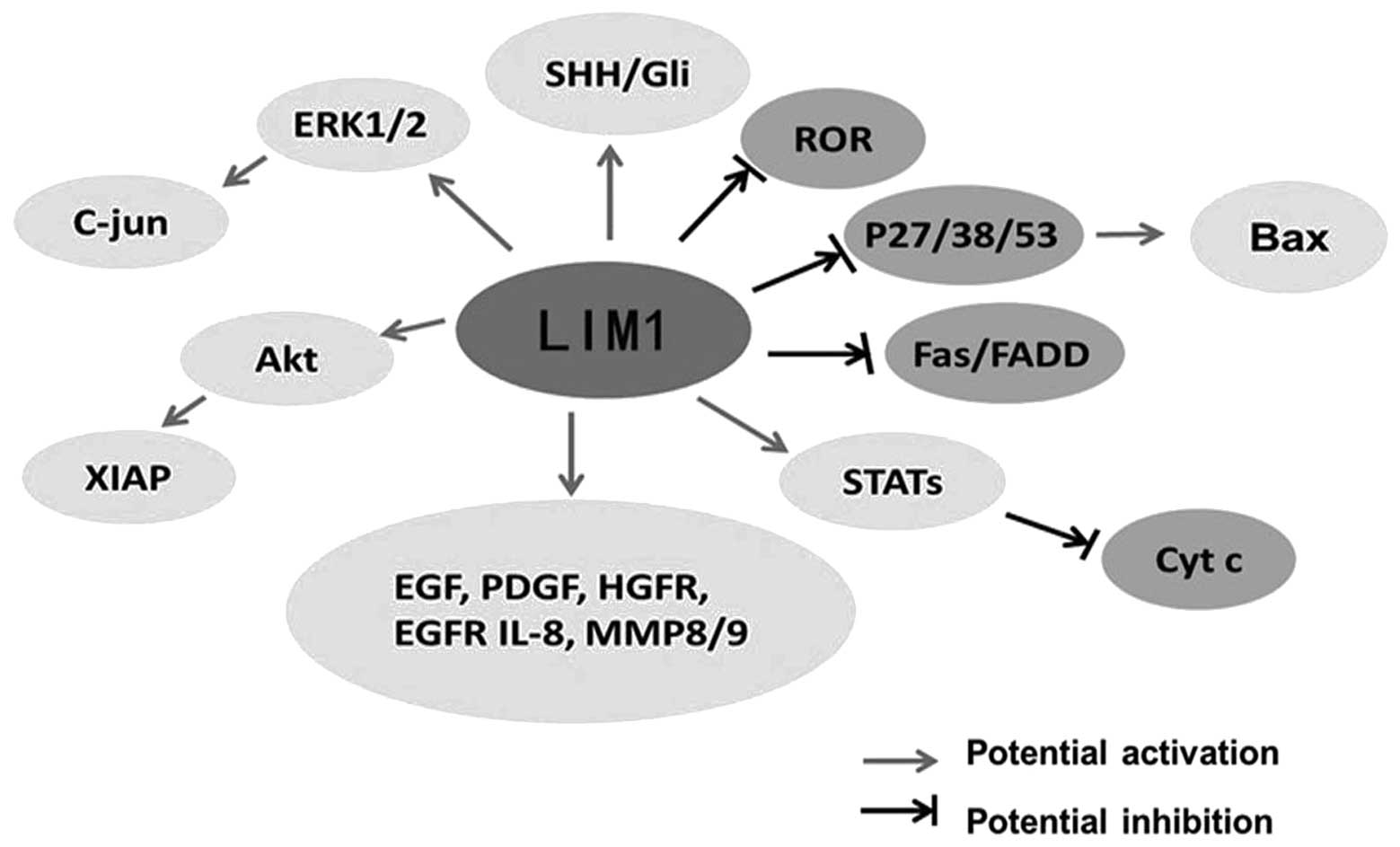
phosphoinositide kinase-3/Akt, MAPK and NK-κB pathways (13). To elucidate additional mechanisms
accounting for the effect of LIM1 on tumor cell growth, western
blot analysis and Proteome Profiler arrays (coated with apoptotic,
phosphoproteins or angiogenesis pathways/markers) were used. The
results showed that the various signaling pathways and molecular
factors involved in cell apoptosis, proliferation, movement,
angiogenesis were regulated at the level of expression and/or
activation directly or indirectly by LIM1, such as phosphorylated
Akt, Stats, cytochrome c, Fas, p38, p53, FGF, PDGF and EGF
receptors as well as markers of cell movement, MMP8 andMMP9. Taken
together, we suggest that LIM1 has a vital role in the expression
and/or activation of various oncogenic and angiogenic pathways in
human CCC (Fig. 3).
Since the developmental pathways involved in the
genesis and the growth of an organ are also responsible for the
development of a tumor, the idea that tumors ‘hijack’ for their own
growth signaling pathways involved in normal development is
emerging (80). The few available
LIM homeobox gene signaling pathway studies in cancer reviewed
above are consisted with this concept. Cancer often arises when
normal cellular growth goes awry due to defects in critical signal
transduction pathways; thus targeting these developemental pathways
to explore the role of LIM homeobox genes in cancer will be
effective.
In the present study, we document various confirmed
signal pathway studies in development involving LIM homeobox
transcription factors, which could provide various clues to explore
the role of LIM homeobox genes in cancer.
Sato et al (81) reported that the kinase activity of
WNKs is required for induction of LHX8 gene expression and the
activation of SPAK/OSR1, and that the kinase-dead form of WNK acts
as an actual dominant-negative form in the signaling pathway.
Furthermore, the expression of LHX8 by either hypertonic or RA
stimulation was required for the expression of both WNK1 and WNK4.
Thus, this study provides initial evidence identifying the LHX8
gene acting downstream in the WNK-SPAK/OSR1 pathway, and
demonstrates the significance of the WNK-OSR1-LHX8 pathway in
neural development. WNK is a family of serine/threonine protein
kinases that are characterized by a typical sequence variation
within the conserved catalytic domain and could phosphorylate and
activate SPAK or OSR1 kinases (82,83).
The WNK-SPAK/OSR1 pathway is known to regulate various ion
co-transporters and is widely conserved among many species
(84,85). WNK1 is also required for cell
division in cultured cells (86),
and proliferation, migration and differentiation of neural
progenitor cells (87). There is
growing evidence for additional roles of WNK kinases in various
signaling cascades related to cancer (88).
Wnt gain of function in cardiac progenitor cells was
found to lead to expansion of ISL1-positive progenitors with a
concomitant increase in FGF signaling through activation of a
specific set of FGF ligands including FGF3, FGF10, FGF16 and FGF20.
These data reveal that Wnt/β-catenin signaling promotes expansion
of ISL1-positive progenitor cells through regulation of FGF
signaling (89).
Roof plate (RP) development has been well studied in
the spinal cord, where its specification relies on interactions
between inductive signals of the TGF-β family produced by the
adjacent epidermal ectoderm and intrinsic homeodomain transcription
factors LMX1A and LMX1B (90–92),
while similar mechanisms are involved in RP differentiation in the
anterior midbrain. Alexandre et al (93) reported that the plasticity of the
midbrain RP derives from two apparently antagonistic influences of
FGF8. On the one hand, FGF8 widens beyond the neural folds the
competence of the neuroepithelium to develop a RP by inducing the
expression of LMX1B and Wnt1. On the other hand, FGF8 exerts a
major destabilizing influence on RP maturation by controlling
signaling by members of the TGF-β superfamily belonging to the BMP,
GDF and activin subgroups.
A hallmark of cancer is reactivation/alteration of
pathways that control cellular differentiation during developmental
processes. Aberrant activation of developmental pathways such as
Wnt, Hedgehog and Notch contributes to cancer development and
progression, and these pathways are even intertwined at the
molecular level. Thus, valuable prognostic biomarkers and the
innovative therapies for cancer may be identified through these
studies.
4. The potential of LIM homeobox genes as
cancer biomarkers and the challenges
It is known that the genes that play important roles
in the various steps of development may later be overexpressed or
downregulated contributing to carcinogenesis. As reviewed above,
several groups have reported the use of LIM homeobox genes as
diagnostic and prognostic biomarkers, as certain gene expression
profiles can be linked to tissue specificity, associated with early
stages of carcinogenesis, and even linked to therapy-resistant
disease resulting in a worse prognosis.
Analysis of the LIM homeobox gene abnormalities in
cancer has demonstrated the presence of overexpression, at times,
due to chromosomal translocations. However, more LIM homeobox genes
are frequently downregulated in cancer due to hypermethylation
(Table III), but not deletion or
mutation.
 | Table IIIThe methylation rate of LIM homeobox
transcription factors in cancer cell lines and/or tumor
tissues. |
Table III
The methylation rate of LIM homeobox
transcription factors in cancer cell lines and/or tumor
tissues.
| LIM-HD | Related cancer | Methylation
rate | Refs. |
|---|
| LHX2 | NHL | B-CLL/SLL, 46.6%;
MCL, 41.6%; FL,73% | (21) |
| Breast cancer | Tumor biopsies,
43.6% | (22,23) |
| Lung tumor | Tumor biopsies,
58% | (24) |
| LHX3 | Breast cancer | Unknown | (29) |
| LHX4 | Lung tumor | Tumor biopsies,
75% | (23) |
| Hepatocellular
carcinoma | Unknown | (38) |
| LHX5 | Breast cancer | Unknown | (23) |
| LHX6 | HNSCC | Cell lines, 92.8%;
tumor biopsies, 65.6% | (43) |
| Colon cancer | Tumor biopsies,
85.7% | (43) |
| Cervical
cancer | Cell lines,
87.5% | (44,45) |
| LHX9 | Pediatric malignant
astrocytomas | High-grade gliomas,
55.6%; | (47) |
| | low-grade gliomas,
29% | (48) |
| Follicular
lymphoma | Unknown | |
| LMX1A | Cervical
cancer | Tumor biopsies,
89.9%; cervical scrapings, 36% | (64–66) |
| Ovarian cancer | Benign, 1.3%;
borderline, 7.1%; malignancy, 34.9% | (67,68) |
| Bladder cancer | Tumor biopsies,
9.43% | (69) |
| Gastric cancer | Tumor biopsies,
82% | (70) |
| Colon cancer | Carcinomas, 55%;
adenomas, 42%; cell lines, 75% | (71) |
Aberrant hypermethylation of the promoter regions of
specific genes is a key event in the formation and progression of
cancer and can be applied to cancer diagnostics in three ways: as a
marker to detect cancer cells or cancer-derived DNA; as a marker to
predict prognosis; and as a biomarker for the assessment of
therapeutic response. Methylation analysis has an advantage in that
it can be performed using chemically stable DNA (compared to RNA).
Moreover, detecting gene methylation is easier than detecting gene
mutation since the exact location of a mutation is usually unknown,
making it difficult to specifically amplify DNA molecules with an
embedded mutation in excess of wild-type molecules. More
interestingly, detection of aberrant methylation can provide
confirmation of the presence of intact cancer cells or
cancer-derived DNA in bodily fluids, such as blood, urine, sputum,
saliva and stool. Thus, their potential as biomarkers is
growing.
Although numerous studies have demonstrated that LIM
homeobox genes may serve as cancer biomarkers for diagnosis and
prognosis, there are still many factors limiting the clinical
application of these biomarkers. First, the majority of these
studies were almost low level clinical reports, and lack of large
sample, multi-center clinical studies exists. In order to confirm
their clinical utility, enlarged sample size, more prospective
studies, detection of the expression of methylated LIM homeobox
genes in various periods of tumorigenesis and testing using an
independent set of collected patient samples must be carried out.
Second, the mechanisms involving LIM homeobox genes and cancer are
not totally understood, thus more in-depth study is required.
Third, the technology analyzing gene methylation is not mature
enough. Quantitative analysis of tumor sample is still a challenge.
In the future, improving research methods and techniques is
necessary. Fourth, as biomarkers, further detailed investigation is
required to determine which markers have high sensitivity and
specificity. Moreover, because of tumor heterogeneity, no single
marker will likely be adequate. The inclusion of more new markers
in a panel of hypermethylated genes in cancer potentially increases
the sensitivity and specificity of tumor detection. Finally LIM
homeobox gene immunohistochemical assessment or methylation
detection should be combined with clinical and radiologic
information to arrive at a definitive diagnosis.
5. Conclusions
LIM homeobox genes are one of the most important
subfamilies of homeobox genes. As reviewed above, they not only
participate in a wide array of developmental events, but also act
as tumor-suppressor genes or oncogenes and are involved in various
signaling pathways. Moreover, the silencing of LIM homeobox genes
caused by hypermethylation is highly correlated with cancer. Yet,
the mechanisms involved in the inhibition or promotion of
tumorigenesis by LIM homeobox genes require further in-depth
research. It has been reported that LIM homeobox genes may be used
as cancer biomarkers for early diagnosis and prognostic evaluation.
However, due to the complexity of clinical tumor samples,
sensitivity and specificity of LIM homeobox genes have become the
main challenges which hinder their clinical application. We believe
that more in-depth research concerning LIM-homeobox genes in cancer
from a signaling network perspective will be highly valuable in
helping to understand tumor profiles, establish biomarkers, and
guide choices for combinatorial drug therapies.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81071651 and
81372622), the Zhejiang Provincial Natural Science Foundation of
China (grant no. R2100213), the Program for Zhejiang Leading Team
of S&T Innovation (grant no. 2012R10046-03), the Major State
Basic Research Development Program (grant no. 2010CB834303), the
National High Technology Research and Development Program of China
(grant no. 2012AA02A601), the Major Projects in Zhejiang Province
(grant no. 2012C13014-1), and the Fundamental Research Funds for
the Central Universities (grant no. 2012FZA7020).
References
|
1
|
Hobert O and Westphal H: Functions of
LIM-homeobox genes. Trends Genet. 16:75–83. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dawid IB: LIM domain proteins. C R Acad
Sci. 3:295–306. 1995.
|
|
3
|
Bach I: The LIM domain: regulation by
association. Mech Dev. 91:5–17. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Glenn DJ and Maurer RA: MRG1 binds to the
LIM domain of Lhx2 and may function as a coactivator to stimulate
glycoprotein hormone alpha-subunit gene expression. J Biol Chem.
274:36159–36167. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Howard PW and Maurer RA: Identification of
a conserved protein that interacts with specific LIM homeodomain
transcription factors. J Biol Chem. 275:13336–13342. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ostendorff HP, Peirano RI, Peters MA, et
al: Ubiquitination-dependent cofactor exchange on LIM homeodomain
transcription factors. Nature. 416:99–103. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shawlot W and Behringer RR: Requirement
for Lim1 in head-organizer function. Nature. 374:425–430. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karavanov AA, Karavanova I, Perantoni A
and Dawid IB: Expression pattern of the rat Lim-1 homeobox gene
suggests a dual role during kidney development. Int J Dev Biol.
42:61–66. 1998.PubMed/NCBI
|
|
9
|
Kobayashi A, Kwan KM, Carroll TJ, McMahon
AP, Mendelsohn CL and Behringer RR: Distinct and sequential
tissue-specific activities of the LIM-class homeobox gene Lim1 for
tubular morphogenesis during kidney development. Development.
132:2809–2823. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen YT, Kobayashi A, Kwan KM, Johnson RL
and Behringer RR: Gene expression profiles in developing nephrons
using Lim1 metanephric mesenchyme-specific conditional mutant mice.
BMC Nephrol. 7:12006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guertl B, Senanayake U, Nusshold E, et al:
Lim1, an embryonal transcription factor, is absent in multicystic
renal dysplasia, but reactivated in nephroblastomas. Pathobiology.
78:210–219. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dormoy V, Béraud C, Lindner V, Thomas L,
Coquard C, Barthelmebs M, Jacqmin D, Lang H and Massfelder T:
LIM-class homeobox gene Lim1, a novel oncogene in human renal cell
carcinoma. Oncogene. 30:1753–1763. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu Y, Baldassare M, Fisher P, et al: LH-2:
a LIM/homeodomain gene expressed in developing lymphocytes and
neural cells. Proc Natl Acad Sci USA. 90:227–231. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Richter K, Pinto do OP, Hägglund AC,
Wahlin A and Carlsson L: Lhx2 expression in hematopoietic
progenitor/stem cells in vivo causes a chronic myeloproliferative
disorder and altered globin expression. Haematologica.
88:1336–1347. 2003.PubMed/NCBI
|
|
15
|
Nadal N, Chapiro E, Flandrin-Gresta P, et
al: LHX2 deregulation by juxtaposition with the IGH
locus in a pediatric case of chronic myeloid leukemia in B-cell
lymphoid blast crisis. Leuk Res. 36:195–198. 2012. View Article : Google Scholar
|
|
16
|
Wu HK, Heng HH, Siderovski DP, et al:
Identification of a human LIM-Hox gene, hLH-2, aberrantly expressed
in chronic myelogenous leukaemia and located on 9q33–34.1.
Oncogene. 12:1205–1212. 1996.PubMed/NCBI
|
|
17
|
Wu HK and Minden MD: Transcriptional
activation of human LIM-HOX gene, hLH-2, in chronic
myelogenous leukemia is due to a cis-acting effect of
Bcr-Abl. Biochem Biophys Res Commun. 233:806–812.
1997.PubMed/NCBI
|
|
18
|
Gorantla B, Asuthkar S, Rao JS, Patel J
and Gondi CS: Suppression of the uPAR-uPA system retards
angiogenesis, invasion, and in vivo tumor development in
pancreatic cancer cells. Mol Cancer Res. 9:377–389. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tiede S and Paus R: Lhx2-decisive role in
epithelial stem cell maintenance, or just the ‘tip of the iceberg’?
Bioessays. 28:1157–1160. 2006.PubMed/NCBI
|
|
20
|
Abate-Shen C: Deregulated homeobox gene
expression in cancer: cause or consequence? Nat Rev Cancer.
2:777–785. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rahmatpanah FB, Carstens S, Guo J, et al:
Differential DNA methylation patterns of small B-cell lymphoma
subclasses with different clinical behavior. Leukemia.
20:1855–1862. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim MS, Lee J, Oh T, et al: Genome-wide
identification of OTP gene as a novel methylation marker of
breast cancer. Oncol Rep. 27:1681–1688. 2012.PubMed/NCBI
|
|
23
|
Kamalakaran S, Varadan V, Giercksky
Russnes HE, et al: DNA methylation patterns in luminal breast
cancers differ from non-luminal subtypes and can identify relapse
risk independent of other clinical variables. Mol Oncol. 5:77–92.
2011. View Article : Google Scholar
|
|
24
|
Rauch T, Li H, Wu X and Pfeifer GP:
MIRA-assisted microarray analysis, a new technology for the
determination of DNA methylation patterns, identifies frequent
methylation of homeodomain-containing genes in lung cancer cells.
Cancer Res. 66:7939–7947. 2006. View Article : Google Scholar
|
|
25
|
Sharma K, Sheng HZ, Lettieri K, et al: LIM
homeodomain factors Lhx3 and Lhx4 assign subtype identities for
motor neurons. Cell. 95:817–828. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sheng HZ, Zhadanov AB, Mosinger B Jr, et
al: Specification of pituitary cell lineages by the LIM homeobox
gene Lhx3. Science. 272:1004–1007. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sheng HZ, Moriyama K, Yamashita T, et al:
Multistep control of pituitary organogenesis. Science.
278:1809–1812. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thaler JP, Lee SK, Jurata LW, Gill GN and
Pfaff SL: LIM factor Lhx3 contributes to the specification of motor
neuron and interneuron identity through cell-type-specific
protein-protein interactions. Cell. 110:237–249. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dietrich D, Lesche R, Tetzner R, et al:
Analysis of DNA methylation of multiple genes in microdissected
cells from formalin-fixed and paraffin-embedded tissues. J
Histochem Cytochem. 57:477–489. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li H, Witte DP, Branford WW, et al: Gsh-4
encodes a LIM-type homeodomain, is expressed in the developing
central nervous system and is required for early postnatal
survival. EMBO J. 13:2876–2885. 1994.PubMed/NCBI
|
|
31
|
Liu Y, Fan M, Yu S, et al: cDNA cloning,
chromosomal localization and expression pattern analysis of human
LIM-homeobox gene LHX4. Brain Res. 928:147–155. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Machinis K, Pantel J, Netchine I, et al:
Syndromic short stature in patients with a germline mutation in the
LIM homeobox LHX4. Am J Hum Genet. 69:961–968. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Raetzman LT, Ward R and Camper SA:
Lhx4 and Prop1 are required for cell survival and
expansion of the pituitary primordia. Development. 129:4229–4239.
2002.PubMed/NCBI
|
|
34
|
Kawamata N, Sakajiri S, Sugimoto KJ, Isobe
Y, Kobayashi H and Oshimi K: A novel chromosomal translocation
t(1;14)(q25;q32) in pre-B acute lymphoblastic leukemia involves the
LIM homeo-domain protein gene, Lhx4. Oncogene. 21:4983–4991. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yamaguchi M, Yamamoto K and Miura O:
Aberrant expression of the LHX4 LIM homeobox gene caused by
t(1;14)(q25;q32) in chronic myelogenous leukemia in biphenotypic
blast crisis. Genes Chromosomes Cancer. 38:269–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
de Bruijn DR, van Dijk AH, Willemse MP and
van Kessel AG: The C terminus of the synovial sarcoma-associated
SSX proteins interacts with the LIM homeobox protein LHX4.
Oncogene. 27:653–662. 2008.PubMed/NCBI
|
|
37
|
Goc A, Kochuparambil ST, Al-Husein B,
Al-Azayzih A, Mohammad S and Somanath PR: Simultaneous modulation
of the intrinsic and extrinsic pathways by simvastatin in mediating
prostate cancer cell apoptosis. BMC Cancer. 12:4092012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hung TM, Hu RH, Ho CM, et al:
Downregulation of alpha-fetoprotein expression by LHX4: a critical
role in hepatocarcinogenesis. Carcinogenesis. 32:1815–1823. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhao Y, Hermesz E, Yarolin MC and Westphal
H: Genomic structure, chromosomal localization and expression of
the human LIM-homeobox gene LHX5. Gene. 260:95–101. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sheng HZ, Bertuzzi S, Chiang C, et al:
Expression of murine Lhx5 suggests a role in specifying the
forebrain. Dev Dyn. 208:266–277. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao Y, Sheng HZ, Amini R, et al: Control
of hippocampal morphogenesis and neuronal differentiation by the
LIM homeobox gene Lhx5. Science. 284:1155–1158. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Grigoriou M, Tucker AS, Sharpe PT and
Pachnis V: Expression and regulation of Lhx6 and
Lhx7, a novel subfamily of LIM homeodomain encoding genes,
suggests a role in mammalian head development. Development.
125:2063–2074. 1998.
|
|
43
|
Estécio MR, Youssef EM, Rahal P, et al:
LHX6 is a sensitive methylation marker in head and neck carcinomas.
Oncogene. 25:5018–5026. 2006.PubMed/NCBI
|
|
44
|
Jung S, Jeong D, Kim J, et al: Epigenetic
regulation of the potential tumor suppressor gene, hLHX6.1,
in human cervical cancer. Int J Oncol. 38:859–869. 2011.PubMed/NCBI
|
|
45
|
Jung S, Jeong D, Kim J, et al: The role of
hLHX6-HMR as a methylation biomarker for early diagnosis of
cervical cancer. Oncol Rep. 23:1675–1682. 2010.
|
|
46
|
Failli V, Rogard M, Mattei MG, Vernier P
and Rétaux S: Lhx9 and Lhx9α LIM-homeodomain factors: genomic
structure, expression patterns, chromosomal localization, and
phylogenetic analysis. Genomics. 64:307–317. 2000.
|
|
47
|
Vladimirova V, Mikeska T, Waha A, et al:
Aberrant methylation and reduced expression of LHX9 in malignant
gliomas of childhood. Neoplasia. 11:700–711. 2009.PubMed/NCBI
|
|
48
|
Bennett LB, Schnabel JL, Kelchen JM, et
al: DNA hypermethylation accompanied by transcriptional repression
in follicular lymphoma. Genes Chromosomes Cancer. 48:828–841. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Karlsson O, Thor S, Norberg T, Ohlsson H
and Edlund T: Insulin gene enhancer binding protein Isl-1 is a
member of a novel class of proteins containing both a homeo-and a
Cys His domain. Nature. 344:879–882. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ahlgren U, Pfaff SL, Jessell TM, Edlund T
and Edlund H: Independent requirement for ISL1 in formation of
pancreatic mesenchyme and islet cells. Nature. 385:257–260. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yamada T, Pfaff SL, Edlund T and Jessell
TM: Control of cell pattern in the neural tube: motor neuron
induction by diffusible factors from notochord and floor plate.
Cell. 73:673–686. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jensen J: Gene regulatory factors in
pancreatic development. Dev Dyn. 229:176–200. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Dong J, Asa SL and Drucker DJ: Islet cell
and extrapancreatic expression of the LIM domain homeobox gene
isl-1. Mol Endocrinol. 5:1633–1641. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Graham RP, Shrestha B, Caron BL, et al:
Islet-1 is a sensitive but not entirely specific marker for
pancreatic neuroendocrine neoplasms and their metastases. Am J Surg
Pathol. 37:399–405. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Koo J, Mertens RB, Mirocha JM, Wang HL and
Dhall D: Value of Islet 1 and PAX8 in identifying metastatic
neuroendocrine neoplasms of pancreatic origin. Mod Pathol.
25:893–902. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Schmitt AM, Riniker F, Anlauf M, et al:
Islet 1 (Isl1) expression is a reliable marker for pancreatic
endocrine neoplasms and their metastases. Am J Surg Pathol.
32:420–425. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hermann G, Konukiewitz B, Schmitt A,
Perren A and Klöppel G: Hormonally defined pancreatic and duodenal
neuroendocrine tumors differ in their transcription factor
signatures: expression of ISL1, PDX1, NGN3, and CDX2. Virchows
Arch. 459:147–154. 2011. View Article : Google Scholar
|
|
58
|
Cheung IY, Feng Y, Gerald W and Cheung NK:
Exploiting gene expression profiling to identify novel minimal
residual disease markers of neuroblastoma. Clin Cancer Res.
14:7020–7027. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chizhikov VV, Lindgren AG, Mishima Y, et
al: Lmx1a regulates fates and location of cells originating from
the cerebellar rhombic lip and telencephalic cortical hem. Proc
Natl Acad Sci USA. 107:10725–10730. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cai J, Donaldson A, Yang M, German MS,
Enikolopov G and Iacovitti L: The role of Lmx1a in the
differentiation of human embryonic stem cells into midbrain
dopamine neurons in culture and after transplantation into a
Parkinson’s disease model. Stem Cell. 27:220–229. 2009.PubMed/NCBI
|
|
61
|
Lin CK, Chao TK, Lai HC and Lee HS: LMX1A
as a prognostic marker in ovarian mucinous cystadenocarcinoma. Am J
Clin Pathol. 137:971–977. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Tsai WC, Lee HS, Lin CK, Chen A, Nieh S
and Ma HI: The association of osteopontin and LMX1A expression with
World Health Organization grade in meningiomas and gliomas.
Histopathology. 61:844–856. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Tsai WC, Lin CK, Yang YS, et al: The
correlations of LMX1A and osteopontin expression to the
clinicopathologic stages in pancreatic adenocarcinoma. Appl
Immunohistochem Mol Morphol. 21:395–400. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Liu CY, Chao TK, Su PH, et al:
Characterization of LMX-1A as a metastasis suppressor in cervical
cancer. J Pathol. 219:222–231. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lai HC, Lin YW, Huang TH, et al:
Identification of novel DNA methylation markers in cervical cancer.
Int J Cancer. 123:161–167. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Lai HC, Lin YW, Huang RL, et al:
Quantitative DNA methylation analysis detects cervical
intraepithelial neoplasms type 3 and worse. Cancer. 116:4266–4274.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Chao TK, Yo YT, Liao YP, et al:
LIM-homeobox transcription factor 1, alpha (LMX1A) inhibits
tumourigenesis, epithelial-mesenchymal transition and stem-like
properties of epithelial ovarian cancer. Gynecol Oncol.
128:475–482. 2013. View Article : Google Scholar
|
|
68
|
Su HY, Lai HC, Lin YW, et al: An
epigenetic marker panel for screening and prognostic prediction of
ovarian cancer. Int J Cancer. 124:387–393. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhao Y, Guo S, Sun J, et al: Methylcap-seq
reveals novel DNA methylation markers for the diagnosis and
recurrence prediction of bladder cancer in a Chinese population.
PLoS One. 7:e351752012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Dong W, Feng L, Xie Y, Zhang H and Wu Y:
Hypermethylation-mediated reduction of LMX1A expression in gastric
cancer. Cancer Sci. 102:361–366. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Paz MF, Wei S, Cigudosa JC, et al: Genetic
unmasking of epigenetically silenced tumor suppressor genes in
colon cancer cells deficient in DNA methyltransferases. Hum Mol
Genet. 12:2209–2219. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chen H, Lun Y, Ovchinnikov D, et al: Limb
and kidney defects in Lmx1b mutant mice suggest an
involvement of LMX1B in human nail patella syndrome. Nat
Genet. 19:51–55. 1998.
|
|
73
|
Riddle RD, Ensini M, Nelson C, Tsuchida T,
Jessell TM and Tabin C: Induction of the LIM homeobox gene
Lmx1 by WNT7a establishes dorsoventral pattern in the
vertebrate limb. Cell. 83:631–640. 1995.PubMed/NCBI
|
|
74
|
Rieger ME, Sims AH, Coats ER, Clarke RB
and Briegel KJ: The embryonic transcription cofactor LBH is a
direct target of the Wnt signaling pathway in epithelial
development and in aggressive basal subtype breast cancers. Mol
Cell Biol. 30:4267–4279. 2010. View Article : Google Scholar
|
|
75
|
Kengaku M, Capdevila J, Rodriguez-Esteban
C, et al: Distinct WNT pathways regulating AER formation and
dorsoventral polarity in the chick limb bud. Science.
280:1274–1277. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Clevers H: Wnt/β-catenin signaling in
development and disease. Cell. 127:469–480. 2006.
|
|
77
|
Millen KJ, Millonig JH and Hatten ME: Roof
plate and dorsal spinal cord dl1 interneuron development in the
dreher mutant mouse. Dev Biol. 270:382–392. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Chizhikov VV and Millen KJ: Control of
roof plate formation by Lmx1a in the developing spinal cord.
Development. 131:2693–2705. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kishigami S and Mishina Y: BMP signaling
and early embryonic patterning. Cytokine Growth Factor Rev.
16:265–278. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Dormoy V, Jacqmin D, Lang H and Massfelder
T: From development to cancer: lessons from the kidney to uncover
new therapeutic targets. Anticancer Res. 32:3609–3617.
2012.PubMed/NCBI
|
|
81
|
Sato A and Shibuya H: WNK signaling is
involved in neural development via Lhx8/Awh expression. PLoS One.
8:e553012013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Moriguchi T, Urushiyama S, Hisamoto N, et
al: WNK1 regulates phosphorylation of cation-chloride-coupled
cotransporters via the STE20-related kinases, SPAK and OSR1. J Biol
Chem. 280:42685–42693. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Vitari AC, Deak M, Morrice NA and Alessi
DR: The WNK1 and WNK4 protein kinases that are mutated in Gordon’s
hypertension syndrome phosphorylate and activate SPAK and OSR1
protein kinases. Biochem J. 391:17–24. 2005.PubMed/NCBI
|
|
84
|
Moniz S and Jordan P: Emerging roles for
WNK kinases in cancer. Cell Mol Life Sci. 67:1265–1276. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Veríssimo F and Jordan P: WNK kinases, a
novel protein kinase subfamily in multi-cellular organisms.
Oncogene. 20:5562–5569. 2001.PubMed/NCBI
|
|
86
|
Tu SW, Bugde A, Luby-Phelps K and Cobb MH:
WNK1 is required for mitosis and abscission. Proc Natl Acad Sci
USA. 108:1385–1390. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Sun X, Gao L, Yu RK and Zeng G:
Down-regulation of WNK1 protein kinase in neural progenitor cells
suppresses cell proliferation and migration. J Neurochem.
99:1114–1121. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Moniz S and Jordan P: Emerging roles for
WNK kinases in cancer. Cell Mol Life Sci. 67:1265–1276. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Cohen ED, Wang Z, Lepore JJ, et al:
Wnt/β-catenin signaling promotes expansion of Isl-1-positive
cardiac progenitor cells through regulation of FGF signaling. J
Clin Invest. 117:1794–1804. 2007.
|
|
90
|
Liem KF Jr, Jessell TM and Briscoe J:
Regulation of the neural patterning activity of sonic hedgehog by
secreted BMP inhibitors expressed by notochord and somites.
Development. 127:4855–4866. 2000.PubMed/NCBI
|
|
91
|
Chizhikov VV and Millen KJ: Control of
roof plate development and signaling by Lmx1b in the caudal
vertebrate CNS. J Neurosci. 24:5694–5703. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Chizhikov VV and Millen KJ: Control of
roof plate formation by Lmx1a in the developing spinal cord.
Development. 131:2693–2705. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Alexandre P, Bachy I, Marcou M and Wassef
M: Positive and negative regulations by FGF8 contribute to midbrain
roof plate developmental plasticity. Development. 133:2905–2913.
2006. View Article : Google Scholar : PubMed/NCBI
|