Introduction
Gallbladder cancer (GBC) represents the most
frequent and aggressive type among biliary tract malignancies.
Although recent advances have been made in the diagnosis and
treatment, GBC has a poor overall prognosis with a 5-year survival
rate <10% (1). Currently,
radical resection remains the mainstay of treatment for GBC.
However, due to lacking typical symptoms and specific biomarkers,
most GBC patients are diagnosed at advanced stages with
unresectable tumors. Less than 30% of GBC patients are considered
surgical candidates (2). Moreover,
effective adjuvant treatments including chemotherapy and
radiotherapy are not well developed. Therefore, there is an urgent
need to develop novel and effective therapy regimens for GBC
patients. To achieve this, it is imperative to have a better
understanding of the molecular biology and carcinogenic mechanisms
underlying the development and progression of GBC.
Similar to other types of cancer, the development
and progression of GBC are facilitated by the epigenetic silencing
of tumor suppressor genes (3).
Ubiquitin-like containing PHD and RING finger domains 1 (UHRF1)
functions as an important regulator in cell proliferation and
epigenetic regulation belonging to the RING-finger type E3
ubiquitin ligase subfamily. As a potential oncogenic factor, UHRF1
contributes to silencing of tumor suppressor genes by recruiting
DNA methyltransferase 1 (DNMT1) to their hemi-methylated promoters
(4,5). The expression of UHRF1 is cell
cycle-regulated in normal cells, but stably high throughout cell
cycle in numerous cancer cell lines including breast, colorectal,
bladder, lung, prostate, esophageal and laryngeal cancer (6–12).
Downregulation of UHRF1 expression showed antitumor activities in
cancer cells (13,14). Therefore, UHRF1 is likely to be a
novel target of pivotal interest for developing new anticancer
drugs.
However, the role of UHRF1 in GBC remains unclear.
In the present study, UHRF1 expression in GBC tissues and cell
lines was examined. The effects of UHRF1 depletion on
proliferation, cell cycle, apoptosis and migration of GBC cell
lines in vitro and in vivo were also
investigated.
Materials and methods
Patients and tissue samples
Tumor samples from 38 patients with GBC (12 men and
26 women; age range, 51–73 years; mean age, 62.4±5.2 years) treated
with radical cholecystectomy (without prior radiotherapy or
chemotherapy) and corresponding non-tumor samples from 12 of these
GBC cases were collected between 2011 and 2013 at the Department of
General Surgery, Xinhua Hospital, School of Medicine, Shanghai
Jiaotong University. The tumors were staged according to the TNM
staging system (AJCC, 7th edition, 2010); 4 were stage I; 10 stage
II; 15 stage III and 9 stage IV. The histological diagnosis of all
GBC cases was confirmed by two pathologists. The present study was
approved by the Ethics Committee of Xinhua Hospital, School of
Medicine, Shanghai Jiaotong University and all patients provided
informed consent.
Immunohistochemistry
Tissue sections (4 μm) were cut from
paraffin-embedded human GBC tissues and xenograft tumors,
deparaffinized within xylene and rehydrated within a graded series
of ethanol solutions. To block endogenous peroxidase activity, the
sections were treated for 30 min with methanol containing 1%
hydrogen peroxide. The sections were incubated with anti-UHRF1
mouse monoclonal antibody (1:50 dilution) followed by incubation
with secondary antibody (1:1,000 dilution) (both from Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA). The score on the
intensity of staining was graded by using a 3-scale system (0,
negative; 1, weak; 2, moderate; and 3, strong).
Cell cultures and transfection
Human GBC cell line (GBC-SD) was obtained from the
Cell Bank of the Chinese Academy of Sciences, Shanghai, China;
human GBC cell line (SGC996) was obtained from the Academy of Life
Science, Tongji University, Shanghai, China; human GBC cell lines
(NOZ and OCUG-1) were obtained from the Health Science Research
Resources Bank, Osaka, Japan. All cell lines were maintained either
in Dulbecco’s modified Eagle’s medium (DMEM) or RPMI (Invitrogen,
Carlsbad, CA, USA) at 37°C in a humidified 5% CO2
incubator. Both media were supplemented with 10% fetal bovine serum
(FBS) and 1% penicillin/streptomycin (both from Invitrogen).
Cells in the exponential phase of growth were
transfected with siRNA or shRNA (Santa Cruz Biotechnology, Inc.)
according to the manufacturer’s protocol. Briefly, cells were
incubated with siRNA or shRNA transfection medium for 6 h at 37°C
in a CO2 incubator. Then, the transfection medium was
replaced with fresh growth medium containing 2 times the normal
serum and antibiotics concentration. Cells were incubated for an
additional 24 h and then cultured under conditions normally used to
culture the cells. To obtain stable transfectants,
UHRF1-shRNA-transfected cells were selected with puromycin (1–2
μg/ml) for 10 days. Stable transfectants were used to establish
xenograft tumor models. At the indicated time points post
transfection, transfected cells were harvested and the silencing of
UHRF1 was examined using western blot assay.
RT-PCR
Total RNA was extracted from GBC-SD, NOZ, SGC996 and
OCUG-1 cells using TRIzol reagent kit (Gibco-BRL, Gaithersburg, MD,
USA). RNA extracts were reverse-transcribed using a Takara RNA PCR
kit (Takara Bio, Inc., Dalian, China) and the obtained cDNA was
subjected to PCR. The sequences of the primers for UHRF1 were:
forward, gtcgagatctttccggcaac and reverse,
tagatgccatcgtagcggtt.
Western blot analysis
Forty-eight hours after transfection with UHRF1
siRNA, total proteins were extracted from GBC cells and the protein
content was measured using the Bradford assay. The amount of
cytosolic cytochrome c was determined using cytosolic
extract. Mitochondrial and cytosolic fractions were extracted from
cultured cells by Mitochondria/Cytosol Fractionation kit (Abcam,
Cambridge, UK). Forty micrograms of proteins were separated by
10–12% SDS-PAGE and electrotransferred to PVDF membranes
(Millipore, Billerica, MA, USA). The blot was blocked with 5%
non-fat dry milk, and incubated with corresponding primary
antibodies, followed by incubation with corresponding secondary
antibodies (both from Santa Cruz Biotechnology, Inc.). The signals
were detected by enhanced chemiluminescence (ECL) assay and
visualized using X-OMAT Blue film.
Cell proliferation and clonogenic
assays
Cell proliferation was determined by the water
soluble tetrazolium (WST)-1 method using WST-1 cell proliferation
and cytotoxicity assay kit (Beyotime, Shanghai, China). Briefly, 24
h after transfection with UHRF1 siRNA, GBC-SD and NOZ cells were
seeded at a density of 5×103/well into 96-well plates
and cultured for 24, 48, 72 or 96 h. The cells were then incubated
with WST-1 reagent for 2 h at 37°C. The optical density was
measured at 450 nm with an automated microplate reader (Bio-Rad
Model 550; Bio-Rad, Hercules, CA, USA).
The clonogenic assay was performed by plating
UHRF1-siRNA-transfected cells (1×103/well) in 6-well
plates. Transfected cells were further cultured in the growth media
at 37°C and allowed to form colonies for 10–14 days. Colonies were
stained with crystal violet and counted using an automated colony
counter system (Alpha Innotech, San Leandro, CA, USA).
Cell cycle analysis
Forty-eight hours after transfection with UHRF1
siRNA, cells were harvested and fixed with 70% methanol overnight
at 4°C. Fixed cells were stained with RNase A and propidium iodide
(PI) followed by flow cytometric analysis (Beckman Coulter, Brea,
CA, USA). The percentage of cells in each phase of the cell cycle
was analyzed using Cylchred version 1.0.2 software (Cardiff
University, Wales, UK).
Cell apoptosis assay
The cell apoptosis was determined by Annexin V-FITC
apoptosis detection kit (BD Biosciences, San Diego, CA, USA)
according to the manufacturer’s protocol. Briefly, 48 h after
transfection with UHRF1 siRNA, cells were collected, washed twice
with cold PBS, and resuspended in 1X binding buffer. Then, the
cells were stained with FITC-Annexin V and PI for 15 min at room
temperature in the dark. The percentage of apoptotic cells was
determined by flow cytometry and the analysis was carried out
within 1 h.
Cell migration assay
The migration of transfected cells was measured
using a Transwell chamber (Corning Inc., Corning, NY, USA) with
8-μm pores. The UHRF1-siRNA-transfected cells with serum-free DMEM
or RPMI-1640 were placed in the upper chamber compartment. The
lower chambers contained DMEM or RPMI-1640 with 10% FBS as
chemoattractant. After incubation for 24 h at 37°C, cells on the
lower side of the insert filter were fixed by 4% paraformaldehyde
for 30 min and then stained with crystal violet for 20 min.
Tumor xenograft experiments
All animal experiments were approved by the Ethics
Committee of Xinhua Hospital, Shanghai Jiaotong University.
Four-week-old female BALB/c nude mice were obtained from Shanghai
Laboratory Animal Center of the Chinese Academy of Sciences and
housed under specific pathogen-free (SPF) conditions. GBC-SD and
NOZ cells which were stably transfected with UHRF1 shRNA or control
shRNA were used to establish subcutaneous tumor models. Briefly,
the transfected GBC-SD or NOZ cells were injected subcutaneously
into the right flank region of nude mice with a concentration of
1×107 cells/0.1 ml. Six weeks later, the mice were
sacrificed. The xenograft tumors were collected for further
investigation. The tumor volumes in each group were evaluated using
the following formula: Volume = length × width2/2.
TUNEL assay
Apoptosis in xenograft tumors was assessed by
terminal deoxynucleotidyl transferase-mediated dUTP nick end
labeling (TUNEL) staining using FragEL DNA Fragmentation Detection
kit (Merck, Darmstadt, Germany). Briefly, after deparaffinization,
rehydration and permeabilization, the endogenous peroxidase
activity was eliminated. Subsequently, the tissue sections were
incubated with TUNEL reaction mixture, and then counterstained with
methyl green. A dark brown 3,3′-diaminobenzidine tetrachloride
(DAB) signal indicated positive-TUNEL staining (apoptotic cell)
while shades of blue to green indicated viable cells. The number of
apoptotic cells was counted under a microscope and the results were
presented as means ± SD.
Statistical analysis
Statistical analysis was performed with SPSS 19.0
(SPSS Inc., Chicago, IL, USA). The numerical data are expressed as
means ± SD. The differences between groups were evaluated using
Student’s t-test or one-way ANOVA or Chi-square test. P<0.05 was
considered to indicate a statistically significant difference.
Results
The expression of UHRF1 in GBC tissues
and cell lines
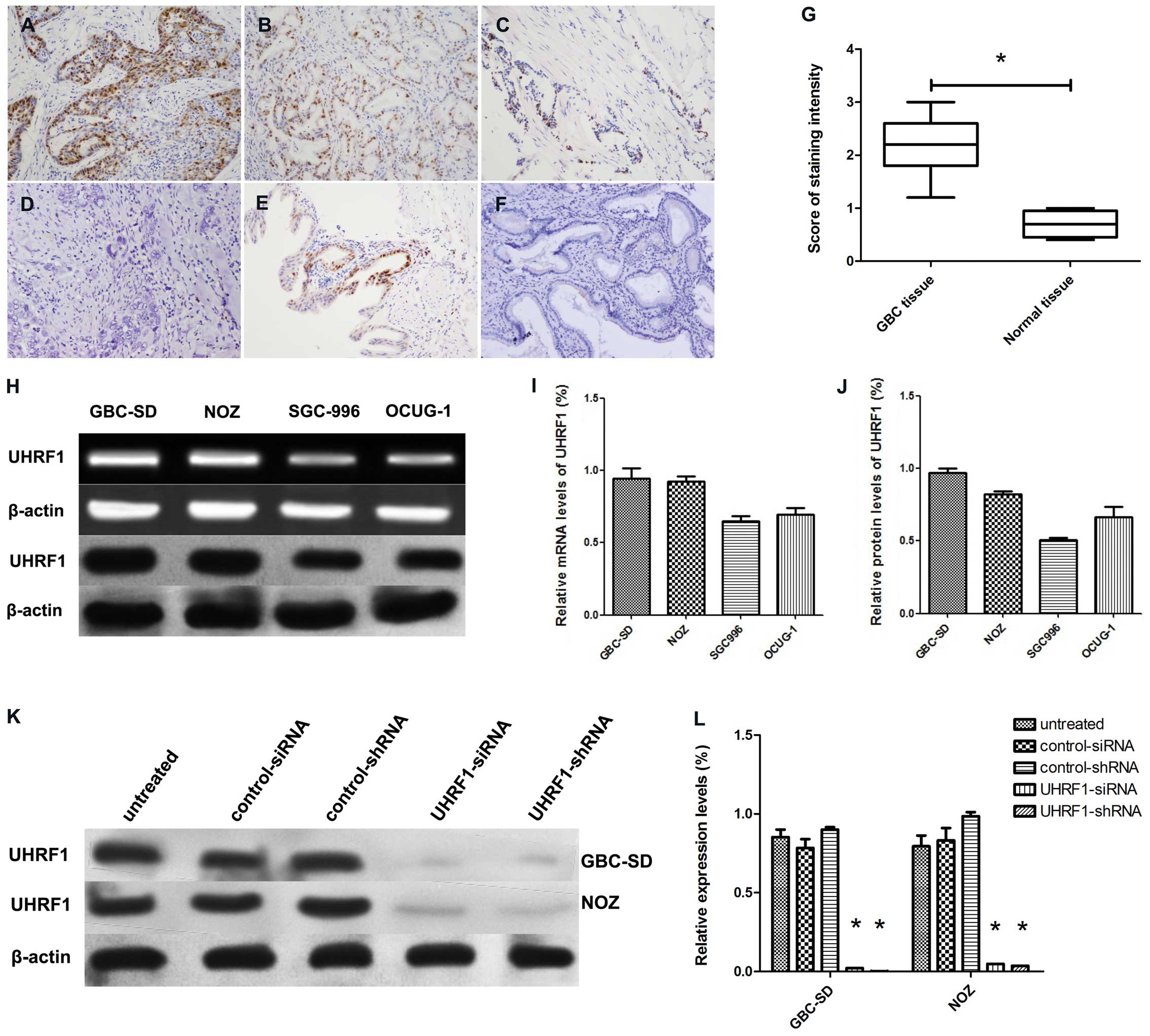
Immunohistochemical staining showed that
positive-staining for UHRF1 was found abundantly in the nuclei of
cancer cells (63.2%, 24/38) and occasionally in the nuclei of
normal epithelial cells (33.3%, 3/12) (P<0.05; Fig. 1A–F). The score of UHRF1-positive
immunostaining in GBC tissues was much higher than that in normal
tissues (P<0.05; Fig. 1G).
Additionally, the expression of UHRF1 correlated with advanced
stage and lymph node metastasis (P<0.05; Table I).
 | Table IAssociation between UHRF1 expression
and clinicopathological characteristics of gallbladder cancer. |
Table I
Association between UHRF1 expression
and clinicopathological characteristics of gallbladder cancer.
| Clinicopathological
characteristics | UHRF1 expression |
|---|
|
|---|
| Total | Positive no. (%) | P-value |
|---|
| Gender | | | >0.05 |
| Male | 12 | 7 (58.3) | |
| Female | 26 | 17 (65.4) | |
| Age (years) | | | >0.05 |
| ≤60 | 10 | 5 (50.0) | |
| >60 | 28 | 19 (67.9) | |
| TNM stage | | | 0.047a |
| I | 4 | 1 (25.0) | |
| II | 10 | 4 (40.0) | |
| III | 15 | 11 (73.3) | |
| IV | 9 | 8 (88.9) | |
| Lymph node
metastasis | | | 0.005a |
| Yes | 22 | 18 (81.2) | |
| No | 16 | 6 (37.5) | |
| Presence of
gallstones | | | >0.05 |
| Yes | 18 | 12 (66.7) | |
| No | 20 | 12 (60.0) | |
UHRF1 mRNA and protein were detected in GBC-SD, NOZ,
SGC996 and OCUG-1 cell lines (Fig.
1H). Since GBC-SD and NOZ cell lines have higher levels of
UHRF1 mRNA and protein than the other two cell lines (Fig. 1I and J), they were chosen for the
subsequent studies. As expected, western blot analysis showed the
expression of UHRF1 protein was decreased in GBC-SD and NOZ cells
after transfection with UHRF1 siRNA or UHRF1 shRNA (Fig. 1K and L).
UHRF1 depletion inhibits the growth of
GBC-SD and NOZ cell lines in vitro and in vivo
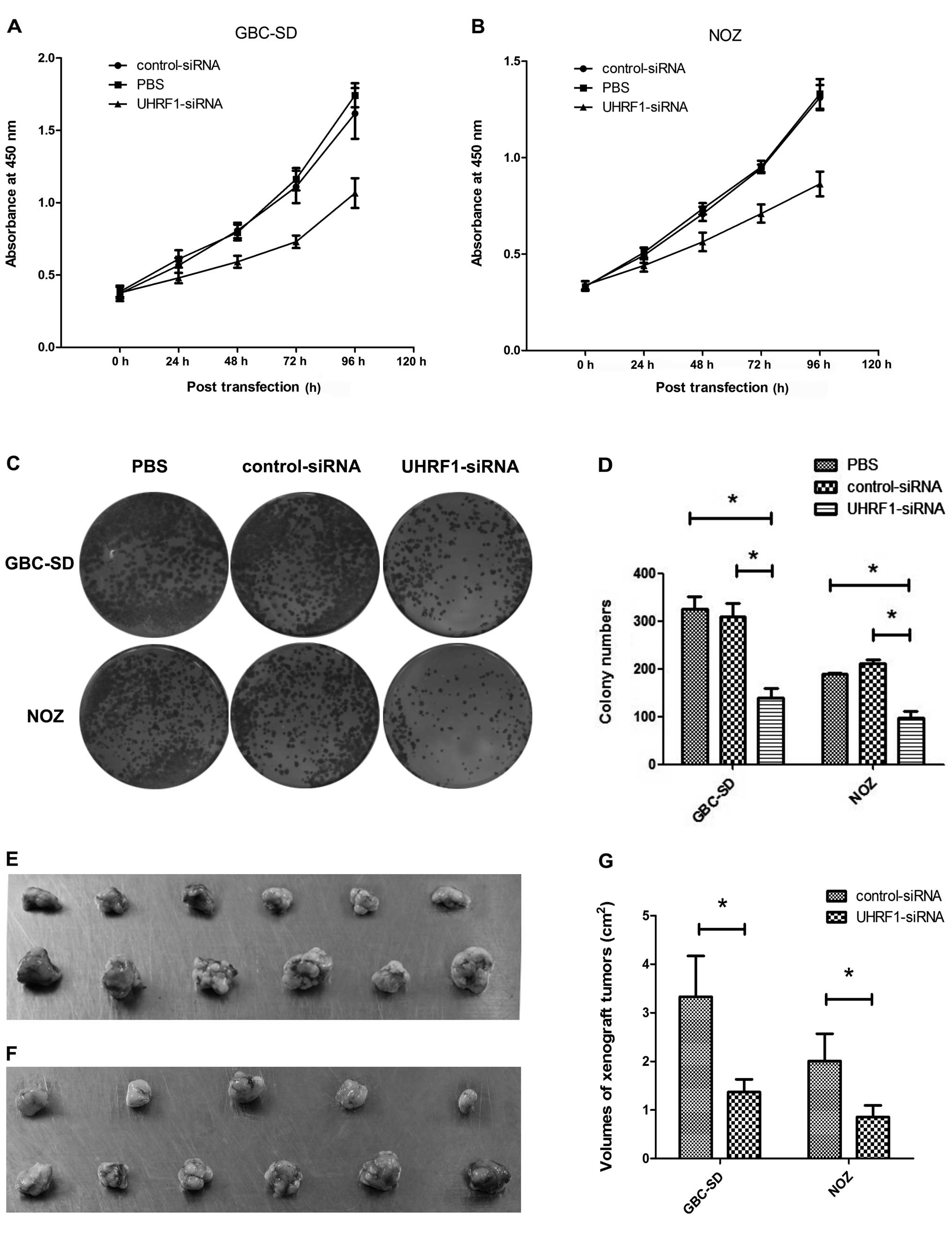
WST-1 assay showed that transfection with UHRF1
siRNA markedly inhibited proliferation of GBC-SD and NOZ cells in a
time-dependent manner (P<0.05 from 24 to 96 h) in comparison
with untransfected and control-siRNA-transfected cells (Fig. 2A and B).
To evaluate the effects of UHRF1 depletion on more
long-term cell proliferation, colony formation assay was also
performed (Fig. 2C). The colony
numbers were significantly reduced in GBC-SD and NOZ cells
transfected with UHRF1 siRNA in comparison with those in GBC-SD and
NOZ cells untransfected or transfected with control siRNA
(P<0.05; Fig. 2D).
All animals injected with GBC-SD or NOZ cells
transfected with control shRNA developed visible tumors within the
first 3 weeks of the experiment, and these xenograft tumors grew
progressively. However, none of the animals that received GBC-SD or
NOZ cells transfected with UHRF1 shRNA developed visible tumors
within the first 3 weeks. Moreover, one of the animals that was
injected with UHRF1-shRNA-transfected NOZ cells did not develop
tumors during 6 weeks of observation (Fig. 2E and F). The volume of the xenograft
tumors formed by UHRF1-shRNA-transfected cells was significantly
smaller than that of the control groups (P<0.05; Fig. 2G).
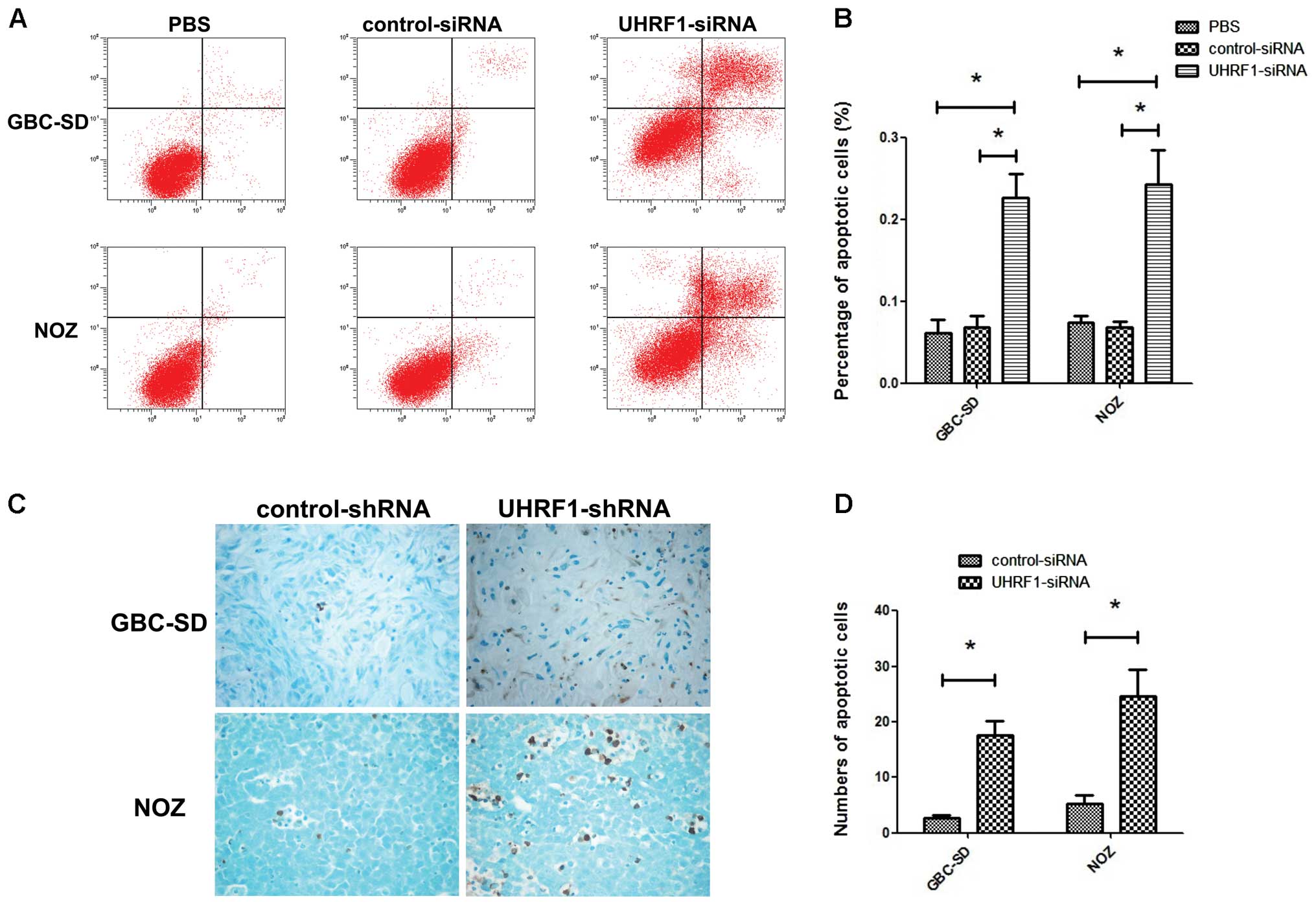
UHRF1 depletion correlates with high
apoptosis in GBC cell lines and xenograft tumors
The effects of UHRF1 depletion on apoptosis in
vitro and in vivo were determined using Annexin V/PI
assay and TUNEL assay, respectively. Annexin V/PI assay showed that
UHRF1 siRNA triggered apoptosis (early apoptosis plus late
apoptosis) in GBC-SD cells and NOZ cells (Fig. 3A). The proportions of apoptotic
GBC-SD cells and NOZ cells after transfection with UHRF1 siRNA were
much higher than those of apoptotic GBC-SD cells and NOZ cells
untransfected or transfected with control siRNA (P<0.05;
Fig. 3B). TUNEL assay showed that
stable transfection with UHRF1 shRNA increased the percentage of
TUNEL-positive cells both in GBC-SD and NOZ cells in vivo
(Fig. 3C). The mean numbers of
apoptotic GBC-SD and NOZ cells transfected with UHRF1 shRNA/field
were 17.7±2.5 and 24.7±4.7, both of which were much more than those
of the control groups (P<0.05; Fig.
3D).
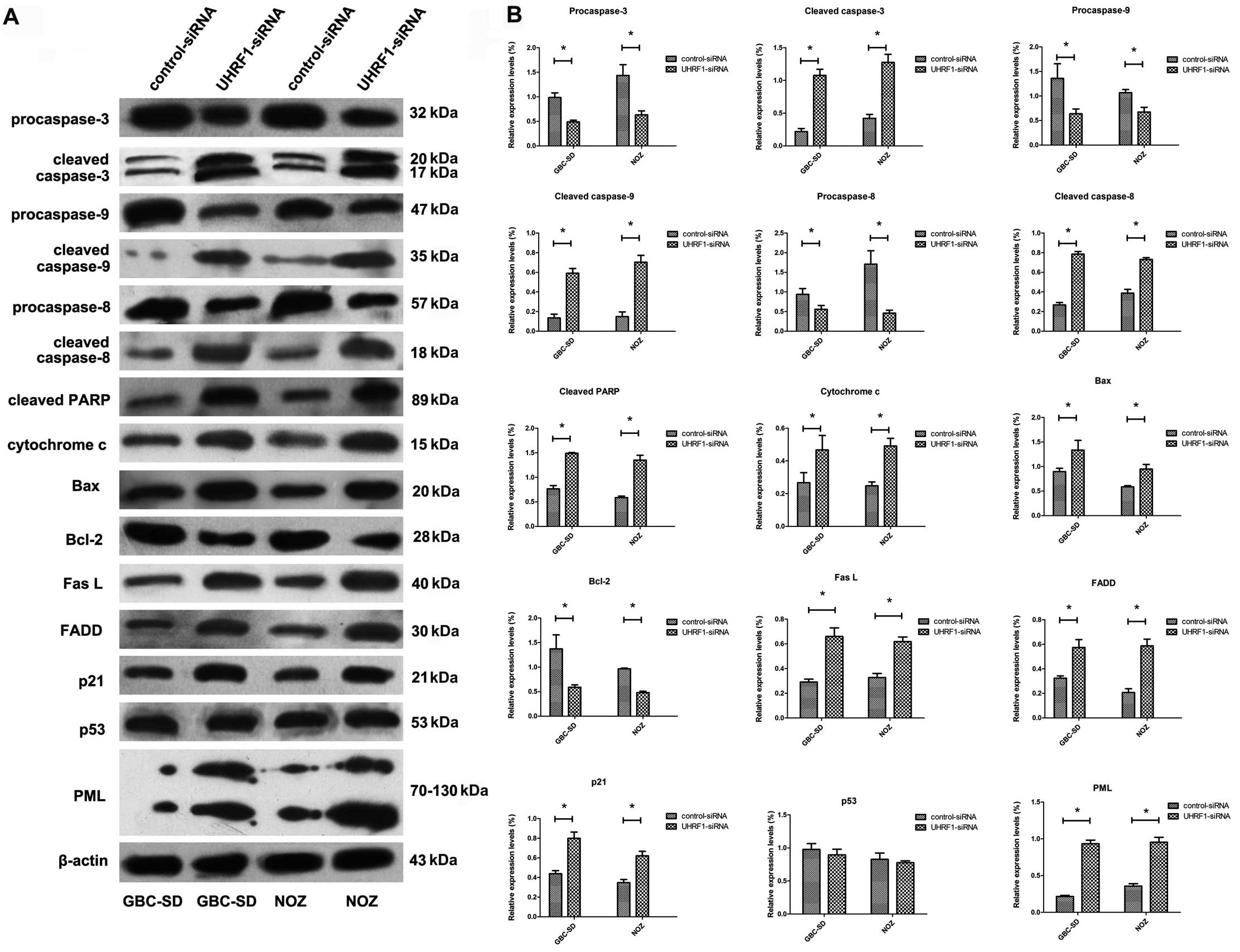
UHRF1 depletion activates extrinsic and
intrinsic apoptotic pathways in GBC-SD and NOZ cells in vitro
The expressions of a series of pro-apoptotic and
anti-apoptotic proteins that may participate in the process of
apoptosis induced by UHRF1 depletion were examined using western
blot assay. As shown in Fig. 4, the
pro-caspase-3, -8 and -9 and bcl-2 were decreased while the cleaved
caspase-3, -8 and -9 and PARP, Bax, cytosolic cytochrome c,
FasL and FADD were increased after transfection with UHRF1 siRNA.
In addition, the expression levels of PML and p21 were increased
after transfection with UHRF1 siRNA, while the expression of p53
was not altered in the same conditions.
UHRF1 depletion arrests GBC-SD and NOZ
cells at G1 phase
Transfection with UHRF1 siRNA induced an obvious
cell cycle arrest at G1 phase in GBC-SD and NOZ cells (P<0.05;
Table II), and the percentages of
cells at S and G2 phase were correspondingly decreased.
 | Table IICell cycle analysis after
transfection. |
Table II
Cell cycle analysis after
transfection.
| | Cell cycle
distribution (G1, S, G2) |
|---|
| |
|
|---|
| Cell line | Transfection | G1 phase (%) | S phase (%) | G2 phase (%) |
|---|
| GBC-SD | UHRF1 siRNA |
75.5±4.5a | 14.4±2.4 | 10.0±6.9 |
| Control siRNA | 55.7±2.6 | 29.1±4.0 | 15.2±4.2 |
| PBS | 57.3±4.3 | 17.9±3.2 | 24.9±3.3 |
| NOZ | UHRF1 siRNA |
78.5±4.3a | 10.9±1.2 | 8.6±1.8 |
| Control siRNA | 58.7±3.9 | 22.8±3.2 | 18.5±1.3 |
| PBS | 60.6±1.7 | 22.0±2.4 | 17.4±3.3 |
UHRF1 depletion reduces the migration of
GBC-SD and NOZ cells in vitro
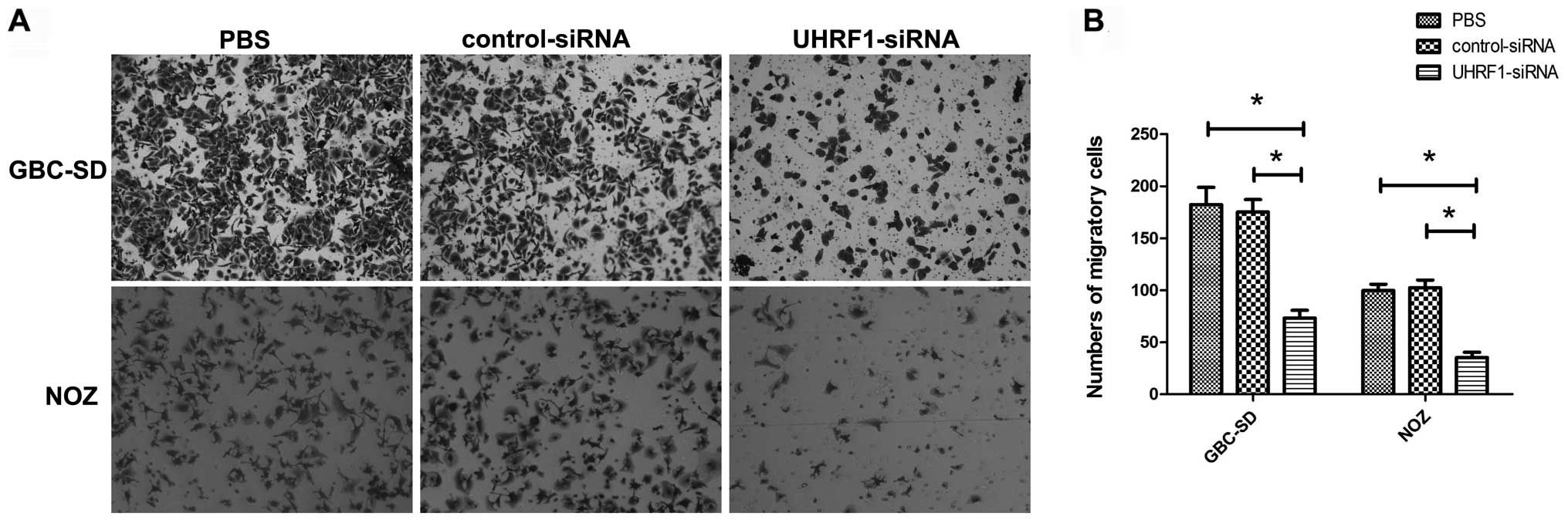
The number of migratory cells stained with crystal
violet reflected their migratory capacity (Fig. 5A). Transfection with UHRF1 siRNA
resulted in a significant decrease in the numbers of migratory
GBC-SD and NOZ cells in comparison with the control groups
(P<0.05; Fig. 5B).
Discussion
In the present study, the role of UHRF1 in GBC was
preliminarily investigated. First, we examined the expression of
UHRF1 in GBC tissues and cell lines. Consistent with other cancer
tissues, immunohistochemical staining showed that UHRF1 protein was
overexpressed in GBC tissues in comparison with non-tumor tissues.
The positive expression of UHRF1 correlated with advanced TNM stage
and lymph node metastasis, but not with age, gender and presence of
gallstones. Since the expression of UHRF1 was regulated by p53, the
frequent abnormality of p53 detected in GBC (15) would be an explanation for the
observed UHRF1 overexpression. However, the 5-year survival rate
and median survival time could not be calculated now, therefore the
relationship between UHRF1 expression and prognosis was not
evaluated in the present study. These observations indicate that
UHRF1 may play a role in the tumorigenesis and progression of
GBC.
To further explore the possible biological function
of UHRF1 in GBC growth and development, UHRF1 expression was
depleted in GBC-SD and NOZ cell lines using siRNA or shRNA. Our
findings showed that UHRF1 depletion significantly inhibited
proliferation in vitro and tumorigenicity in vivo,
accompanied by cell cycle arrest and induction of apoptosis.
Comparable to the results of other studies that UHRF1 depletion may
cause cell cycle arrest either at G1 (14,16) or
G2/M phase (13), our results
showed that the UHRF1 depletion induced cell cycle arrest at G1
phase in GBC cell lines. These findings indicate that UHRF1 may
play a critical role during multiple phases of the cell cycle and
the effects of UHRF1 depletion on cell cycle arrest may be
cell-type specific.
Apoptosis induction by UHRF1 depletion has been
reported in several studies, but the exact molecular mechanisms are
not fully understood. As is known, caspase family members are
crucial mediators during the process of apoptosis. Among them,
caspase-3 is a key effector in the execution of the apoptotic
program and is primarily responsible for the cleavage of
poly-(ADP-ribose) polymerase (PARP) (17). The latter serves as a sensitive
parameter for identification of different types of cell death and
as a marker for activation of different death proteases. Caspase-8
activation is typically associated with the extrinsic or death
receptor pathway of apoptosis, whereas caspase-9 activation
following mitochondrial disruption and cytochrome c release
is associated with the intrinsic or mitochondrial pathway of
apoptosis (18). In the present
study, we found the expressions of cleaved caspase-3, -8 and -9,
PARP, FasL, FADD, bax and cytosolic cytochrome c were
significantly increased, while the expressions of pro-caspase-3, -8
and -9 and bcl-2 were significantly decreased. These findings
suggested that UHRF1 siRNA induced both extrinsic and intrinsic
apoptotic pathways in GBC-SD an NOZ cells.
In addition to growth inhibition, UHRF1 siRNA also
inhibited the migration of GBC-SD and NOZ cells, which indicated
that UHRF1 may play a role in the migration of GBC cells. This
result also supported the aforementioned finding that the
overexpression of UHRF1 correlated with late TNM stage and lymph
node metastasis of GBC.
UHRF1 depletion could reactivate the expressions of
these tumor suppressor genes such as PML (19), which was frequently downregulated in
GBC tissues (20). In the present
study, we found PML expression was markedly increased after
transfection with UHRF1 siRNA. Considering that PML was
functionally linked to various fundamental cellular processes
including caspase-dependent apoptosis in response to Fas L/Fas
signals and control of cell migration (21,22),
we suggested that reactivation of PML by UHRF1 depletion may lead
to induction of apoptosis and inhibition of migration in GBC cell
lines. In addition, the expressions of two other tumor suppressor
genes p53 and p21 were also examined, both of which play important
roles in apoptosis and cell cycle regulation. The level of p21
protein was significantly increased, but the level of p53 protein
which acts as a transcriptional activator of p21 was not altered,
suggesting UHRF1 depletion results in an accumulation of p21 in a
p53-independent manner. Induction of p53-independent p21 which was
also observed in other studies (23,24)
may be a reasonable explanation for the G1 arrest caused by UHRF1
siRNA. These findings further indicated that UHRF1 induced
p53-independent, Fas mediated apoptosis in GBC-SD and NOZ cell
lines.
Taken together, we found UHRF1 was overexpressed in
GBC tissues and UHRF1 depletion by siRNA or shRNA inhibited
proliferation and migration in GBC cell lines. In addition, UHRF1
depletion induced cell cycle arrest and apoptosis in GBC cell
lines. Based on our findings, siRNA or shRNA against UHRF1 therapy
may be applied to the treatment of GBC patients. However, due to
rapid degradation by serum nucleases, siRNA is very unstable in
blood. Moreover, due to its small molecular weight, siRNA is
excreted from the body quite rapidly. Establishment of good
delivery platforms to deliver siRNA to a targeted location remains
the biggest hurdle to overcome prior to using siRNA as a
therapeutic strategy and has become an active research area. In
recent years, notable advances have been made for the delivery of
siRNA to tumors and several promising siRNA delivery platforms have
been investigated in order to carry the siRNA into target cells,
including liposomes, polyplexes, liposome-polycation-DNA (LPD) and
siRNA conjugates (25). In
addition, chemical modifications and alternative backbones were
used to improve the stability of siRNAs (26). It seems that the need for efficient
delivery platforms and stable siRNAs may be met in the near future,
and siRNA therapy will provide considerable benefits to cancer
patients, including GBC patients.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (nos. 81272747 and 81372642).
References
|
1
|
Nagorney DM and McPherson GA: Carcinoma of
the gallbladder and extrahepatic bile ducts. Semin Oncol.
15:106–115. 1988.PubMed/NCBI
|
|
2
|
Zhu AX, Hong TS, Hezel AF and Kooby DA:
Current manage- ment of gallbladder carcinoma. Oncologist.
15:168–181. 2010. View Article : Google Scholar
|
|
3
|
Takahashi T, Shivapurkar N, Riquelme E, et
al: Aberrant promoter hypermethylation of multiple genes in
gallbladder carcinoma and chronic cholecystitis. Clin Cancer Res.
10:6126–6133. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Unoki M, Nishidate T and Nakamura Y:
ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG
through its SRA domain. Oncogene. 23:7601–7610. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jeanblanc M, Mousli M, Hopfner R, et al:
The retinoblastoma gene and its product are targeted by ICBP90: a
key mechanism in the G1/S transition during the cell cycle.
Oncogene. 24:7337–7345. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Geng Y, Gao Y, Ju H and Yan F: Diagnostic
and prognostic value of plasma and tissue ubiquitin-like,
containing PHD and RING finger domains 1 in breast cancer patients.
Cancer Sci. 104:194–199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang F, Zhang P, Ma Y, et al: NIRF is
frequently upregulated in colorectal cancer and its oncogenicity
can be suppressed by let-7a microRNA. Cancer Lett. 314:223–231.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Unoki M, Kelly JD, Neal DE, Ponder BA,
Nakamura Y and Hamamoto R: UHRF1 is a novel molecular marker for
diagnosis and the prognosis of bladder cancer. Br J Cancer.
101:98–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Unoki M, Daigo Y, Koinuma J, Tsuchiya E,
Hamamoto R and Nakamura Y: UHRF1 is a novel diagnostic marker of
lung cancer. Br J Cancer. 103:217–222. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Babbio F, Pistore C, Curti L, et al: The
SRA protein UHRF1 promotes epigenetic crosstalks and is involved in
prostate cancer progression. Oncogene. 31:4878–4887. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang C, Wang Y, Zhang F, et al: Inhibiting
UHRF1 expression enhances radiosensitivity in human esophageal
squamous cell carcinoma. Mol Biol Rep. 40:5225–5235. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pi JT, Lin Y, Quan Q, Chen LL, Jiang LZ,
Chi W and Chen HY: Overexpression of UHRF1 is significantly
associated with poor prognosis in laryngeal squamous cell
carcinoma. Med Oncol. 30:6132013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tien AL, Senbanerjee S, Kulkarni A, et al:
UHRF1 depletion causes a G2/M arrest, activation of DNA
damage response and apoptosis. Biochem J. 435:175–185.
2011.PubMed/NCBI
|
|
14
|
Arima Y, Hirota T, Bronner C, et al:
Down-regulation of nuclear protein ICBP90 by
p53/p21Cip1/WAF1-dependent DNA damage checkpoint signals
contributes to cell cycle arrest at G1/S transition. Genes Cells.
9:131–142. 2004.PubMed/NCBI
|
|
15
|
Lazcano-Ponce EC, Miquel JF, Muñoz N, et
al: Epidemiology and molecular pathology of gallbladder cancer. CA
Cancer J Clin. 51:349–364. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jenkins Y, Markovtsov V, Lang W, et al:
Critical role of the ubiquitin ligase activity of UHRF1, a nuclear
RING finger protein, in tumor cell growth. Mol Biol Cell.
16:5621–5629. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Porter AG and Jänicke RU: Emerging roles
of caspase-3 in apoptosis. Cell Death Differ. 6:99–104. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vermeulen K, Van Bockstaele DR and
Berneman ZN: Apoptosis: mechanisms and relevance in cancer. Ann
Hematol. 84:627–639. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guan D, Factor D, Liu Y, Wang Z and Kao
HY: The epigenetic regulator UHRF1 promotes ubiquitination-mediated
degradation of the tumor-suppressor protein promyelocytic leukemia
protein. Oncogene. 32:3819–3828. 2013. View Article : Google Scholar
|
|
20
|
Chang HJ, Yoo BC, Kim SW, Lee BL and Kim
WH: Significance of PML and p53 protein as molecular prognostic
markers of gallbladder carcinomas. Pathol Oncol Res. 13:326–335.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Krieghoff-Henning E and Hofmann TG: Role
of nuclear bodies in apoptosis signalling. Biochim Biophys Acta.
1783:2185–2194. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Reineke EL, Liu Y and Kao HY:
Promyelocytic leukemia protein controls cell migration in response
to hydrogen peroxide and insulin-like growth factor-1. J Biol Chem.
285:9485–9492. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim WH, Kang KH, Kim MY and Choi KH:
Induction of p53-independent p21 during ceramide-induced G1 arrest
in human hepatocarcinoma cells. Biochem Cell Biol. 78:127–135.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zuo S, Liu C, Wang J, et al: IGFBP-rP1
induces p21 expression through a p53-independent pathway, leading
to cellular senescence of MCF-7 breast cancer cells. J Cancer Res
Clin Oncol. 138:1045–1055. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen Y: Advances in the development of
siRNA-based therapeutics for cancer. IDrugs. 11:572–578.
2008.PubMed/NCBI
|
|
26
|
Unoki M, Kumamoto K and Harris CC: ING
proteins as potential anticancer drug targets. Curr Drug Targets.
10:442–454. 2009. View Article : Google Scholar : PubMed/NCBI
|