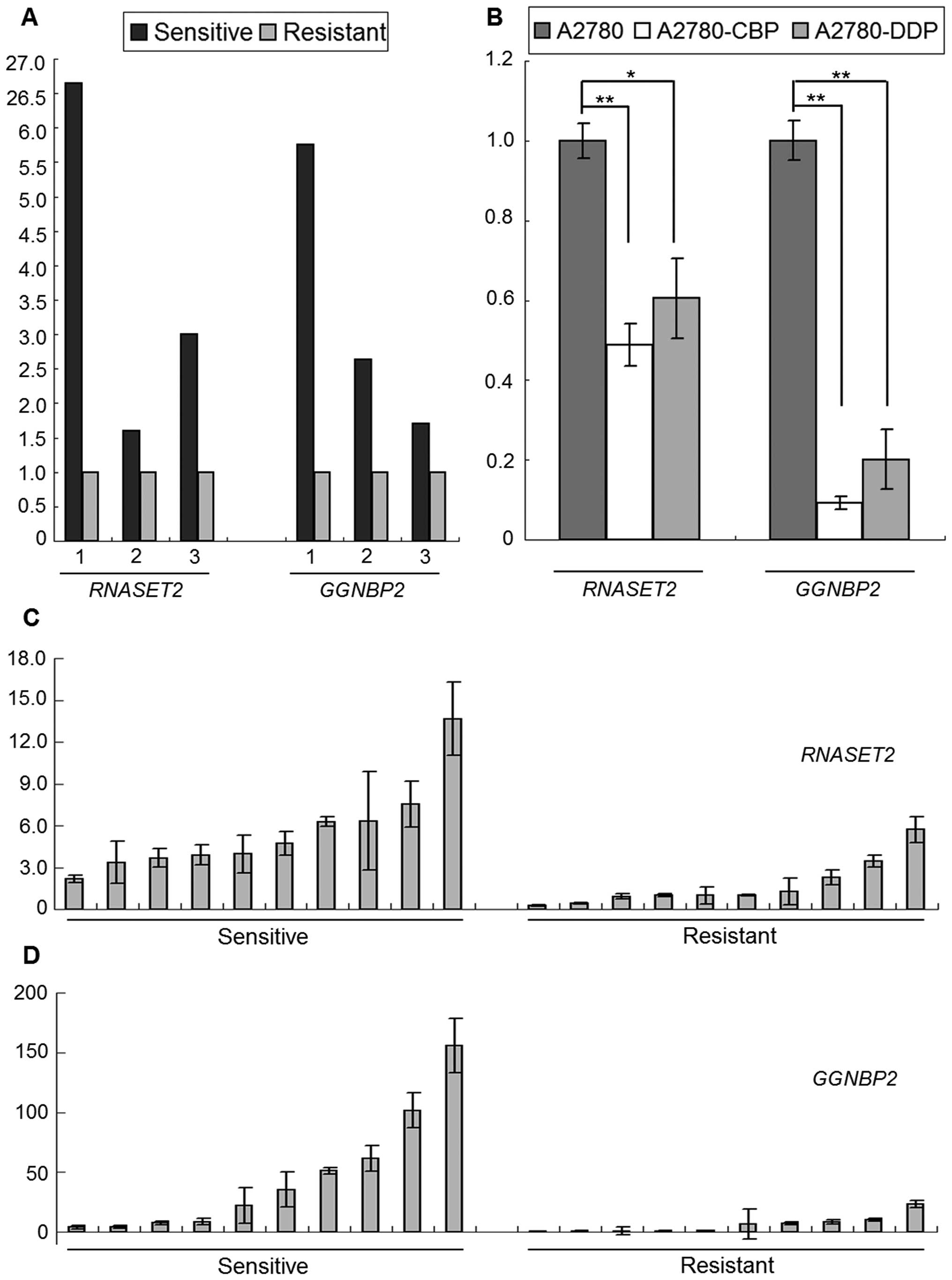
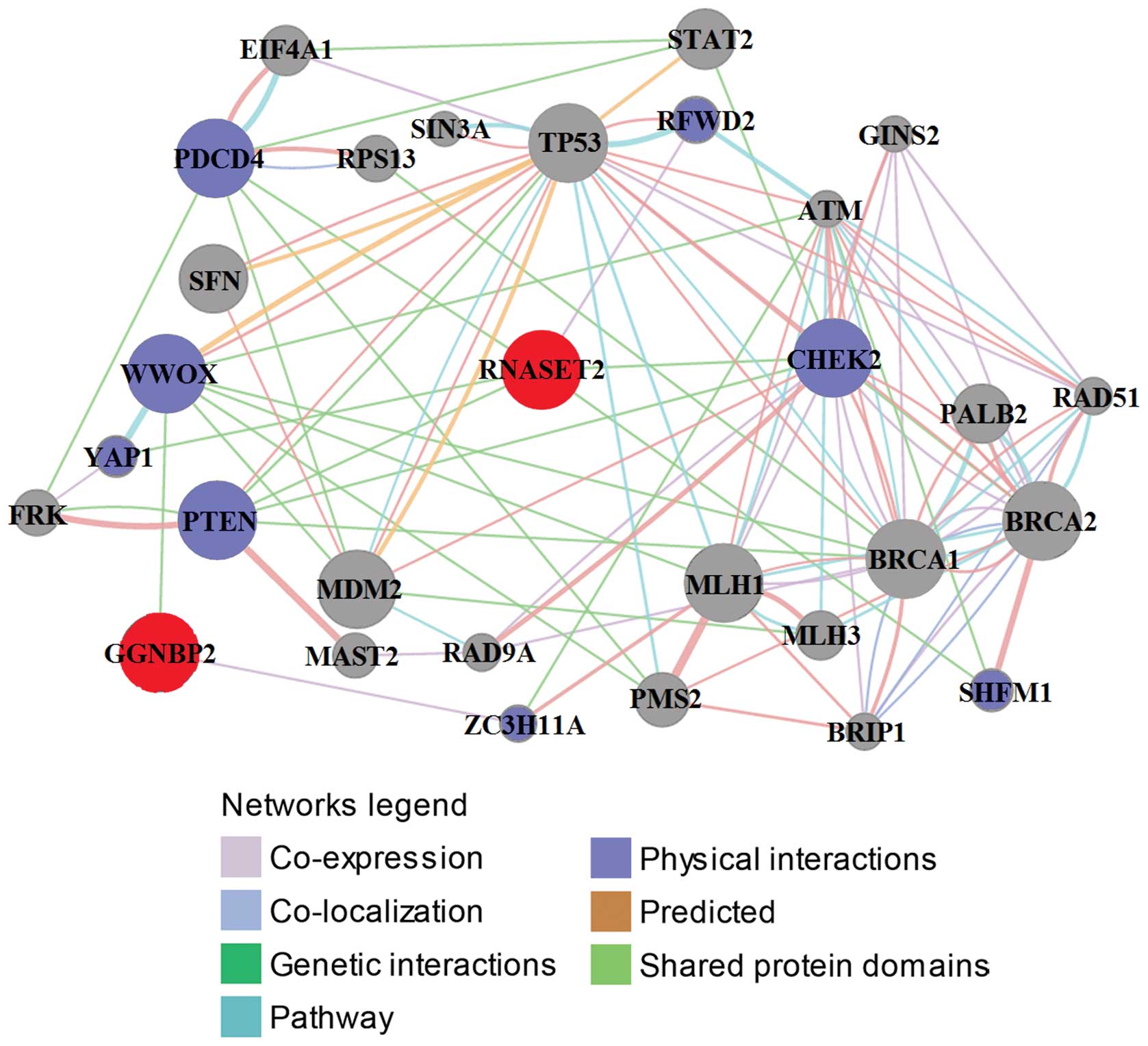
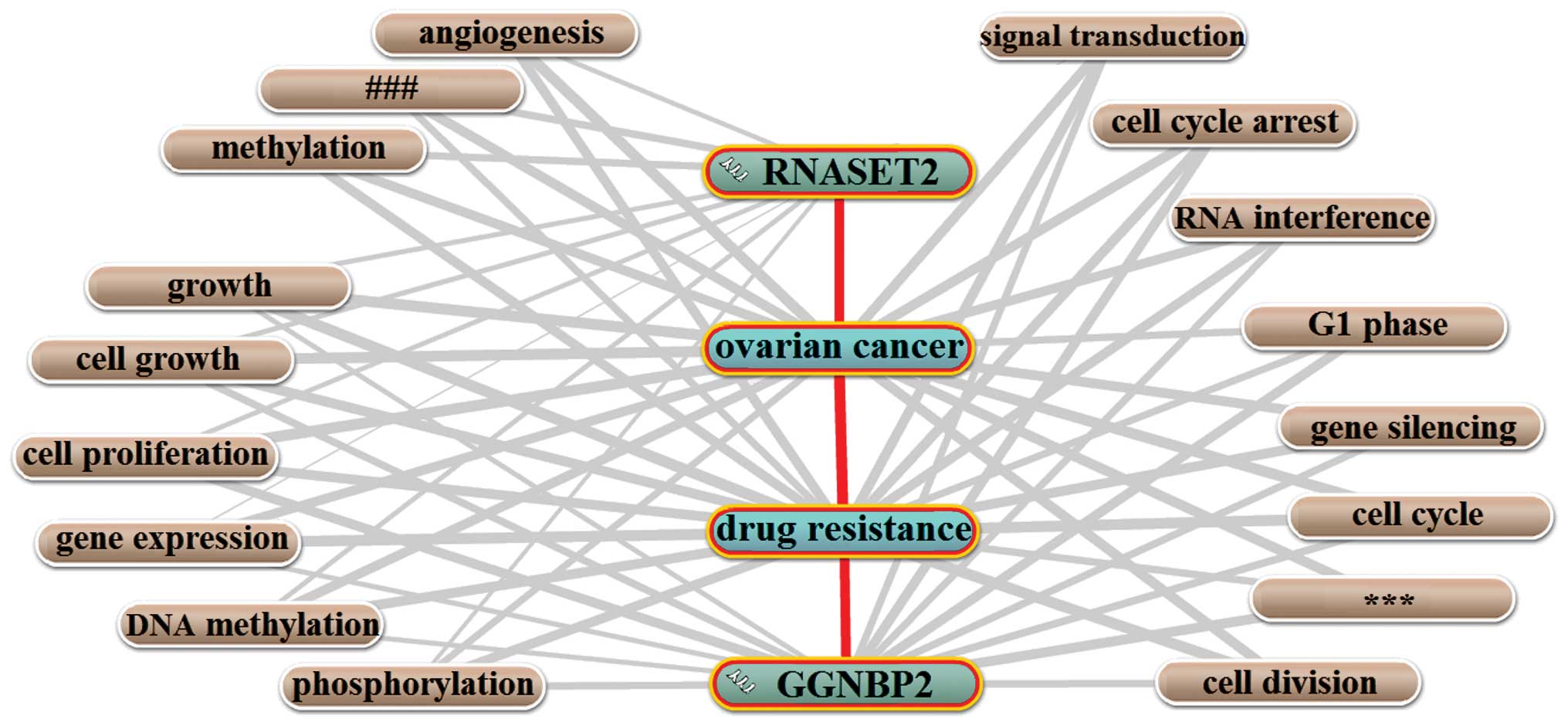
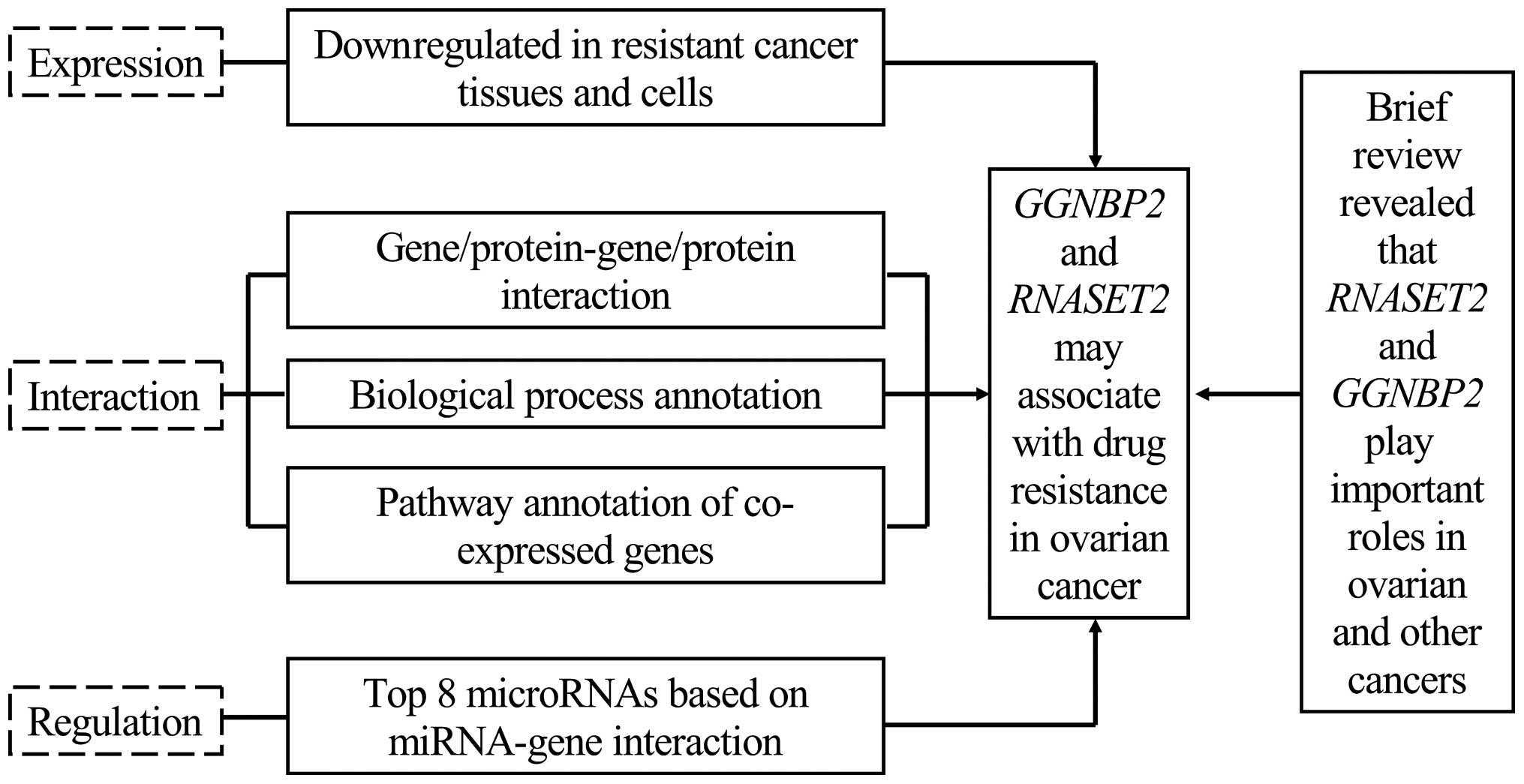
|
1
|
Balch C, Huang TH, Brown R and Nephew KP:
The epigenetics of ovarian cancer drug resistance and
resensitization. Am J Obstet Gynecol. 191:1552–1572. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
3
|
Suh DH, Kim MK, No JH, Chung HH and Song
YS: Metabolic approaches to overcoming chemoresistance in ovarian
cancer. Ann N Y Acad Sci. 1229:53–60. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Matsuo K, Eno ML, Im DD, Rosenshein NB and
Sood AK: Clinical relevance of extent of extreme drug resistance in
epithelial ovarian carcinoma. Gynecol Oncol. 116:61–65. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gottesman MM: Mechanisms of cancer drug
resistance. Annu Rev Med. 53:615–627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Johnson SW, Ozols RF and Hamilton TC:
Mechanisms of drug resistance in ovarian cancer. Cancer.
71:644–649. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNAs in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sager R: Tumor suppressor genes: the
puzzle and the promise. Science. 246:1406–1412. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sherr CJ: Principles of tumor suppression.
Cell. 116:235–246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yin F, Liu X, Li D, Wang Q, Zhang W and Li
L: Tumor suppressor genes associated with drug resistance in
ovarian cancer (Review). Oncol Rep. 30:3–10. 2013.PubMed/NCBI
|
|
11
|
Wu H, Cao Y, Weng D, Xing H, Song X, Zhou
J, Xu G, Lu Y, Wang S and Ma D: Effect of tumor suppressor gene
PTEN on the resistance to cisplatin in human ovarian cancer cell
lines and related mechanisms. Cancer Lett. 271:260–271. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Swisher EM, Sakai W, Karlan BY, Wurz K,
Urban N and Taniguchi T: Secondary BRCA1 mutations in
BRCA1-mutated ovarian carcinomas with platinum resistance.
Cancer Res. 68:2581–2586. 2008.PubMed/NCBI
|
|
13
|
Haber D and Harlow E: Tumour-suppressor
genes: evolving definitions in the genomic age. Nat Genet.
16:320–322. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jones PA and Laird PW: Cancer epigenetics
comes of age. Nat Genet. 21:163–167. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Acquati F, Morelli C, Cinquetti R, Bianchi
MG, Porrini D, Varesco L, Gismondi V, Rocchetti R, Talevi S,
Possati L, Magnanini C, Tibiletti MG, Bernasconi B, Daidone MG,
Shridhar V, Smith DI, Negrini M, Barbanti-Brodano G and Taramelli
R: Cloning and characterization of a senescence inducing and class
II tumor suppressor gene in ovarian carcinoma at chromosome region
6q27. Oncogene. 20:980–988. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Trubia M, Sessa L and Taramelli R:
Mammalian Rh/T2/S-glycoprotein ribonuclease family genes: cloning
of a human member located in a region of chromosome 6 (6q27)
frequently deleted in human malignancies. Genomics. 42:342–344.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McClure BA, Haring V, Ebert PR, Anderson
MA, Simpson RJ, Sakiyama F and Clarke AE: Style
self-incompatibility gene products of Nicotlana alata are
ribonucleases. Nature. 342:955–957. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Steinemann D, Gesk S, Zhang Y, Harder L,
Pilarsky C, Hinzmann B, Martin-Subero JI, Calasanz MJ, Mungall A,
Rosenthal A, Siebert R and Schlegelberger B: Identification of
candidate tumor-suppressor genes in 6q27 by combined deletion
mapping and electronic expression profiling in lymphoid neoplasms.
Genes Chromosomes Cancer. 37:421–426. 2003. View Article : Google Scholar
|
|
19
|
Kim TY, Zhong S, Fields CR, Kim JH and
Robertson KD: Epigenomic profiling reveals novel and frequent
targets of aberrant DNA methylation-mediated silencing in malignant
glioma. Cancer Res. 66:7490–7501. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Monti L, Rodolfo M, Lo Russo G, Noonan D,
Acquati F and Taramelli R: RNASET2 as a tumor antagonizing
gene in a melanoma cancer model. Oncol Res. 17:69–74. 2008.
|
|
21
|
Acquati F, Possati L, Ferrante L,
Campomenosi P, Talevi S, Bardelli S, Margiotta C, Russo A,
Bortoletto E, Rocchetti R, Calza R, Cinquetti R, Monti L, Salis S,
Barbanti-Brodano G and Taramelli R: Tumor and metastasis
suppression by the human RNASET2 gene. Int J Oncol. 26:1159–1168.
2005.PubMed/NCBI
|
|
22
|
Acquati F, Bertilaccio S, Grimaldi A,
Monti L, Cinquetti R, Bonetti P, Lualdi M, Vidalino L, Fabbri M,
Sacco MG, van Rooijen N, Campomenosi P, Vigetti D, Passi A, Riva C,
Capella C, Sanvito F, Doglioni C, Gribaldo L, Macchi P, Sica A,
Noonan DM, Ghia P and Taramelli R: Microenvironmental control of
malignancy exerted by RNASET2, a widely conserved extracellular
RNase. Proc Natl Acad Sci USA. 108:1104–1109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang X, Xiao Z, Chen Z, Li C, Li J,
Yanhui Y, Yang F, Yang Y and Oyang Y: Comparative proteomics
analysis of the proteins associated with laryngeal
carcinoma-related gene 1. Laryngoscope. 116:224–230. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Y and Chen Z: Molecular cloning and
characterization of LCRG1 a novel gene localized to the
tumor suppressor locus D17S800-D17S930. Cancer Lett. 209:75–85.
2004.
|
|
25
|
Guan R, Wen XY, Wu J, Duan R, Cao H, Lam
S, Hou D, Wang Y, Hu J and Chen Z: Knockdown of ZNF403 inhibits
cell proliferation and induces G2/M arrest by modulating cell-cycle
mediators. Mol Cell Biochem. 365:211–222. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang Z: Verification of differential
expression gene and its epigenic and genic study between
carbo-resistance cell line and its parental cell line of ovarian
carcinoma (unpublished PhD dissertation). Guangxi Medical
University. 2010.
|
|
27
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk - database: prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011.
|
|
28
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2008.PubMed/NCBI
|
|
29
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009.PubMed/NCBI
|
|
30
|
Zhang P, Gao W, Li H, Reed E and Chen F:
Inducible degradation of checkpoint kinase 2 links to
cisplatin-induced resistance in ovarian cancer cells. Biochem
Biophys Res Commun. 328:567–572. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jansen AP, Camalier CE, Stark C and
Colburn NH: Characterization of programmed cell death 4 in multiple
human cancers reveals a novel enhancer of drug sensitivity. Mol
Cancer Ther. 3:103–110. 2004.PubMed/NCBI
|
|
32
|
Shiota M, Izumi H, Tanimoto A, Takahashi
M, Miyamoto N, Kashiwagi E, Kidani A, Hirano G, Masubuchi D,
Fukunaka Y, Yasuniwa Y, Naito S, Nishizawa S, Sasaguri Y and Kohno
K: Programmed cell death protein 4 down-regulates Y-box binding
protein-1 expression via a direct interaction with Twist1 to
suppress cancer cell growth. Cancer Res. 69:3148–3156. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang X, Wang X, Song X, Liu C, Shi Y,
Wang Y, Afonja O, Ma C, Chen YH and Zhang L: Programmed cell death
4 enhances chemosensitivity of ovarian cancer cells by activating
death receptor pathway in vitro and in vivo. Cancer Sci.
101:2163–2170. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chetram MA and Hinton CV: PTEN regulation
of ERK1/2 signaling in cancer. J Recept Signal Transduct Res.
32:190–195. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Steelman LS, Chappell WH, Abrams SL, Kempf
RC, Long J, Laidler P, Mijatovic S, Maksimovic-Ivanic D, Stivala F,
Mazzarino MC, Donia M, Fagone P, Malaponte G, Nicoletti F, Libra M,
Milella M, Tafuri A, Bonati A, Bäsecke J, Cocco L, Evangelisti C,
Martelli AM, Montalto G, Cervello M and McCubrey JA: Roles of the
Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR pathways in controlling growth
and sensitivity to therapy-implications for cancer and aging.
Aging. 3:192–222. 2011.PubMed/NCBI
|
|
36
|
Chalhoub N and Baker SJ: PTEN and the
PI3-kinase pathway in cancer. Annu Rev Pathol. 4:127–150. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Okumura N, Yoshida H, Kitagishi Y,
Nishimura Y and Matsuda S: Alternative splicings on p53, BRCA1 and
PTEN genes involved in breast cancer. Biochem Biophys Res Commun.
413:395–399. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Selvendiran K, Tong L, Vishwanath S,
Bratasz A, Trigg NJ, Kutala VK, Hideg K and Kuppusamy P: EF24
induces G2/M arrest and apoptosis in cisplatin-resistant human
ovarian cancer cells by increasing PTEN expression. J Biol Chem.
282:28609–28618. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lee S, Choi EJ, Jin C and Kim DH:
Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA
mRNA amplification contributes to cisplatin resistance in an
ovarian cancer cell line. Gynecol Oncol. 97:26–34. 2005.PubMed/NCBI
|
|
40
|
Yuan M, Tomlinson V, Lara R, Holliday D,
Chelala C, Harada T, Gangeswaran R, Manson-Bishop C, Smith P,
Danovi SA, Pardo O, Crook T, Mein CA, Lemoine NR, Jones LJ and Basu
S: Yes-associated protein (YAP) functions as a tumor suppressor in
breast. Cell Death Differ. 15:1752–1759. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li J, Zou C, Bai Y, Wazer DE, Band V and
Gao Q: DSS1 is required for the stability of BRCA2. Oncogene.
25:1186–1194. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sakai W, Swisher EM, Jacquemont C,
Chandramohan KV, Couch FJ, Langdon SP, Wurz K, Higgins J, Villegas
E and Taniguchi T: Functional restoration of BRCA2 protein by
secondary BRCA2 mutations in BRCA2-mutated ovarian
carcinoma. Cancer Res. 69:6381–6386. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vitari AC, Leong KG, Newton K, Yee C,
O’Rourke K, Liu J, Phu L, Vij R, Ferrando R, Couto SS, Mohan S,
Pandita A, Hongo JA, Arnott D, Wertz IE, Gao WQ, French DM and
Dixit VM: COP1 is a tumour suppressor that causes degradation of
ETS transcription factors. Nature. 474:403–406. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Migliorini D, Bogaerts S, Defever D, Vyas
R, Denecker G, Radaelli E, Zwolinska A, Depaepe V, Hochepied T,
Skarnes WC and Marine JC: Cop1 constitutively regulates c-Jun
protein stability and functions as a tumor suppressor in mice. J
Clin Invest. 121:1329–1343. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li YF, Wang DD, Zhao BW, Wang W, Huang CY,
Chen YM, Zheng Y, Keshari RP, Xia JC and Zhou ZW: High level of
COP1 expression is associated with poor prognosis in primary
gastric cancer. Int J Biol Sci. 8:1168–1177. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liang J, Song W, Tromp G, Kolattukudy PE
and Fu M: Genome-wide survey and expression profiling of CCCH-zinc
finger family reveals a functional module in macrophage activation.
PLoS One. 3:e28802008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shih JY and Yang PC: The EMT regulator
slug and lung carcinogenesis. Carcinogenesis. 32:1299–1304. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Paige AJ, Taylor KJ, Taylor C, Hillier SG,
Farrington S, Scott D, Porteous DJ, Smyth JF, Gabra H and Watson
JE: WWOX: a candidate tumor suppressor gene involved in
multiple tumor types. Proc Natl Acad Sci USA. 98:11417–11422. 2001.
View Article : Google Scholar
|
|
49
|
Chiang MF, Yeh ST, Liao HF, Chang NS and
Chen YJ: Overexpression of WW domain-containing oxidoreductase WOX1
preferentially induces apoptosis in human glioblastoma cells
harboring mutant p53. Biomed Pharmacother. 66:433–438. 2012.
View Article : Google Scholar
|
|
50
|
Liu YY, Li L, Li DR, Zhang W and Wang Q:
Suppression of WWOX gene by RNA interference reverses platinum
resistance acquired in SKOV3/SB cells. Zhonghua Fu Chan Ke Za Zhi.
43:854–858. 2008.(In Chinese).
|
|
51
|
de Leeuw N1, Dijkhuizen T, Hehir-Kwa JY,
Carter NP, Feuk L, Firth HV, Kuhn RM, Ledbetter DH, Martin CL, van
Ravenswaaij-Arts CM, Scherer SW, Shams S, Van Vooren S, Sijmons R,
Swertz M and Hastings R: Diagnostic interpretation of array data
using public databases and internet sources. Hum Mutat. 33:930–940.
2012.PubMed/NCBI
|
|
52
|
Moitra K, Im K, Limpert K, Borsa A,
Sawitzke J, Robey R, Yuhki N, Savan R, Huang da W, Lempicki RA,
Bates S and Dean M: Differential gene and microRNA expression
between etoposide resistant and etoposide sensitive MCF7 breast
cancer cell lines. PLoS One. 7:e452682012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Boren T, Xiong Y, Hakam A, Wenham R, Apte
S, Chan G, Kamath SG, Chen DT, Dressman H and Lancaster JM:
MicroRNAs and their target messenger RNAs associated with ovarian
cancer response to chemotherapy. Gynecol Oncol. 113:249–255. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ko MA, Zehong G, Virtanen C, Guindi M,
Waddell TK, Keshavjee S and Darling GE: MicroRNA expression
profiling of esophageal cancer before and after induction
chemoradiotherapy. Ann Thorac Surg. 94:1094–1103. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Eitan R, Kushnir M, Lithwick-Yanai G,
David MB, Hoshen M, Glezerman M, Hod M, Sabah G, Rosenwald S and
Levavi H: Tumor microRNA expression patterns associated with
resistance to platinum based chemotherapy and survival in ovarian
cancer patients. Gynecol Oncol. 114:253–259. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jung EJ, Santarpia L, Kim J, Esteva FJ,
Moretti E, Buzdar AU, Di Leo A, Le XF, Bast RC Jr, Park ST, Pusztai
L and Calin GA: Plasma microRNA 210 levels correlate with
sensitivity to trastuzumab and tumor presence in breast cancer
patients. Cancer. 118:2603–2614. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Guo L, Liu Y, Bai Y, Sun Y, Xiao F and Guo
Y: Gene expression profiling of drug-resistant small cell lung
cancer cells by combining microRNA and cDNA expression analysis.
Eur J Cancer. 46:1692–1702. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wu XM, Shao XQ, Meng XX, Zhang XN, Zhu L,
Liu SX, Lin J and Xiao HS: Genome-wide analysis of microRNA and
mRNA expression signatures in hydroxycamptothecin-resistant gastric
cancer cells. Acta Pharmacol Sin. 32:259–269. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sun JY, Huang Y, Li JP, Zhang X, Wang L,
Meng YL, Yan B, Bian YQ, Zhao J, Wang WZ, Yang AG and Zhang R:
MicroRNA-320a suppresses human colon cancer cell proliferation by
directly targeting β-catenin. Biochem Biophys Res Commun.
420:787–792. 2012.PubMed/NCBI
|
|
60
|
Zhang Y, He X, Liu Y, Ye Y, Zhang H, He P,
Zhang Q, Dong L, Liu Y and Dong J: microRNA-320a inhibits tumor
invasion by targeting neuropilin 1 and is associated with liver
metastasis in colorectal cancer. Oncol Rep. 27:685–694. 2012.
|
|
61
|
Zhang H, Li W, Nan F, Ren F, Wang H, Xu Y
and Zhang F: MicroRNA expression profile of colon cancer stem-like
cells in HT29 adenocarcinoma cell line. Biochem Biophys Res Commun.
404:273–278. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yang N, Kaur S, Volinia S, Greshock J,
Lassus H, Hasegawa K, Liang S, Leminen A, Deng S, Smith L,
Johnstone CN, Chen XM, Liu CG, Huang Q, Katsaros D, Calin GA, Weber
BL, Bützow R, Croce CM, Coukos G and Zhang L: MicroRNA microarray
identifies Let-7i as a novel biomarker and therapeutic
target in human epithelial ovarian cancer. Cancer Res.
68:10307–10314. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hisaoka M, Matsuyama A and Nakamoto M:
Aberrant calreticulin expression is involved in the
dedifferentiation of dedifferentiated liposarcoma. Am J Pathol.
180:2076–2083. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li M, Balch C, Montgomery JS, Jeong M,
Chung JH, Yan P, Huang TH, Kim S and Nephew KP: Integrated analysis
of DNA methylation and gene expression reveals specific signaling
pathways associated with platinum resistance in ovarian cancer. BMC
Med Genomics. 2:342009. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang Y, Niu XL, Qu Y, Wu J, Zhu YQ, Sun WJ
and Li LZ: Autocrine production of interleukin-6 confers cisplatin
and paclitaxel resistance in ovarian cancer cells. Cancer Lett.
295:110–123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wang Y, Qu Y, Niu XL, Sun WJ, Zhang XL and
Li LZ: Autocrine production of interleukin-8 confers cisplatin and
paclitaxel resistance in ovarian cancer cells. Cytokine.
56:365–375. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lian L, Qu LJ, Sun HY, Chen YM, Lamont SJ,
Liu CJ and Yang N: Gene expression analysis of host spleen
responses to Marek’s disease virus infection at late tumor
transformation phase. Poult Sci. 91:2130–2138. 2012.
|
|
68
|
Chang MY, Yu YP, Tsai JR, Sheu CC, Chong
IW and Lin SR: Combined oligonucleotide microarray-bioinformatics
and constructed membrane arrays to analyze the biological pathways
in the carcinogenesis of human lung adenocarcinoma. Oncol Rep.
18:569–579. 2007.
|
|
69
|
Morales M, Planet E, Arnal-Estape A,
Pavlovic M, Tarragona M and Gomis RR: Tumor-stroma interactions a
trademark for metastasis. Breast. 20(Suppl 3): S50–S55. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Che CL, Zhang YM, Zhang HH, Sang YL, Lu B,
Dong FS, Zhang LJ and Lv FZ: DNA microarray reveals different
pathways responding to paclitaxel and docetaxel in non-small cell
lung cancer cell line. Int J Clin Exp Pathol. 6:1538–1548.
2013.PubMed/NCBI
|
|
71
|
Wang JM, Wu JT, Sun DK, Zhang P and Wang
L: Pathway crosstalk analysis based on protein-protein network
analysis in prostate cancer. Eur Rev Med Pharmacol Sci.
16:1235–1242. 2012.PubMed/NCBI
|
|
72
|
Wilson C, Purcell C, Seaton A, Oladipo O,
Maxwell PJ, O’Sullivan JM, Wilson RH, Johnston PG and Waugh DJ:
Chemotherapy-induced CXC-chemokine/CXC-chemokine receptor signaling
in metastatic prostate cancer cells confers resistance to
oxaliplatin through potentiation of nuclear factor-κB transcription
and evasion of apoptosis. J Pharmacol Exp Ther. 327:746–759.
2008.PubMed/NCBI
|
|
73
|
He D, Xu Q, Yan M, Zhang P, Zhou X, Zhang
Z, Duan W, Zhong L, Ye D and Chen W: The NF-kappaB inhibitor,
celastrol, could enhance the anti-cancer effect of gambogic acid on
oral squamous cell carcinoma. BMC Cancer. 9:3432009. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Choi BH, Kim CG, Lim Y, Shin SY and Lee
YH: Curcumin down-regulates the multidrug-resistance mdr1b
gene by inhibiting the PI3K/Akt/NFκB pathway. Cancer Lett.
259:111–118. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Godwin P, Baird AM, Heavey S, Barr MP,
O’Byrne KJ and Gately K: Targeting nuclear factor-kappa B to
overcome resistance to chemotherapy. Front Oncol. 3:1202013.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yang G, Xiao X, Rosen DG, Cheng X, Wu X,
Chang B, Liu G, Xue F, Mercado-Uribe I, Chiao P, Du X and Liu J:
The biphasic role of NF-κB in progression and chemoresistance of
ovarian cancer. Clin Cancer Res. 17:2181–2194. 2011.
|
|
77
|
Lee SC, Xu X, Lim YW, Iau P, Sukri N, Lim
SE, Yap HL, Yeo WL, Tan P, Tan SH, McLeod H and Goh BC:
Chemotherapy-induced tumor gene expression changes in human breast
cancers. Pharmacogenet Genomics. 19:181–192. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Arafa el-SA, Zhu Q, Barakat BM, Wani G,
Zhao Q, El-Mahdy MA and Wani AA: Tangeretin sensitizes
cisplatin-resistant human ovarian cancer cells through
downregulation of phosphoinositide 3-kinase/Akt signaling pathway.
Cancer Res. 69:8910–8917. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Li QQ, Yunmbam MK, Zhong X, Yu JJ,
Mimnaugh EG, Neckers L and Reed E: Lactacystin enhances cisplatin
sensitivity in resistant human ovarian cancer cell lines via
inhibition of DNA repair and ERCC-1 expression. Cell Mol Biol.
47:OL61–OL72. 2001.PubMed/NCBI
|
|
80
|
Menendez JA, Vellon L, Mehmi I, Teng PK,
Griggs DW and Lupu R: A novel CYR61-triggered
‘CYR61-αvβ3 integrin loop’
regulates breast cancer cell survival and chemosensitivity through
activation of ERK1/ERK2 MAPK signaling pathway. Oncogene.
24:761–779. 2005.PubMed/NCBI
|
|
81
|
Galan-Moya EM, de la Cruz-Morcillo MA,
Llanos Valero M, Callejas-Valera JL, Melgar-Rojas P, Hernadez Losa
J, Salcedo M, Fernández-Aramburo A, Ramon y Cajal S and
Sánchez-Prieto R: Balance between MKK6 and MKK3 mediates p38 MAPK
associated resistance to cisplatin in NSCLC. PLoS One.
6:e284062011. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang J, Wang Y, Zhen P, Luo X, Zhang C,
Zhou L, Lu Y, Yang Y, Zhang W and Wan J: Genome-wide analysis of
miRNA signature differentially expressed in doxorubicin-resistant
and parental human hepatocellular carcinoma cell lines. PLoS One.
8:e541112013. View Article : Google Scholar
|
|
83
|
Lange TS, Stuckey AR, Robison K, Kim KK,
Singh RK, Raker CA and Brard L: Effect of a vitamin D3
derivative (B3CD) with postulated anti-cancer activity in an
ovarian cancer animal model. Invest New Drugs. 28:543–553.
2010.PubMed/NCBI
|
|
84
|
Zhang P, Ling G, Pan X, Sun J, Zhang T, Pu
X, Yin S and He Z: Novel nanostructured lipid-dextran sulfate
hybrid carriers overcome tumor multidrug resistance of mitoxantrone
hydrochloride. Nanomedicine. 8:185–193. 2012. View Article : Google Scholar
|
|
85
|
Calzolari A, Papucci A, Baroni G, Ficarra
G, Porfirio B, Chiarelli I and Di Lollo S: Epstein-Barr virus
infection and P53 expression in HIV-related oral large B cell
lymphoma. Head Neck. 21:454–460. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Longley DB and Johnston PG: Molecular
mechanisms of drug resistance. J Pathol. 205:275–292. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Yin F, Liu X, Li D, Wang Q, Zhang W and Li
L: Bioinformatic analysis of chemokine (C-C motif) ligand 21 and
SPARC-like protein 1 revealing their associations with drug
resistance in ovarian cancer. Int J Oncol. 42:1305–1316.
2013.PubMed/NCBI
|
|
88
|
Kong KA, Yoon H and Kim MH: Akt1 as a
putative regulator of Hox genes. Gene. 513:287–291. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Sharan R, Ulitsky I and Shamir R:
Network-based prediction of protein function. Mol Syst Biol.
3:882007. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Mostafavi S, Ray D, Warde-Farley D,
Grouios C and Morris Q: GeneMANIA: a real-time multiple association
network integration algorithm for predicting gene function. Genome
Biol. 9(Suppl 1): S42008. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Jenssen TK, Laegreid A, Komorowski J and
Hovig E: A literature network of human genes for high-throughput
analysis of gene expression. Nat Genet. 28:21–28. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Behm-Ansmant I, Rehwinkel J and Izaurralde
E: MicroRNAs silence gene expression by repressing protein
expression and/or by promoting mRNA decay. Cold Spring Harb Symp
Quant Biol. 71:523–530. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Tili E, Michaille JJ, Gandhi V, Plunkett
W, Sampath D and Calin GA: miRNAs and their potential for use
against cancer and other diseases. Future Oncol. 3:521–537. 2007.
View Article : Google Scholar : PubMed/NCBI
|