Introduction
Activating mutations of the epidermal growth factor
receptor (EGFR) gene are characteristic genetic alterations
in non-small lung cancer (NSCLC) patients, particularly in those
with lung adenocarcinomas (1–3).
EGFR exon 19 deletions or the exon 21 L858R mutation account
for more than 90% of all EGFR mutations (4,5). Of
particular importance, these mutations are known as predictors of a
favorable clinical outcome in response to treatment with
EGFR-tyrosine kinase inhibitors (EGFR-TKIs) (1,2).
Although ~80% of NSCLC patients with drug-sensitive EGFR
mutations such as EGFR exon 19 deletions or the exon 21
L858R mutation initially show satisfactory responsiveness to
EGFR-TKI treatment (1,2), acquired resistance to EGFR-TKIs occurs
in most cases (6). Half of all
resistance to EGFR-TKIs is caused by an acquired T790M mutation in
exon 20 of the EGFR gene (7–9). The
T790M mutation has been reported to cause EGFR-TKI resistance by
sterically hindering the binding site of gefitinib and erlotinib,
two first-generation EGFR-TKIs (7),
thereby causing a relative decrease in binding with EGFR-TKIs
(10).
The T790M mutation has been detected in some
patients who have not been treated with EGFR-TKIs (11–16).
Since the incidence of a T790M mutation has been reported to range
from 0.02 to 0.05% in all surgically resected NSCLC patients based
on studies using direct sequencing (2,17), it
is difficult to clarify the association due to the T790M mutation
and clinicopathological factors because of the limitation in the
number of cases. Using highly sensitive assays, the clinical impact
of the minor T790M mutation in NSCLC patients with EGFR-TKI
treatment has been previously investigated (11,14,15).
However, the impact on EGFR-TKI-naive NSCLC patients has not been
adequately investigated (12,13,16).
Recently, various therapies to overcome the T790M mutation are
being developed. These include next generation EGFR-TKIs (18) and combination therapies (19). Minor T790M mutated clones are
enriched by EGFR-TKI treatment (12,16);
thus, early detection of a minor T790M mutation using a highly
sensitive assay may be useful for predicting the cause of
resistance to EGFR-TKIs and for selecting optimum therapeutic
strategies.
In the present study, we determined the T790M
mutational status using high-resolution melting (HRM) analysis
combined with mutant-enriched (12,20)
and co-amplification at lower denaturation temperature
(COLD)-polymerase chain reactions (PCRs) (21–24) to
investigate the relationship between the presence of minor T790M
mutations and clinicopathological factors in EGFR-TKI-naive lung
adenocarcinoma patients with pulmonary resection.
Materials and methods
Clinical samples, cell lines and DNA
extraction
We studied 146 patients with lung adenocarcinomas
who underwent surgical resection without a preoperative history of
EGFR-TKI treatment at our institute between 2006 and 2009. Approval
of the Institutional Review Board and the informed consent of each
patient were obtained. After pulmonary resection, fresh tissue was
immediately frozen and stored at −80°C. DNA was extracted from the
frozen tissue using proteinase K treatment followed by
phenol-chloroform extraction (25).
We also used a human bronchial epithelial cell line (HBEC-5KT)
harboring the wild-type EGFR gene and the NCI-H1975 cell
line (H1975) harboring the EGFR mutations L858R and T790M as
negative and positive controls, respectively. These cell lines were
kindly provided by Dr Adi F. Gazdar (The University of Texas
Southwestern Medical Center at Dallas, Dallas, TX, USA). DNA of the
cell lines was extracted using the DNeasy Blood & Tissue kit
(Qiagen, Hilden, Germany).
Plasmids containing exon 20 of the EGFR
gene
We used a plasmid containing EGFR exon 20
with the T790M mutation, which is one of the standardized plasmids
containing each of the EGFR mutations occurring in the
exons, and was used in a study by Goto et al (26). We also constructed a plasmid
containing wild-type EGFR exon 20 using the TOPO TA Cloning
Kit (Invitrogen, Carlsbad, CA, USA) as previously reported by us
(27). The sequences of each
plasmid were confirmed by direct sequencing.
Detection of EGFR exon 19 deletions and
the L858R mutation
EGFR exon 19 deletions and the exon 21 L858R
mutation were examined using a restriction fragment length
polymorphism (RFLP) assay without the enrichment of mutant alleles,
as previously reported by us (20).
We defined these EGFR exon 19 deletions and the exon 21
L858R mutation as drug-sensitive EGFR mutations.
Detection of the EGFR T790M mutation
Mutant-enriched COLD-HRM (MEC-HRM) is a two-step
PCR-based assay combining a standard HRM assay with a
mutant-enriched PCR (12,20) that enriches the mutant allele by the
intermittent restriction digestion of the wild-type allele and a
COLD-PCR (21) that enriches the
mutant allele by means of the difference in melting temperatures
between the mutant and wild-type alleles. The principle of the
MEC-HRM assay is shown in Fig.
1A.
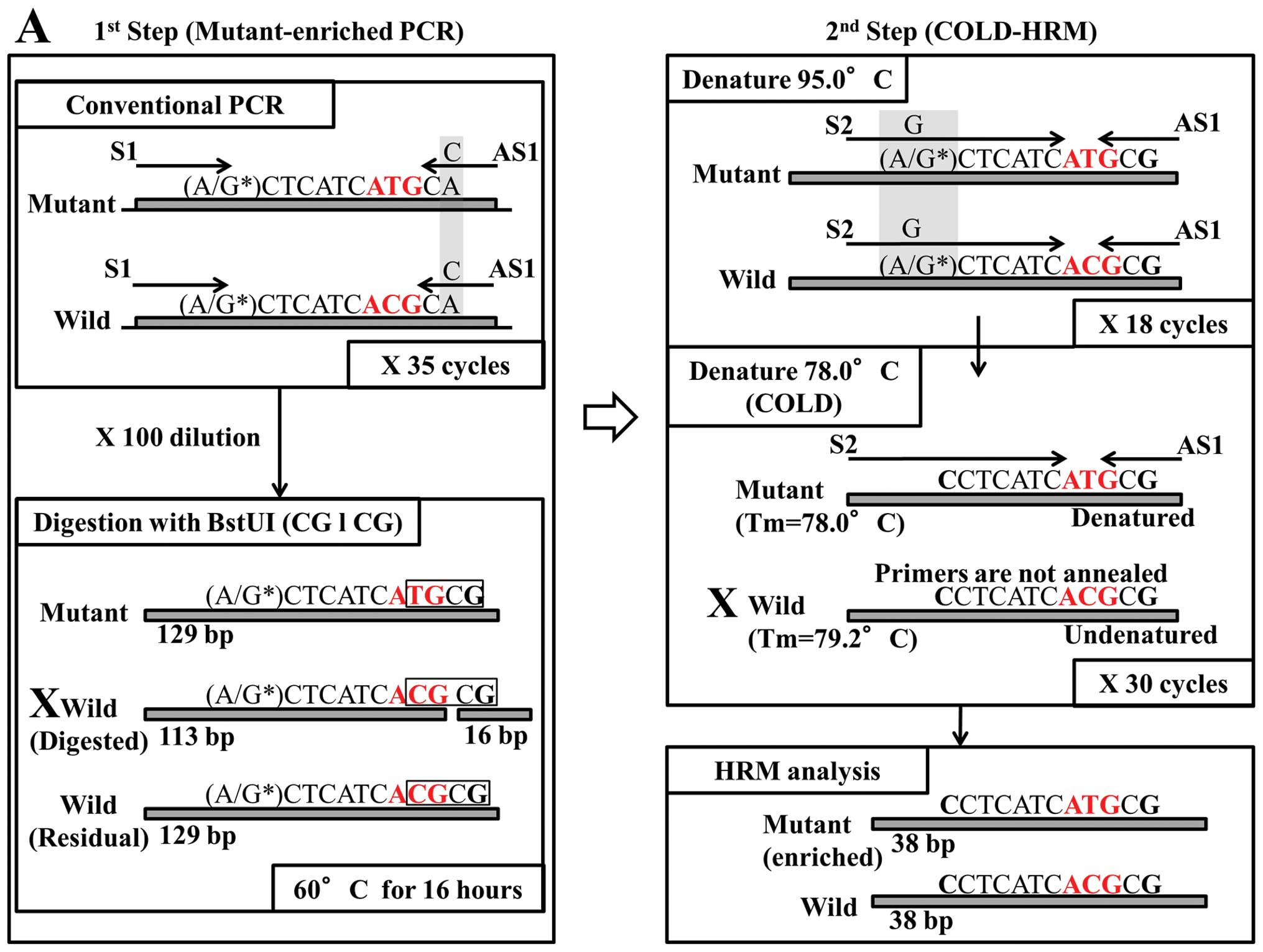 | Figure 1(A) Principle of the mutant-enriched
COLD-HRM (MEC-HRM) assay. In the first step (left box), the upper
box indicates the conventional PCR step for the amplification of
EGFR exon 20, including codon 790. The mismatched site of
the AS1 primer to introduce the CGCG sequence (framed) is
emphasized with light gray shading. The exchanged bases are
emphasized with bold print. An asterisk indicates the SNP site. The
lower box indicates the step for the selective digestion of the
wild-type amplicon. The crosses are applied on reduced amplicons.
After digestion, the product that includes the mutant amplicon and
residual undigested wild-type amplicon is used for the second step.
The upper box indicates the first step of the real-time PCR for the
amplification of the template product. The mismatched site of the
S2 primer to diminish the influence of SNP is emphasized in light
gray shading. The lower box indicates the second step, which is a
COLD-PCR step for the selective amplification of the mutant. The Td
(78.0°C) was lower than Tm of the wild-type amplicon; thus, they
were not denatured and the primers were not annealed. The final
products were analyzed using an HRM analysis. (B) Principle of
standard HRM assay. The box indicates the real-time PCR step using
the S2 primer (forward) and the AS1 primer (reverse). The real-time
PCR conditions are as follows: an initial denaturation step at 95°C
for 10 min, and an amplification step for 55 cycles (95°C for 15
sec, 60°C for 60 sec). The final products are analyzed using HRM
analysis. (C) Principle of COLD-HRM assay. The upper box indicates
the first step of 18 cycles of real-time PCR for the amplification
of the template product, and the lower box indicates the second
step, which is a COLD-PCR step, for the selective amplification of
the mutant allele. In the second step, the denaturing temperature
(Td) is decreased to 78°C and amplification is performed for 30
cycles. The final products are analyzed using HRM analysis. (D)
Principle of mutant-enriched HRM assay. In the first step (left
box), conventional PCR and digestion are performed, similar to that
shown in the left box of (A). The second step (right box) is
performed in the same manner as standard HRM (B) using the products
from the first step. Forty cycles are used for the mutant-enriched
HRM assay, instead of 55 cycles for the standard HRM assay. |
As the first step, conventional PCR was performed to
amplify the target region (129 base pairs), including codon 790 of
the EGFR gene, using the GeneAmp® 9700 thermal
cycler (Applied Biosystems, Foster City, CA, USA). The forward
primer sequence was 5′-ACTGACGTGCCTCTCCCTCC-3′ (S1). The reverse
primer sequence was 5′-CGAAGGGCATGAGCC*GC-3′ (AS1), harboring one
mismatched site (* T to C) to introduce the CGCG sequence into the
amplicon of the wild-type. The final volume of the PCR mixture was
10 μl contained 100 ng of sample DNA, 150 nmol of deoxynucleotide
triphosphate, 2 pmol of each primer, and 1 unit of HotStarTaq DNA
Polymerase Plus (Qiagen). The PCR conditions were as follows: an
initial denaturation step at 95°C for 5 min, followed by 35 cycles
of 94°C for 20 sec, 60°C for 30 sec, and 72°C for 20 sec. Diluted
PCR products (100-fold dilution with distilled water) were treated
with the restriction enzyme BstUI (New England BioLabs,
Ipswich, MA, USA), which digests the wild-type allele but not the
mutant allele, for 16 h at 60°C, resulting in the enrichment of the
mutant alleles, as previously reported by us (12).
After the first step, the second step (COLD-PCR,
including melting curve analysis) was performed using the
StepOnePlus™ real-time PCR system (Applied Biosystems). The forward
primer sequence was 5′-CCTCCACCGTGCAC*CTCATC-3′ (S2). Since one SNP
site (rs1050171 A/G) exists in the S2 primer sequence, we designed
a mismatched-base (* C) at the SNP position to diminish the
influence of the SNP (28). We used
the AS1 primer as a reverse primer. The final volume of the PCR
mixture was 20 μl, containing 10 μl of MeltDoctor™ Master Mix
(Applied Biosystems), 12 pmol of each primer and 1 μl of the
first-step product. The real-time PCR conditions were as follows:
an initial denaturation step at 95°C for 10 min, 18 cycles for the
first round of standard amplification (95°C for 15 sec, 60°C for 60
sec), and 30 cycles for the second round of amplification to enrich
the mutant-amplicons (78°C for 15 sec and 60°C for 60 sec). The
denaturing temperature (Td) of the COLD-PCR step (78°C) was
determined using a melting curve analysis of the standard HRM [the
melting temperatures (Tm) of the mutant-type and wild-type
amplicons were 78°C and 79.2°C, respectively] and an evaluation of
the sensitivity of COLD-PCR from Td 79.2 to 78°C every 0.2°C. After
the amplification step, a melting curve was generated and analyzed
using HRM software ver. 3.0.1 (Applied Biosystems).
We also performed standard-HRM, COLD-HRM and
mutant-enriched HRM assays. The principles of these three assays
are provided in Fig. 1B–D. All the
samples, including the standard DNA mixtures, were analyzed in
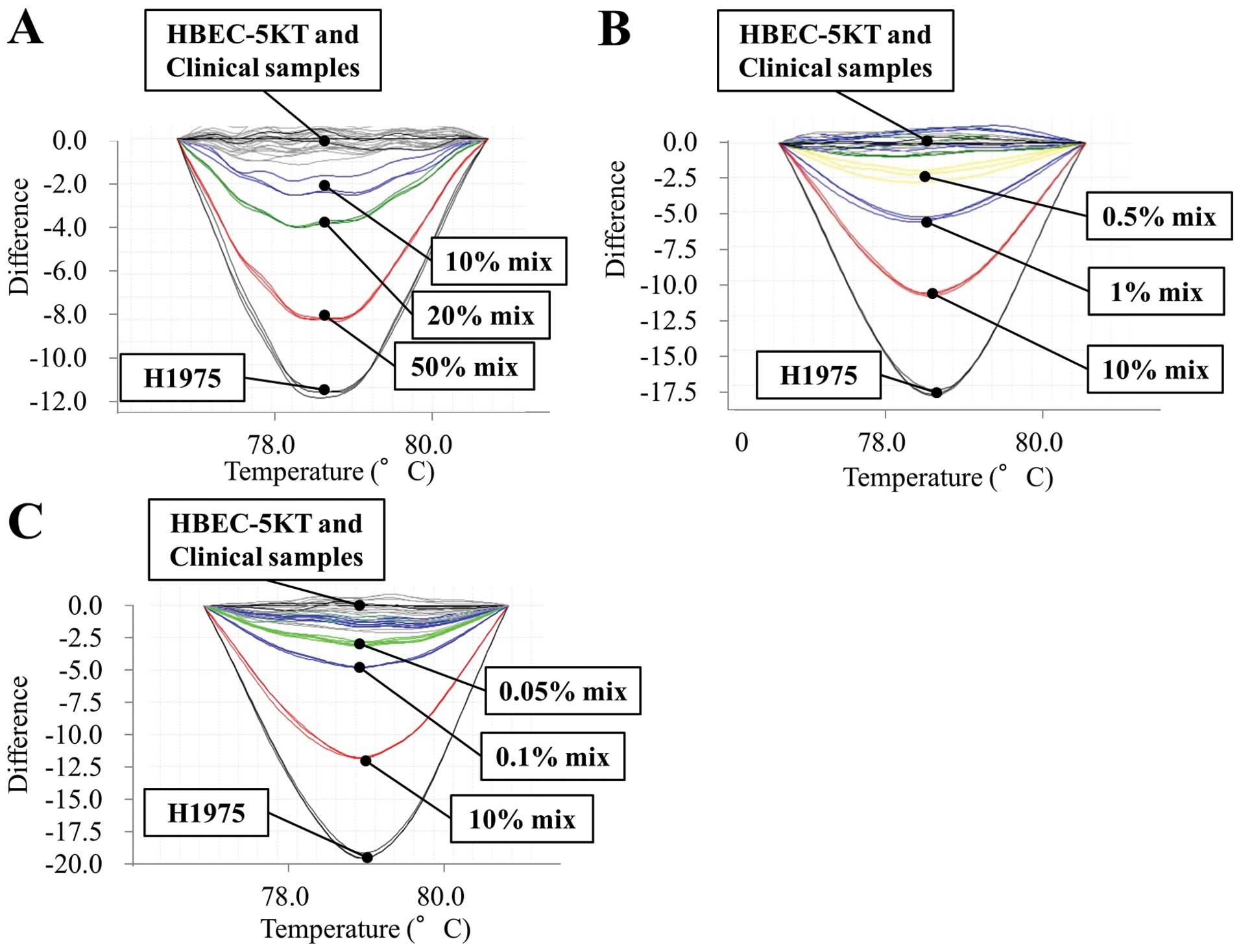
triplicate in all the assays. The sensitivities of all four assays
were determined using multiple DNA mixtures of T790M mutant and
wild-type alleles (0.01, 0.05, 0.1, 1, 10, 20 or 50% of T790M
mutant allele).
Statistical analysis
Chi-square tests or Fisher’s exact tests were used
to examine the differences in categorical factors across groups, as
appropriate. The multivariate logistic regression model was used to
identify clinicopathological factors that might independently
predict the presence of T790M mutations.
After pulmonary resection, imaging studies were
repeated every 3 months for at least 2 years and every 6 months
thereafter for 3 years. After 5 years, medical examinations were
repeated every year. The progression-free survival (PFS) was
calculated from the date of surgery until confirmed disease
recurrence or death. The overall survival (OS) was calculated from
the date of surgery until the date of death or the last follow-up.
The survival curve was calculated using the Kaplan-Meier method,
and the difference between groups was compared using the log-rank
test. Multivariate analyses were performed using the Cox
proportional hazard model. All the data were analyzed using JMP,
version 9.0.0 (SAS Institute Inc., Cary, NC, USA). All the
statistical tests were two-sided, and probability values (P)
<0.05 were considered statistically significant.
Results
Sensitivity of each assay for the
detection of the T790M mutation
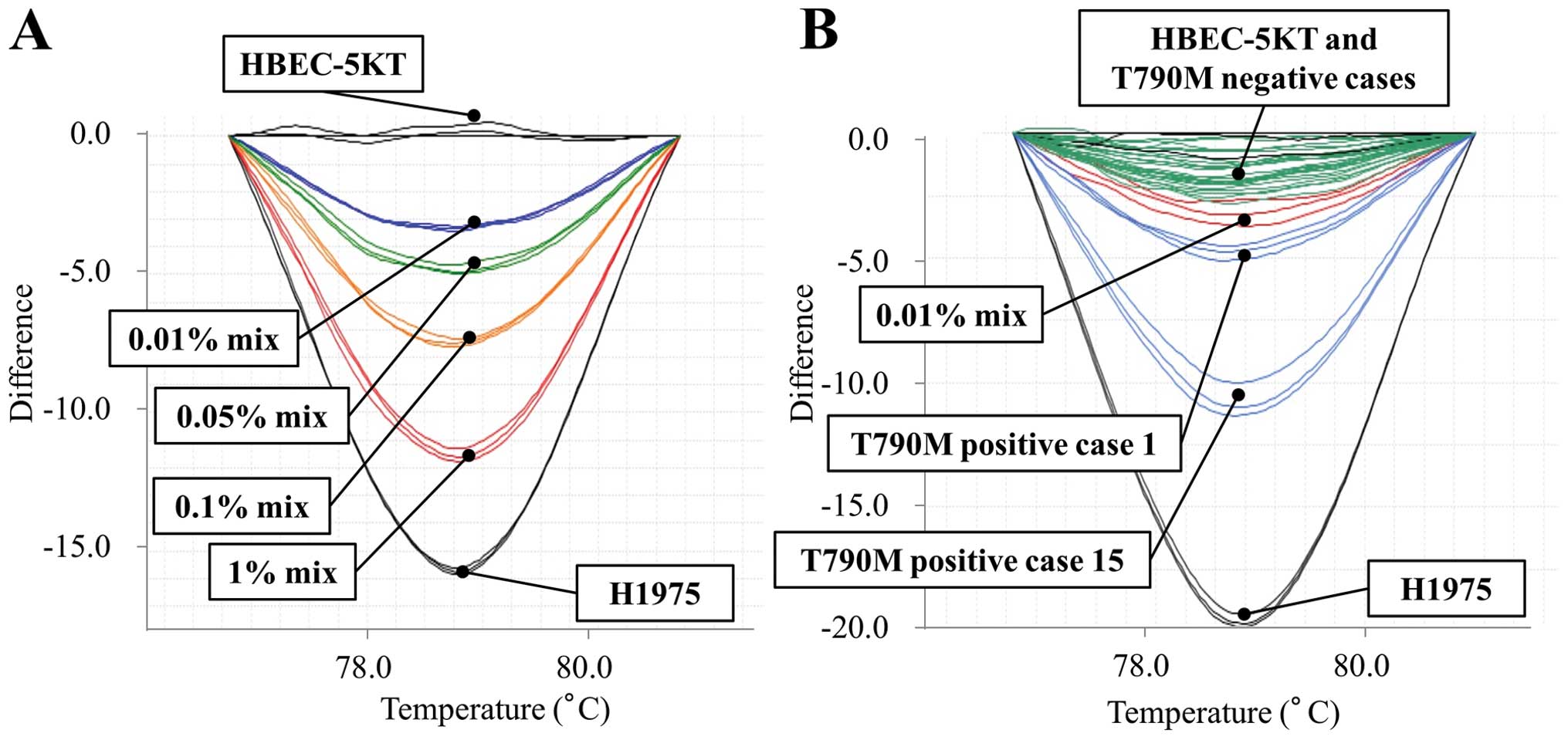
The MEC-HRM assay was able to detect the T790M
mutant allele in the sample with a mutant allele content of 0.01%.
In other words, it detected the mutant allele in the presence of a
10,000-fold background of wild-type allele (Figs. 2 and 3). Meanwhile, the detection limits of
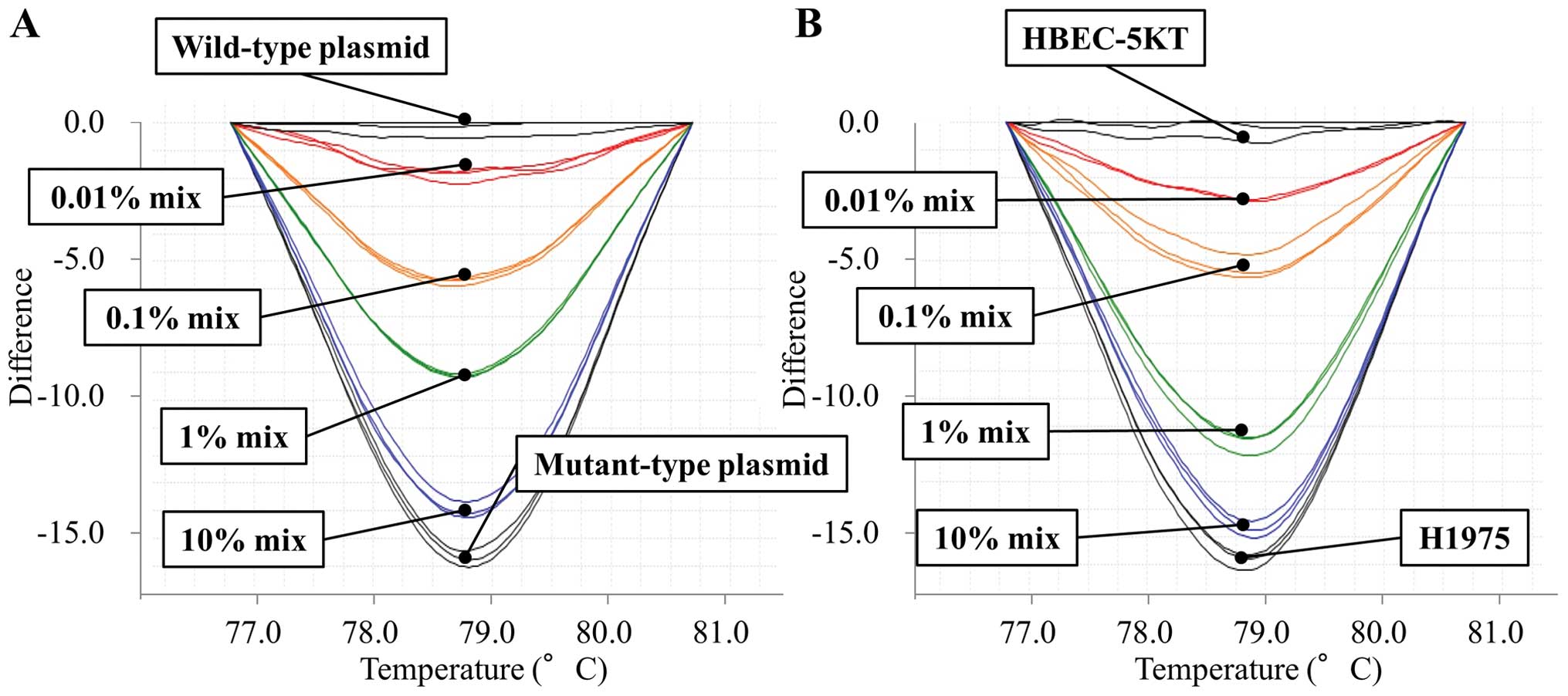
standard HRM, COLD-HRM and mutant-enriched HRM assays were a mutant
allele content of 10, 1 and 0.1%, respectively (Fig. 4). Based on these data, we
investigated the T790M mutation in 146 clinical samples using the
standard HRM using a DNA mixture with T790M content of 10% as a
positive control, and the MEC-HRM assays using a DNA mixture with
T790M content of 0.01% as a positive control. Clinical samples and
positive controls were investigated in the same plate in
triplicate. When all three signals from a clinical sample exceeded
the maximal signal from the control, the sample was identified to
be positive for the T790M mutation (Fig. 2).
EGFR mutations in the clinical
samples
Drug-sensitive EGFR mutations were found in
54 (37%) of the 146 lung adenocarcinomas (26 exon 19 deletions and
28 L858R mutations). Drug-sensitive EGFR mutations were
significantly associated with females (P<0.01) and never-smokers
(P<0.01).
Although standard HRM did not detect any EGFR
T790M mutations, EGFR T790M mutations were detected in 19
(13%) of the 146 lung adenocarcinomas using the MEC-HRM assay.
Thus, these 19 patients were defined as harboring minor T790M
mutations. We confirmed that these 19 tumor samples had a tumor
cell composition of at least 20% by examining the proportion of
tumor cells in the tissue using hematoxylin and eosin staining.
Next, we determined the dosage of the T790M mutant allele in 19
mutant samples by reanalyzing these samples and comparing them with
10, 1, 0.1 and 0.01% standard T790M mutant DNA mixtures. We
subcategorized the 19 minor T790M mutations into groups with T790M
mutant DNA levels corresponding to 0.01–0.1%, 0.1–1% and 1–10%. As
shown in Table I, 17 of the 19
patients (84%) had a T790M mutant allele proportion of
<0.1%.
 | Table IDetails of the 19 T790M-positive
patients. |
Table I
Details of the 19 T790M-positive
patients.
| Case | Age (years) | Gender | Smoking
history | Pathological
stage | Drug-sensitive
EGFR mutation | Proportion of T790M
clones |
|---|
| 1 | 59 | Male | Never-smoker | I | ex19 del. | 0.01–0.1% |
| 2 | 75 | Male | Never-smoker | I | ex19 del. | 0.01–0.1% |
| 3 | 75 | Female | Never-smoker | I | L858R | 0.01–0.1% |
| 4 | 75 | Female | Never-smoker | I | L858R | 0.01–0.1% |
| 5 | 79 | Female | Never-smoker | I | L858R | 0.01–0.1% |
| 6 | 60 | Female | Never-smoker | I | WT | 0.01–0.1% |
| 7 | 60 | Male | Never-smoker | III | WT | 0.01–0.1% |
| 8 | 66 | Male | Smoker | I | ex19 del. | 0.01–0.1% |
| 9 | 57 | Female | Smoker | I | ex19 del. | 0.1–1% |
| 10 | 76 | Male | Smoker | I | ex19 del. | 0.01–0.1% |
| 11 | 82 | Male | Smoker | I | L858R | 0.01–0.1% |
| 12 | 58 | Male | Smoker | I | L858R | 0.01–0.1% |
| 13 | 56 | Male | Smoker | I | WT | 0.01–0.1% |
| 14 | 32 | Female | Smoker | I | WT | 0.1–1% |
| 15 | 59 | Female | Smoker | I | WT | 0.01–0.1% |
| 16 | 78 | Female | Smoker | I | WT | 0.01–0.1% |
| 17 | 68 | Male | Smoker | I | WT | 0.01–0.1% |
| 18 | 74 | Male | Smoker | I | WT | 0.01–0.1% |
| 19 | 84 | Female | Smoker | II | L858R | 1–10% |
The details of the 19 patients harboring minor T790M
mutations and the relationships between a minor T790M mutational
status and clinicopathological factors are shown in Tables I and II, respectively. Minor T790M mutation was
not significantly associated with age, gender, pathological stage,
smoking history or drug-sensitive EGFR mutations in
univariate analyses. However, drug-sensitive EGFR mutations
are considered to be associated with age, gender and smoking status
(27,29,30);
thus, a multivariate analysis was performed to identify independent
factors associated with a minor T790M mutation. Consequently, the
minor T790M mutation was found to be independently associated with
drug-sensitive EGFR mutations (OR, 3.0; 95% CI, 1.0–9.0;
P=0.04) (Table III).
 | Table IIRelationship between minor T790M
mutations and clinicopathological factors. |
Table II
Relationship between minor T790M
mutations and clinicopathological factors.
| Subsets
(n=146) | T790M-positive
(n=19)
n, (%) | T790M-negative
(n=127)
n, (%) | P-value |
|---|
| Age (median; range)
(68; 32–87 years) |
| <68 (n=72) | 9 (47.4) | 63 (49.6) | 0.9 |
| ≥68 (n=74) | 10 (52.6) | 64 (50.4) | |
| Gender (male vs.
female) |
| Male (n=71) | 10 (52.6) | 61 (48.0) | 0.7 |
| Female (n=75) | 9 (47.4) | 66 (52.0) | |
| Smoking
history |
| Smoker (n=74) | 12 (63.2) | 62 (48.8) | 0.2 |
| Never-smoker
(n=72) | 7 (36.8) | 65 (51.2) | |
| Pathological
stage |
| I (n=103) | 17 (89.5) | 86 (67.7) | 0.06 |
| II (n=12) | 1 (5.3) | 11 (8.7) | (I vs. others) |
| III (n=21) | 1 (5.3) | 20 (15.7) | |
| IVa (n=10) | 0 (0.0) | 10 (7.9) | |
| Drug-sensitive
EGFR mutation |
| Mutant (n=54) | 10 (52.6) | 44 (34.6) | 0.1 |
| Wild (n=92) | 9 (47.4) | 83 (65.4) | |
 | Table IIIMultivariate analysis of the minor
T790M mutation-related factors. |
Table III
Multivariate analysis of the minor
T790M mutation-related factors.
| Variables | OR (95% CI) | P-value |
|---|
| Age (years) | | |
| (<68 vs.
≥68) | 1.0 (0.4–2.6) | 1.0 |
| Gender |
| (Male vs.
female) | 0.6 (0.2–2.5) | 0.5 |
| Smoking
history |
| (Smoker vs.
never-smoker) | 3.8 (0.9–18) | 0.08 |
| Drug-sensitive
EGFR mutation |
| (Mutant vs.
wild) | 3.0 (1.0–9.0) | 0.04 |
Regarding the prognostic impact of the minor T790M
mutation, it was not associated with either the OS or the PFS in
our cohort (Table IV).
 | Table IVMultivariate analysis for recurrence
and mortality. |
Table IV
Multivariate analysis for recurrence
and mortality.
| PFS | OS |
|---|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (<68 vs. ≥68
years) | 0.3 (0.1–0.6) | <0.01 | 0.3 (0.1–0.9) | 0.02 |
| Gender (Male vs.
female) | 0.7 (0.2–1.9) | 0.4 | 1.0 (0.3–3.6) | 0.9 |
| Smoking history
(Smoker vs. never-smoker) | 2.3 (0.8–7.3) | 0.1 | 2.1 (0.6–8.5) | 0.3 |
| Pathological stage
(I vs. II–IV) | 0.2 (0.09–0.4) | <0.01 | 0.2 (0.05–0.4) | <0.01 |
| Drug-sensitive
EGFR mutation (Mutant vs. wild) | 1.0 (0.4–2.2) | 1.0 | 0.5 (0.1–1.4) | 0.2 |
| T790M mutation
(Mutant vs. wild) | 0.5 (0.08–1.8) | 0.3 | 1.7 (0.4–5.9) | 0.4 |
Discussion
In the present study, we determined the T790M
mutational status using our newly developed, highly sensitive
assay, the MEC-HRM assay. We previously developed a mutant-enriched
PCR assay (sensitivity, 0.1%) and found that the T790M mutation was
detected in 3.8% of NSCLC patients without a history of treatment
with EGFR-TKIs (12). Su et
al (16) found that the T790M
mutation was present in 25.2% of EGFR-TKI-naive NSCLC patients
using matrix-assisted laser desorption ionization-time of flight
mass spectrometry (MALDI-TOF MS) (sensitivity, 1.5%), and Oh et
al (13) found that the T790M
mutation was present in 8.2% of EGFR-TKI-naive NSCLC patients using
peptide nucleic acid (PNA)-clamping PCR with a melting curve
analysis (sensitivity, 0.01%). The present study revealed that the
MEC-HRM assay could detect the T790M mutation in 13% of
EGFR-TKI-naive adenocarcinomas. Of note, our standard HRM assay
(10% sensitivity) did not detect any T790M-positive specimens, and
we considered all the T790M mutations that were detected using the
MEC-HRM assay to be minor mutations (2,17).
As novel findings, the minor T790M mutation was
significantly associated with drug-sensitive EGFR mutations
in a multivariate analysis. Previous reports have described that
drug-sensitive EGFR mutations and the T790M mutation
generally occur in cis (31–33),
although the statistical relationship between minor T790M mutation
and drug-sensitive EGFR mutations has never been revealed in
clinical samples. Our present study is, to the best of our
knowledge, the first report to confirm that the minor T790M
mutation was significantly associated with drug-sensitive EGFR
mutations in lung adenocarcinomas. To understand this situation,
two issues should be discussed: i) the relationship between
drug-sensitive EGFR mutations and the T790M mutation, and
ii) the presence of the T790M mutation as a minor population.
Regarding the first issue, while the reason for this association is
unknown, lung cancers with germ-line EGFR mutations such as
T790M (31), V843I (34), and R776H (35) are accompanied by additional
EGFR mutations in the development of lung cancer, suggesting
that EGFR mutations themselves cause genetic instability,
thereby predisposing cells to additional mutations within the gene
(34). The second issue can be
explained by the following hypothesis. During the carcinogenic
process of EGFR-mutant-related lung cancer, drug-sensitive
EGFR mutations occur first in the progenitor cells of lung
cancer, followed by the T790M mutation. At this stage, the tumor
consists of a heterogeneous population of drug-sensitive
EGFR mutant cells with or without the T790M mutation.
Previous studies have reported that drug-sensitive EGFR
mutant cells with an additional T790M mutation exhibit an indolent
progression, indicating that the possession of the T790M mutation
is a disadvantage for cell proliferation (36). This fact suggests that cancer cells
with only drug-sensitive EGFR mutations may be dominant in
tumors, in addition to those with T790M and drug-sensitive
EGFR mutations as a minor population.
Our study also suggested that the T790M mutation
tended to be associated with a smoking history in this study.
Drug-sensitive EGFR mutations are known to be frequent in
never-smokers with NSCLCs. However, drug-sensitive EGFR
mutations themselves have not been reported to be associated with a
never-smoking habit (30). Matsuo
et al (30) indicated that
EGFR mutations presumably occur in both smokers and
never-smokers with a similar incidence, but other smoking-related
mutations such as TP53 or KRAS mutations preferentially occur in
smokers resulting in the enrichment of the prevalence of
EGFR mutations in never-smokers. In fact, the average
mutation frequency has been reported to be >10-fold higher in
smokers than in never-smokers (37). Considering these observations, the
EGFR mutation at codon 790, unlike drug-sensitive
EGFR mutations, might be susceptible to tobacco-related
carcinogens.
In conclusion, we revealed an association between
the minor T790M mutation and clinicopathological factors in
surgically resected lung adenocarcinoma patients without a history
of EGFR-TKI treatment using our novel, highly sensitive assay.
Minor T790M mutations are independently associated with
drug-sensitive EGFR mutations.
Acknowledgements
The authors thank Ms. Fumiko Isobe (Department of
Thoracic, Breast and Endocrinological Surgery, Okayama University
Graduate School of Medicine, Dentistry and Pharmaceutical Sciences,
Okayama, Japan) for her technical support. The present study was
supported by a Grant-in Aid for Scientific Research from the
Ministry of Education, Culture, Sports, Science and Technology of
Japan (S.T.).
Abbreviations:
|
HRM
|
high resolution melting
|
|
EGFR
|
epidermal growth factor receptor
|
|
TKI
|
tyrosine kinase inhibitor
|
|
PCR
|
polymerase chain reaction
|
|
RFLP
|
restriction fragment length
polymorphism
|
|
COLD-PCR
|
CO-amplification at lower denaturation
temperature PCR
|
|
MEC-HRM
|
mutant-enriched COLD-HRM
|
References
|
1
|
Mitsudomi T, Morita S, Yatabe Y, et al:
Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer harbouring mutations of the epidermal
growth factor receptor (WJTOG3405): an open label, randomised phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar
|
|
2
|
Sequist LV, Martins RG, Spigel D, et al:
First-line gefitinib in patients with advanced non-small-cell lung
cancer harboring somatic EGFR mutations. J Clin Oncol.
26:2442–2449. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yamamoto H, Toyooka S and Mitsudomi T:
Impact of EGFR mutation analysis in non-small cell lung
cancer. Lung Cancer. 63:315–321. 2009.
|
|
5
|
Kosaka T, Yatabe Y, Endoh H, Kuwano H,
Takahashi T and Mitsudomi T: Mutations of the epidermal growth
factor receptor gene in lung cancer: biological and clinical
implications. Cancer Res. 64:8919–8923. 2004. View Article : Google Scholar
|
|
6
|
Araki T, Yashima H, Shimizu K, et al:
Review of the treatment of non-small cell lung cancer with
gefitinib. Clin Med Insights Oncol. 6:407–421. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kobayashi S, Boggon TJ, Dayaram T, et al:
EGFR mutation and resistance of non-small-cell lung cancer
to gefitinib. N Engl J Med. 352:786–792. 2005. View Article : Google Scholar
|
|
8
|
Sequist LV, Waltman BA, Dias-Santagata D,
et al: Genotypic and histological evolution of lung cancers
acquiring resistance to EGFR inhibitors. Sci Transl Med.
3:75ra262011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Oxnard GR, Arcila ME, Sima CS, et al:
Acquired resistance to EGFR tyrosine kinase inhibitors in
EGFR-mutant lung cancer: distinct natural history of patients with
tumors harboring the T790M mutation. Clin Cancer Res. 17:1616–1622.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eck MJ and Yun CH: Structural and
mechanistic underpinnings of the differential drug sensitivity of
EGFR mutations in non-small cell lung cancer. Biochim Biophys Acta.
1804:559–566. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maheswaran S, Sequist LV, Nagrath S, et
al: Detection of mutations in EGFR in circulating
lung-cancer cells. N Engl J Med. 359:366–377. 2008.
|
|
12
|
Inukai M, Toyooka S, Ito S, et al:
Presence of epidermal growth factor receptor gene T790M mutation as
a minor clone in non-small cell lung cancer. Cancer Res.
66:7854–7858. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oh JE, An CH, Yoo NJ and Lee SH: Detection
of low-level EGFR T790M mutation in lung cancer tissues.
APMIS. 119:403–411. 2011.PubMed/NCBI
|
|
14
|
Fujita Y, Suda K, Kimura H, et al: Highly
sensitive detection of EGFR T790M mutation using colony
hybridization predicts favorable prognosis of patients with lung
cancer harboring activating EGFR mutation. J Thorac Oncol.
7:1640–1644. 2012. View Article : Google Scholar
|
|
15
|
Rosell R, Molina MA, Costa C, et al:
Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in
erlotinib-treated advanced non-small-cell lung cancer patients with
EGFR mutations. Clin Cancer Res. 17:1160–1168. 2011. View Article : Google Scholar
|
|
16
|
Su KY, Chen HY, Li KC, et al: Pretreatment
epidermal growth factor receptor (EGFR) T790M mutation
predicts shorter EGFR tyrosine kinase inhibitor response duration
in patients with non-small-cell lung cancer. J Clin Oncol.
30:433–440. 2012.PubMed/NCBI
|
|
17
|
Toyooka S, Kiura K and Mitsudomi T:
EGFR mutation and response of lung cancer to gefitinib. N
Engl J Med. 352:21362005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sakuma Y, Yamazaki Y, Nakamura Y, et al:
WZ4002, a third-generation EGFR inhibitor, can overcome
anoikis resistance in EGFR-mutant lung adenocarcinomas more
efficiently than Src inhibitors. Lab Invest. 92:371–383.
2012.PubMed/NCBI
|
|
19
|
Regales L, Gong Y, Shen R, et al: Dual
targeting of EGFR can overcome a major drug resistance mutation in
mouse models of EGFR mutant lung cancer. J Clin Invest.
119:3000–3010. 2009.PubMed/NCBI
|
|
20
|
Asano H, Toyooka S, Tokumo M, et al:
Detection of EGFR gene mutation in lung cancer by
mutant-enriched polymerase chain reaction assay. Clin Cancer Res.
12:43–48. 2006.PubMed/NCBI
|
|
21
|
Li J, Wang L, Mamon H, Kulke MH, Berbeco R
and Makrigiorgos GM: Replacing PCR with COLD-PCR enriches variant
DNA sequences and redefines the sensitivity of genetic testing. Nat
Med. 14:579–584. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luthra R and Zuo Z: COLD-PCR finds hot
application in mutation analysis. Clin Chem. 55:2077–2078. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Milbury CA, Li J and Makrigiorgos GM:
COLD-PCR-enhanced high-resolution melting enables rapid and
selective identification of low-level unknown mutations. Clin Chem.
55:2130–2143. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Song C, Milbury CA, Li J, Liu P, Zhao M
and Makrigiorgos GM: Rapid and sensitive detection of KRAS mutation
after fast-COLD-PCR enrichment and high-resolution melting
analysis. Diagn Mol Pathol. 20:81–89. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fan H and Gulley ML: DNA extraction from
fresh or frozen tissues. Methods Mol Med. 49:5–10. 2001.PubMed/NCBI
|
|
26
|
Goto K, Satouchi M, Ishii G, et al: An
evaluation study of EGFR mutation tests utilized for
non-small-cell lung cancer in the diagnostic setting. Ann Oncol.
23:2914–2919. 2012.
|
|
27
|
Tokumo M, Toyooka S, Kiura K, et al: The
relationship between epidermal growth factor receptor mutations and
clinicopathologic features in non-small cell lung cancers. Clin
Cancer Res. 11:1167–1173. 2005.PubMed/NCBI
|
|
28
|
Do H, Krypuy M, Mitchell PL, Fox SB and
Dobrovic A: High resolution melting analysis for rapid and
sensitive EGFR and KRAS mutation detection in
formalin fixed paraffin embedded biopsies. BMC Cancer. 8:1422008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ueno T, Toyooka S, Suda K, et al: Impact
of age on epidermal growth factor receptor mutation in lung cancer.
Lung Cancer. 78:207–211. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Matsuo K, Ito H, Yatabe Y, et al: Risk
factors differ for non-small-cell lung cancers with and without
EGFR mutation: assessment of smoking and sex by a
case-control study in Japanese. Cancer Sci. 98:96–101. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bell DW, Gore I, Okimoto RA, et al:
Inherited susceptibility to lung cancer may be associated with the
T790M drug resistance mutation in EGFR. Nat Genet. 37:1315–1316.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Soh J, Okumura N, Lockwood WW, et al:
Oncogene mutations, copy number gains and mutant allele specific
imbalance (MASI) frequently occur together in tumor cells. PLoS
One. 4:e74642009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pao W, Miller VA, Politi KA, et al:
Acquired resistance of lung adenocarcinomas to gefitinib or
erlotinib is associated with a second mutation in the EGFR kinase
domain. PLoS Med. 2:e732005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ohtsuka K, Ohnishi H, Kurai D, et al:
Familial lung adenocarcinoma caused by the EGFR V843I
germ-line mutation. J Clin Oncol. 29:e191–e192. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
van Noesel J, van der Ven WH, van Os TA,
et al: Activating germline R776H mutation in the epidermal growth
factor receptor associated with lung cancer with squamous
differentiation. J Clin Oncol. 31:e161–e164. 2013.PubMed/NCBI
|
|
36
|
Chmielecki J, Foo J, Oxnard GR, et al:
Optimization of dosing for EGFR-mutant non-small cell lung cancer
with evolutionary cancer modeling. Sci Transl Med. 3:90ra592011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Govindan R, Ding L, Griffith M, et al:
Genomic landscape of non-small cell lung cancer in smokers and
never-smokers. Cell. 150:1121–1134. 2012. View Article : Google Scholar : PubMed/NCBI
|