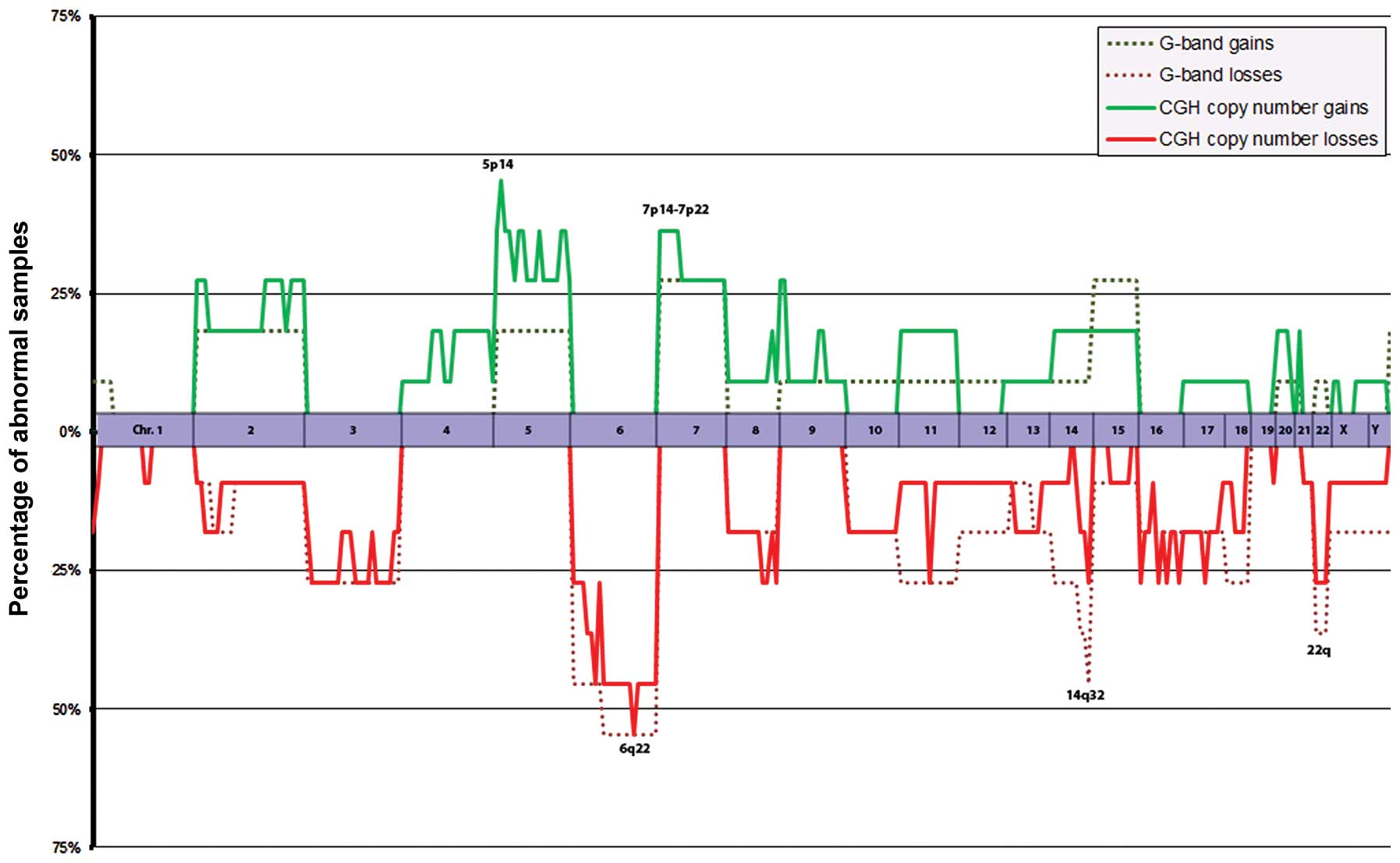
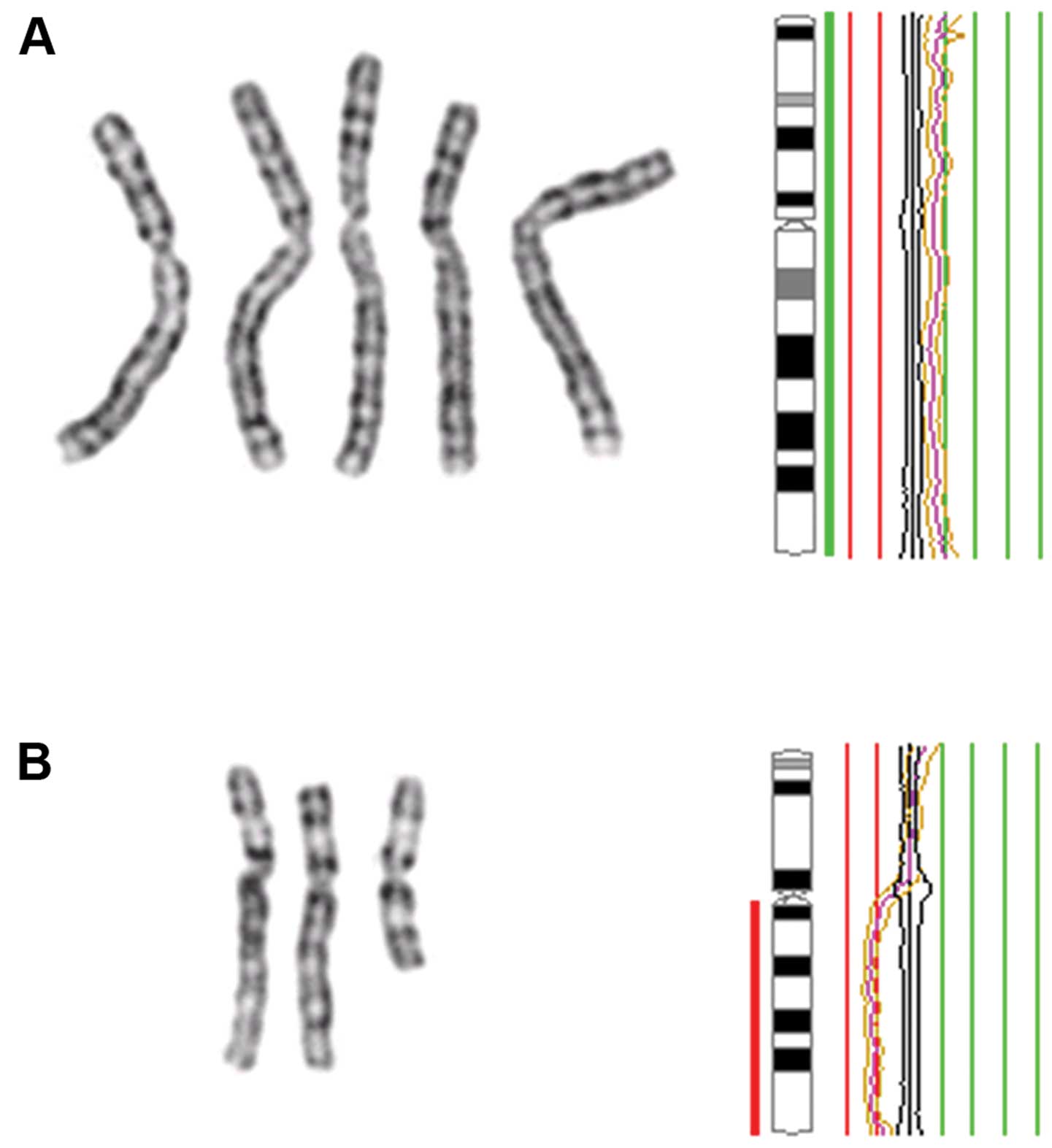
|
1
|
McGuire CS, Sainani KL and Fisher PG:
Incidence patterns for ependymoma: a Surveillance, Epidemiology,
and End Results study. J Neurosurg. 110:725–729. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McLendon RE, Schiffer D, Rosenblum MK,
Wiestler OD, Kros JM, Korshunov A and Ng HK: Ependymal tumours. WHO
Classification of Tumours of the Central Nervous System. 4th
edition. Louis DN, Ohgaki H, Wiestler OD and Cavenee WK:
International Agency for Research on Cancer (IARC); Lyon: pp.
69–80. 2007
|
|
3
|
Reni M, Gatta G, Mazza E and Vecht C:
Ependymoma. Crit Rev Oncol Hematol. 63:81–89. 2007. View Article : Google Scholar
|
|
4
|
Dolecek TA, Propp JM, Stroup NE and
Kruchko C: CBTRUS statistical report: primary brain and central
nervous system tumors diagnosed in the United States in 2005–2009.
Neuro Oncol. 14(Suppl 5): v1–v49. 2012.PubMed/NCBI
|
|
5
|
Sant M, Minicozzi P, Lagorio S, Borge
Johannesen T, Marcos-Gragera R and Francisci S: Survival of
European patients with central nervous system tumors. Int J Cancer.
131:173–185. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gilbert MR, Ruda R and Soffietti R:
Ependymomas in adults. Curr Neurol Neurosci Rep. 10:240–247. 2010.
View Article : Google Scholar
|
|
7
|
Surawicz TS, McCarthy BJ, Kupelian V,
Jukich PJ, Bruner JM and Davis FG: Descriptive epidemiology of
primary brain and CNS tumors: results from the Central Brain Tumor
Registry of the United States 1990–1994. Neuro Oncol. 1:14–25.
1999.PubMed/NCBI
|
|
8
|
Chamberlain MC: Ependymomas. Curr Neurol
Neurosci Rep. 3:193–199. 2003. View Article : Google Scholar
|
|
9
|
Zacharoulis S and Moreno L: Ependymoma: an
update. J Child Neurol. 24:1431–1438. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ruda R, Gilbert M and Soffietti R:
Ependymomas of the adult: molecular biology and treatment. Curr
Opin Neurol. 21:754–761. 2008. View Article : Google Scholar
|
|
11
|
Crocetti E, Trama A, Stiller C, et al:
Epidemiology of glial and non-glial brain tumours in Europe. Eur J
Cancer. 48:1532–1542. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barnholtz-Sloan JS, Sloan AE and Schwartz
AG: Relative survival rates and patterns of diagnosis analyzed by
time period for individuals with primary malignant brain tumor,
1973–1997. J Neurosurg. 99:458–466. 2003.PubMed/NCBI
|
|
13
|
Rodriguez D, Cheung MC, Housri N,
Quinones-Hinojosa A, Camphausen K and Koniaris LG: Outcomes of
malignant CNS ependymomas: an examination of 2408 cases through the
Surveillance, Epidemiology, and End Results (SEER) database
1973–2005. J Surg Res. 156:340–351. 2009.PubMed/NCBI
|
|
14
|
Villano JL, Parker CK and Dolecek TA:
Descriptive epidemiology of ependymal tumours in the United States.
Br J Cancer. 108:2367–2371. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang I, Nagasawa DT, Kim W, Spasic M,
Trang A, Lu DC and Martin NA: Chromosomal anomalies and prognostic
markers for intracranial and spinal ependymomas. J Clin Neurosci.
19:779–785. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Metellus P, Guyotat J, Chinot O, et al:
Adult intracranial WHO grade II ependymomas: long-term outcome and
prognostic factor analysis in a series of 114 patients. Neuro
Oncol. 12:976–984. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Korshunov A, Golanov A, Sycheva R and
Timirgaz V: The histologic grade is a main prognostic factor for
patients with intracranial ependymomas treated in the
microneurosurgical era: an analysis of 258 patients. Cancer.
100:1230–1237. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Reni M, Brandes AA, Vavassori V, et al: A
multicenter study of the prognosis and treatment of adult brain
ependymal tumors. Cancer. 100:1221–1229. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Metellus P, Barrie M, Figarella-Branger D,
et al: Multicentric French study on adult intracranial ependymomas:
prognostic factors analysis and therapeutic considerations from a
cohort of 152 patients. Brain. 130:1338–1349. 2007. View Article : Google Scholar
|
|
20
|
Godfraind C: Classification and
controversies in pathology of ependymomas. Childs Nerv Syst.
25:1185–1193. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Massimino M, Buttarelli FR, Antonelli M,
Gandola L, Modena P and Giangaspero F: Intracranial ependymoma:
factors affecting outcome. Future Oncol. 5:207–216. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Andreiuolo F, Ferreira C, Puget S and
Grill J: Current and evolving knowledge of prognostic factors for
pediatric ependymomas. Future Oncol. 9:183–191. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kilday JP, Rahman R, Dyer S, Ridley L,
Lowe J, Coyle B and Grundy R: Pediatric ependymoma: biological
perspectives. Mol Cancer Res. 7:765–786. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mitelman F, Johansson B and Mertens F:
Mitelman Database of Chromosome Aberrations and Gene Fusions in
Cancer. Available at: http://cgap.nci.nih.gov/Chromosomes/Mitelman.
Accessed May 13, 2013
|
|
25
|
Mazewski C, Soukup S, Ballard E, Gotwals B
and Lampkin B: Karyotype studies in 18 ependymomas with literature
review of 107 cases. Cancer Genet Cytogenet. 113:1–8. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Heim S and Mitelman F: Cancer
Cytogenetics. 3rd edition. John Wiley & Sons; Hoboken, NJ:
2009
|
|
27
|
Mack SC and Taylor MD: The genetic and
epigenetic basis of ependymoma. Childs Nerv Syst. 25:1195–1201.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mandahl N: Methods in solid tumor
cytogenetics. Human Cytogenetics - Malignancy and Acquired
Abnormalities. 3rd edition. Rooney DE: Oxford University Press;
Oxford; pp. 165–203. 2001
|
|
29
|
Shaffer LG, Slovak ML and Campbell LJ:
ISCN: An International System for Human Cytogenetic Nomenclature.
Karger; Basel: 2009
|
|
30
|
Kallioniemi OP, Kallioniemi A, Piper J,
Isola J, Waldman FM, Gray JW and Pinkel D: Optimizing comparative
genomic hybridization for analysis of DNA sequence copy number
changes in solid tumors. Genes Chromosomes Cancer. 10:231–243.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kirchhoff M, Gerdes T, Maahr J, Rose H,
Bentz M, Dohner H and Lundsteen C: Deletions below 10 megabasepairs
are detected in comparative genomic hybridization by standard
reference intervals. Genes Chromosomes Cancer. 25:410–413. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kirchhoff M, Gerdes T, Rose H, Maahr J,
Ottesen AM and Lundsteen C: Detection of chromosomal gains and
losses in comparative genomic hybridization analysis based on
standard reference intervals. Cytometry. 31:163–173. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kraggerud SM, Szymanska J, Abeler VM, et
al: DNA copy number changes in malignant ovarian germ cell tumors.
Cancer Res. 60:3025–3030. 2000.PubMed/NCBI
|
|
34
|
Ribeiro FR, Jeronimo C, Henrique R,
Fonseca D, Oliveira J, Lothe RA and Teixeira MR: 8q gain is an
independent predictor of poor survival in diagnostic needle
biopsies from prostate cancer suspects. Clin Cancer Res.
12:3961–3970. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dahlback HS, Brandal P, Krossnes BK, et
al: Multiple chromosomal monosomies are characteristic of giant
cell ependymoma. Hum Pathol. 42:2042–2046. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hirose Y, Aldape K, Bollen A, et al:
Chromosomal abnormalities subdivide ependymal tumors into
clinically relevant groups. Am J Pathol. 158:1137–1143. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang B, Starostik P, Schraut H, Krauss J,
Sorensen N and Roggendorf W: Human ependymomas reveal frequent
deletions on chromosomes 6 and 9. Acta Neuropathol. 106:357–362.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Korshunov A, Witt H, Hielscher T, et al:
Molecular staging of intracranial ependymoma in children and
adults. J Clin Oncol. 28:3182–3190. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Reardon DA, Entrekin RE, Sublett J, et al:
Chromosome arm 6q loss is the most common recurrent autosomal
alteration detected in primary pediatric ependymoma. Genes
Chromosomes Cancer. 24:230–237. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zeller C, Hinzmann B, Seitz S, et al:
SASH1: a candidate tumor suppressor gene on chromosome 6q24.3 is
downregulated in breast cancer. Oncogene. 22:2972–2983. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Rimkus C, Martini M, Friederichs J, et al:
Prognostic significance of downregulated expression of the
candidate tumour suppressor gene SASH1 in colon cancer. Br J
Cancer. 95:1419–1423. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Suarez-Merino B, Hubank M, Revesz T, et
al: Microarray analysis of pediatric ependymoma identifies a
cluster of 112 candidate genes including four transcripts at
22q12.1–q13.3. Neuro Oncol. 7:20–31. 2005.PubMed/NCBI
|
|
43
|
Korshunov A, Neben K, Wrobel G, et al:
Gene expression patterns in ependymomas correlate with tumor
location, grade, and patient age. Am J Pathol. 163:1721–1727. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
COSMIC: Catalogue of somatic mutations in
cancer - Cancer gene census. Available at: http://cancer.sanger.ac.uk/cancergenome/projects/census/.
Accessed August 20, 2013
|
|
45
|
Monoranu CM, Huang B, Zangen IL, et al:
Correlation between 6q25.3 deletion status and survival in
pediatric intracranial ependymomas. Cancer Genet Cytogenet.
182:18–26. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rajaram V, Gutmann DH, Prasad SK, Mansur
DB and Perry A: Alterations of protein 4.1 family members in
ependymomas: a study of 84 cases. Mod Pathol. 18:991–997. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Witt H, Korshunov A, Pfister SM and Milde
T: Molecular approaches to ependymoma: the next step(s). Curr Opin
Neurol. 25:745–750. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Puget S, Grill J, Valent A, et al:
Candidate genes on chromosome 9q33–34 involved in the progression
of childhood ependymomas. J Clin Oncol. 27:1884–1892. 2009.
|
|
49
|
Dahlback HS, Brandal P, Meling TR,
Gorunova L, Scheie D and Heim S: Genomic aberrations in 80 cases of
primary glioblastoma multiforme: Pathogenetic heterogeneity and
putative cytogenetic pathways. Genes Chromosomes Cancer.
48:908–924. 2009. View Article : Google Scholar
|