Introduction
Ovarian cancer is the most common cause of mortality
from a gynecologic malignancy in the world, posing a serious risk
to women’s health. A previous report showed that, 28,082 women were
diagnosed with epithelial ovarian cancer (EOC) from 1988 to 2001.
The largest histology subgroup, 49.3% (13,835) of patients, had
ovarian papillary serous carcinoma (OPSC) (1). Due to a lack of effective biomarkers
as screening tests and the absence of symptoms at the early stage
of this disease, more than two thirds of women with ovarian cancer
have advanced disease (stage III or IV) at the time of diagnosis.
Platinum (cisplatin or carboplatin) has been used as the first-line
chemotherapeutic drug for ovarian cancer patients since its
introduction in the mid-1970s (2),
and platinum-based chemotherapy has been the standard mainstay
treatment for advanced EOC (2);
however, successful long-term treatment is prevented by the
development of drug resistance (3).
Moreover, empirically-based treatment strategies generally result
in many chemotherapy-resistant patients receiving significant
toxicities during multiple cycles of therapy. Therefore, it is
clinically important to identify biomarkers that may assist in
detecting and predicting which patients with ovarian cancer will
respond to platinum-based chemotherapy and which patients will
remain refractory to this standard treatment.
MicroRNAs (miRNAs) are short non-coding RNAs that
act as post-transcriptional regulators; they can effectively
silence target genes by binding to the 3′-untranslated region
(3′-UTR) of target mRNAs, causing mRNA degradation or inhibiting
translation (4). Increasing studies
have indicated that miRNAs are not only involved in the development
and progression of human cancers (5,6), but
they also play a vital role in tumor cell response to
chemotherapeutic agents by acting as oncogenes and tumor
suppressors (7). From the advances
in the use of the miRNA microarray technique, which can provide
gene phenotyping that identifies distinct classifications exceeding
traditional histopathologic methods (8–11),
another promising approach has emerged, the use of miRNAs encoded
by the human genome as diagnostic tools to predict drug
response.
In the present study, we applied this approach to
identify miR-224-5p expression patterns strongly associated with
the response to primary platinum-based chemotherapy. To understand
the mechanism of miR-224-5p in regulating chemoresistance, we chose
TargetScan, MicroCosm Targets and miRNA Target Visualization to
find the possible target genes. After finding the common target
genes from them, only the top significantly predicted targets were
obtained, and PRKCD, whose gene product plays a role in both
regulating chemoresistance and promoting apoptosis (4), became a possible target gene of
miR-224-5p.
The purpose of the present study was to validate our
microarray results and then couple this analysis with biofunctional
variations that reflect and identify the regulations of various
oncogenic-miRNA signaling pathways to identify a unique mechanism
of platinum resistance that can guide the use of chemotherapy drugs
in platinum-resistant EOC patients.
Materials and methods
Patients and samples
Patients who were surgically treated for ovarian
cancer at the Obstetrics and Gynecology Hospital, Dalian, China,
between July 2004 and November 2010, were identified. We evaluated
a total of 41 OPSC samples that were resected at the time of
primary surgery from patients who went on to receive platinum-based
chemotherapy. The clinicopathological characteristics of the
patients who contributed the ovarian cancer samples are listed in
Table I. All pathological specimens
were reviewed by two independent pathologists with no knowledge of
patients’ clinical data. The diagnosis of the cases was based on
criteria of the International Federation of Gynecology and
Obstetrics (FIGO) staging system.
 | Table IClinicopathological characteristics of
ovarian cancer patients. |
Table I
Clinicopathological characteristics of
ovarian cancer patients.
| Characteristics | Clinical complete
responders (n=22) | Clinical incomplete
responders (n=19) |
|---|
| Mean age, years | 52.6 | 49.3 |
| Stage |
| I | 1 | 2 |
| II | 3 | 2 |
| III | 18 | 12 |
| IV | 0 | 3 |
| Grade | | |
| 1 | 2 | 1 |
| 2 | 12 | 10 |
| 3 | 8 | 8 |
| Mean serum CA-125,
m/ml |
| Before platinum | 851.3 | 1638.5 |
| After platinum | 15.2 | 316.7 |
| Mean survival time,
months | 50.8 | 32.8a |
| Mortality rate
(%) | 31.8 | 57.9a |
Therapeutic response was evaluated as previously
reported (12) from the medical
record by a single gynecologic oncologist using standard criteria
for patients with measurable disease, based on WHO guidelines
(12,13). A complete response (CR) was
classified as a complete disappearance of all measurable and
assessable disease or, in the absence of measurable lesions, a
normalization of CA-125 levels following platinum-based therapy. An
incomplete response (IR) described patients who exhibited only a
partial response, had no response, or progressed during primary
therapy (12). CA-125 response
criteria were based on established guidelines and were used only in
the cases with the absence of a measurable lesion (12,14).
The experiments were performed with two different
groups of human ovarian cancer cell lines, each group including one
cisplatin-sensitive parental cell line (OV2008 and A2780S) and its
cisplatin-resistant variant (C13 and A2780CP), respectively).
Cell culture, transfection and
treatment
Ovarian cancer cells (A2780CP/A2780S and C13/OV2008)
were cultured in RPMI-1640 medium (Invitrogen, Burlington, ON,
Canada) supplemented with 10% fetal bovine serum (FBS) and
maintained at 37°C, 5% CO2. All tissue culture reagents
were obtained from Sigma-Aldrich (St. Louis, MO, USA). The
miR-224-5p mimics and inhibitors were designed and chemically
synthesized by Ambion (cat. no. 4427975; Life Technologies
Corporation, Denmark).
Lipofectamine 2000 (Invitrogen) was incubated with
pre-miR-224-5p (miRNA mimic), anti-miR-224-5p (inhibitor) or their
scrambled negative controls (Ambion; mock) at a concentration of 90
nmol/l and incubated in serum-free RPMI-1640 for 20 min before
being added to OV2008/A2780S or C13/A2780CP cells, respectively.
Cells were incubated at 37°C for 4 h before 10% FBS was replaced.
Protein and RNA were harvested 48 h after transfection. For
cisplatin treatment, cells were maintained in medium with the
desired doses of cisplatin (cat. no. P4394; Sigma, USA).
miRNA microarray and data analysis
A microarray platform optimized for the analysis of
a panel of 768 human miRNAs was used to analyze and compare the
patterns of miRNA expression between CR and IR (n=4 for all) to
platinum-based chemotherapy in OPSC patients. Total RNA that was
enriched for miRNAs was extracted from the FFPE tissue by using the
Ambion mirVana microRNA Isolation kit (Ambion, USA). The quality of
total RNA was assessed using the Agilent Bioanalyzer (Agilent
Technologies, Santa Clara, CA, USA). Individual quantitative
real-time polymerase chain reaction (qRT-PCR) assays were formatted
into a TaqMan low-density array (TLDA; Applied Biosystems), which
was performed at the Shannon McCormack Advanced Molecular
Diagnostics Laboratory Research Services, the Dana Farber Cancer
Institute, the Harvard Clinic and Translational Science Center. The
normalized microarray data were managed and analyzed by StatMiner
version 3.0 (Integromics™).
RNA isolation and qRT-PCR
Total RNA was prepared using TRIzol reagent,
following the manufacturer’s instructions. qRT-PCR was performed
using the TaqMan MicroRNA Reverse Transcription kit (Applied
Biosystems, Foster City CA, USA) with ABI miRNA specific primers
and primer kits on an Agilent Technologies Stratagene Mx3000P
(USA). Specific kits used were: hsa-miR-224-5p; ABI no. 4427975.
The products were detected with SYBR-Green I, and their relative
miRNA or mRNA levels were calculated using the comparative cycle
threshold [Ct, 2−ΔΔCt] method with U6 and GAPDH as the
endogenous controls, respectively. Samples from at least three
independent experiments, each measured in duplicate, were analyzed
and the data are expressed as the averages ± SD.
MTS cell viability assay
The cytotoxic effect on cell survival was quantified
using the CellTiter 96® AQueous
Non-Radioactive Cell Proliferation Assay kit (cat. no. P9625;
Promega Co., USA). Briefly, cells were cultured in 96-well plates
at a density of 1×103/well for 48 h after transfection,
and then treated with 0, 5, 10, 15, 20, 30, 40, 50, 60, 70, 85 or
100 μmol/l cisplatin (cat. no. P4394; Sigma) for 96 h. MTS/PMS
solution composed of 20 μl of two reagents, a novel tetrazolium
compound (MTS) and an electron coupling reagent (PMS), was added
into one 96-well plate at a volume of 100 μl with cultured cells,
and further incubated at 37°C in a humidified, 5% CO2
atmosphere for 1 h. The absorbance at 490 nm was recorded using an
ELISA plate reader (PowerWavex 340; Bio-Tek Instruments Inc.,
Winooski, VT, USA).
Cellular apoptosis assay by assessing the
activation of caspase-3 and -7, and with TUNEL
Ovarian cancer cells were initially seeded at a
concentration of 1×105 cells/ml, in 6-well plates, and
incubated at 37°C in a humidified atmosphere with 5% CO2
for 48 h after transfection, then treated with 20 μmol/l cisplatin
for C13 and OV2008, respectively. After exposure to the drugs for
48 h, cell apoptosis was first determined by assessing the
activation of caspase-3 and -7 using the Caspase-Glo 3/7 Assay kit
(Promega™ Co., Shanghai, China). After the cells were incubated in
normal medium containing the Caspase-Glo reagent for 1 h, caspase-3
and -7 activities were detected with a luminometer. At the same
time, the nuclear morphology of apoptotic cells was determined
using the TUNEL In Situ Apoptosis Detection kit (KeyGen Biotech
Inc., China), following the manufacturer’s instructions. The
apoptotic cells (brown staining) were counted under a
microscope.
miRNA target prediction and pathway
analysis
In order to understand the mechanism of miR-224-5p
in regulating chemoresistance, several computational approaches
were used to analyze target prediction of miRNAs, including miRDB
(http://mirdb.org/miRDB/), MicroCosm Targets
(http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/)
and miRNA Target Visualization (https://cm.jefferson.edu/rna22v1.0-homo_sapiens/GetInputs.jsp).
Functional analysis of these predicted targets was performed to
identify biologic pathways, according to significant gene
expression. The target prediction algorithm used here is estimated
to have a 20–30% false positive rate. This level of false discovery
is unlikely to affect the overall network findings obviously,
although the number of top predicted gene targets is large.
Furthermore, functional analysis of these predicted gene targets
may assist in identifying biologic pathways with significant
involvement for gene expression.
Western blot analysis
Western blotting was performed as follows: harvested
cells were lysed in lysis buffer (Beyotime Institute of
Biotechnology, Shanghai, China) and the proteins (20 μg) were
separated on 10% SDS-PAGE gels and transferred to nitrocellulose
membranes. Membranes were blocked in PBS containing 0.05% Tween-20
(TBST)-5% non-fat milk. Then, the membrane was incubated with
antibodies for PRKCD or GAPDH. After secondary antibody incubation,
the signal analysis was performed by exposure of the blots to
films, which were then scanned and band intensities were measured
with Labworks-Analyst (GeneCo) software.
Statistical analysis
The results were analyzed by SPSS 17.0 (Chicago, IL,
USA). The data are expressed as arithmetic mean ± SD of the number
(n) of experiments. Samples were analyzed with repeated measures
analysis of variance, and differences in the incidences were
analyzed using ANOVA. Statistical analysis of percent values was
performed by the Pearson’s χ2 test. The overall survival
duration was defined as the interval (in months) between the date
of initial cytoreductive surgery to date of last follow-up or
death. The survival time courses were studied using the
Kaplan-Meier method, and groups were compared using the log-rank
test; p<0.05 was considered to indicate a statistically
significant difference.
Results
Patient characteristics
The clinicopathological characteristics of the
patients who contributed the OPSC samples are listed in Table I. Forty-one patients were identified
matching the study criteria. Twenty-two OPSC patient samples
demonstrated a CR, and 19 showed an IR to primary platinum-based
therapy following the surgery. FFPE blocks were obtained after
surgery.
miRNA microarray results and qRT-PCR
validation
To further characterize the unique miRNAs in OPSC
differentiation, specimens from the CR and IR OPSC patients were
initially analyzed by miRNA microarray, respectively. Of the 768
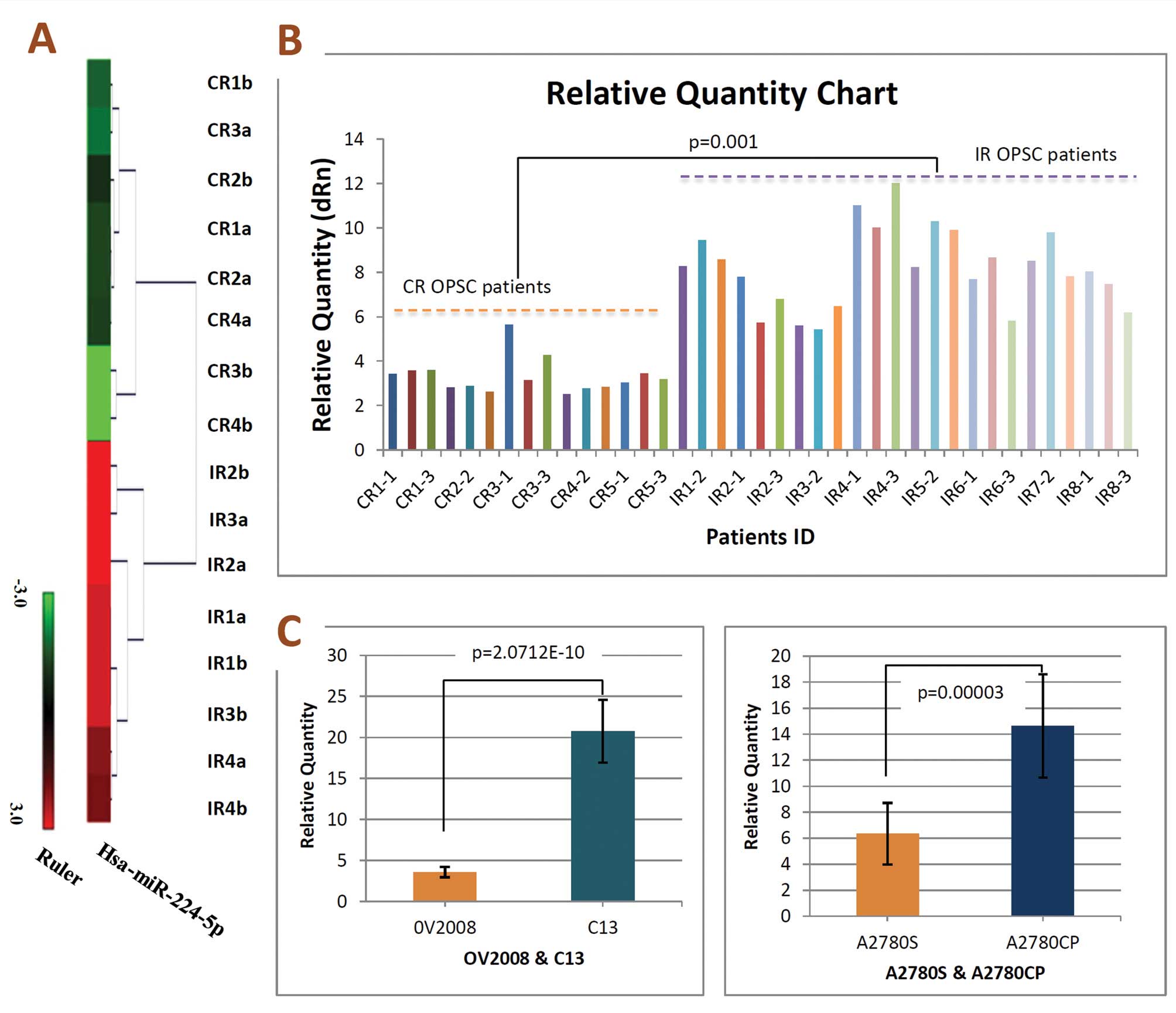
miRNAs analyzed by microarray, we identified miR-224-5p expression
patterns were strongly downregulated in CR OPSC when compared with
IR OPSC patients (Fig. 1A). In
order to confirm microarray results, qRT-PCR validation was
performed. RNA was isolated from a new set of FFPE tissues to
increase the possibility that the observed differences in miRNA
expression profiles represented biologically significant changes.
In keeping with microarray results, miR-224-5p was low-expressed in
CR OPSC with significance, and representative analysis is shown in
Fig. 1B.
miR-224-5p expression in different
chemoresistant ovarian cancer cell lines, and upregulation of
miR-224-5p increases cell survival of chemosensitive cell lines to
cisplatin
In the previous results, we demonstrated that
miR-224-5p expression levels were significantly higher in IR tumor
specimens than in CR tumors specimens (Fig. 1A and B). Thus, we sought to further
verify the relationship between miR-224-5p expression and
chemosensitivity using the human ovarian cancer cell lines OV2008
and A2780S, and their cisplatin-resistant variants, C13 and
A2780CP, respectively. As indicated in Fig. 1C, all significant downregulation of
miR-224-5p in OV2008 and A2780S cell lines compared with their
cisplatin-resistant variants, respectively. These observations are
consistent with the results in clinical ovarian cancer patient
specimens.
Furthermore, we studied the effects of miR-224-5p
expression patterns on chemosensitivity to cisplatin in
vitro. After transfection with pre-miR-224-5p, the cell
viabilities of OV2008 and A2780S appeared obviously higher than the
mock or negative control group (p<0.01 for all, Fig. 2A and B).
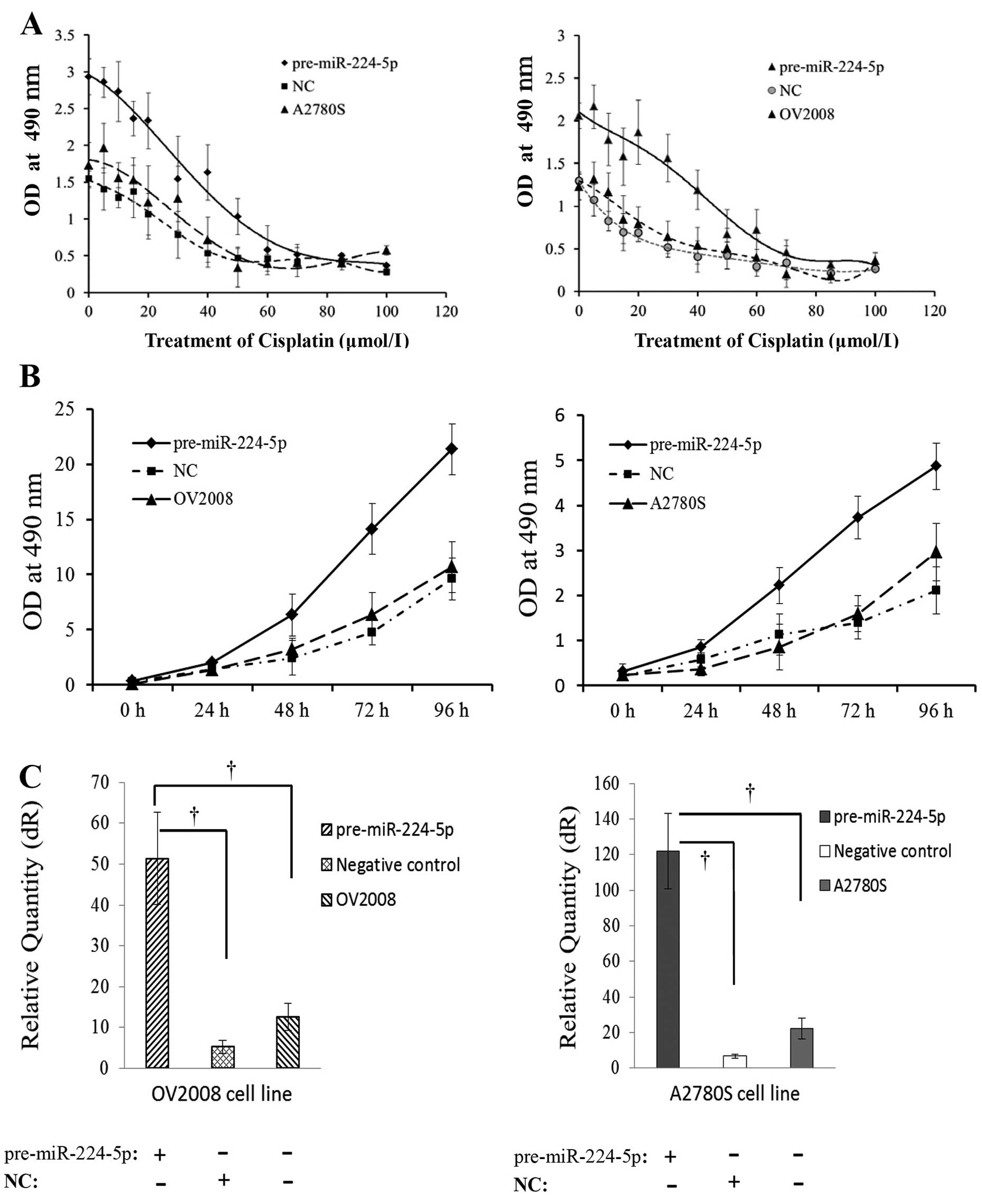 | Figure 2Upregulation of miR-224-5p increases
ovarian cancer cell survival of chemosensitive cell line to
cisplatin. (A) After transfection with pre-miR-224-5p, negative
control, and then treatment with gradient concentrations of
cisplatin, the proliferation curves of both cisplatin-sensitive
parental cell lines (OV2008 and A2780S) were all shifted to the
right (n=10/each point). (B) After transfection with pre-miR-224-5p
for 48 h, and then treatment with 20 μmol/l cisplatin for 0, 24, 48
and 96 h, the cell viabilities of OV2008 and A2780S were all
clearly increased, and their proliferation curves all shifted to
the left compared with their negative control and mock cells
(n=10/each point). (C) Real-time RT-PCR validated that
pre-miR-224-5p transfections were well performed. †,
statistically significant difference of miR-224-5p, compared with
either negative control or mock cells (p<0.01 for all). |
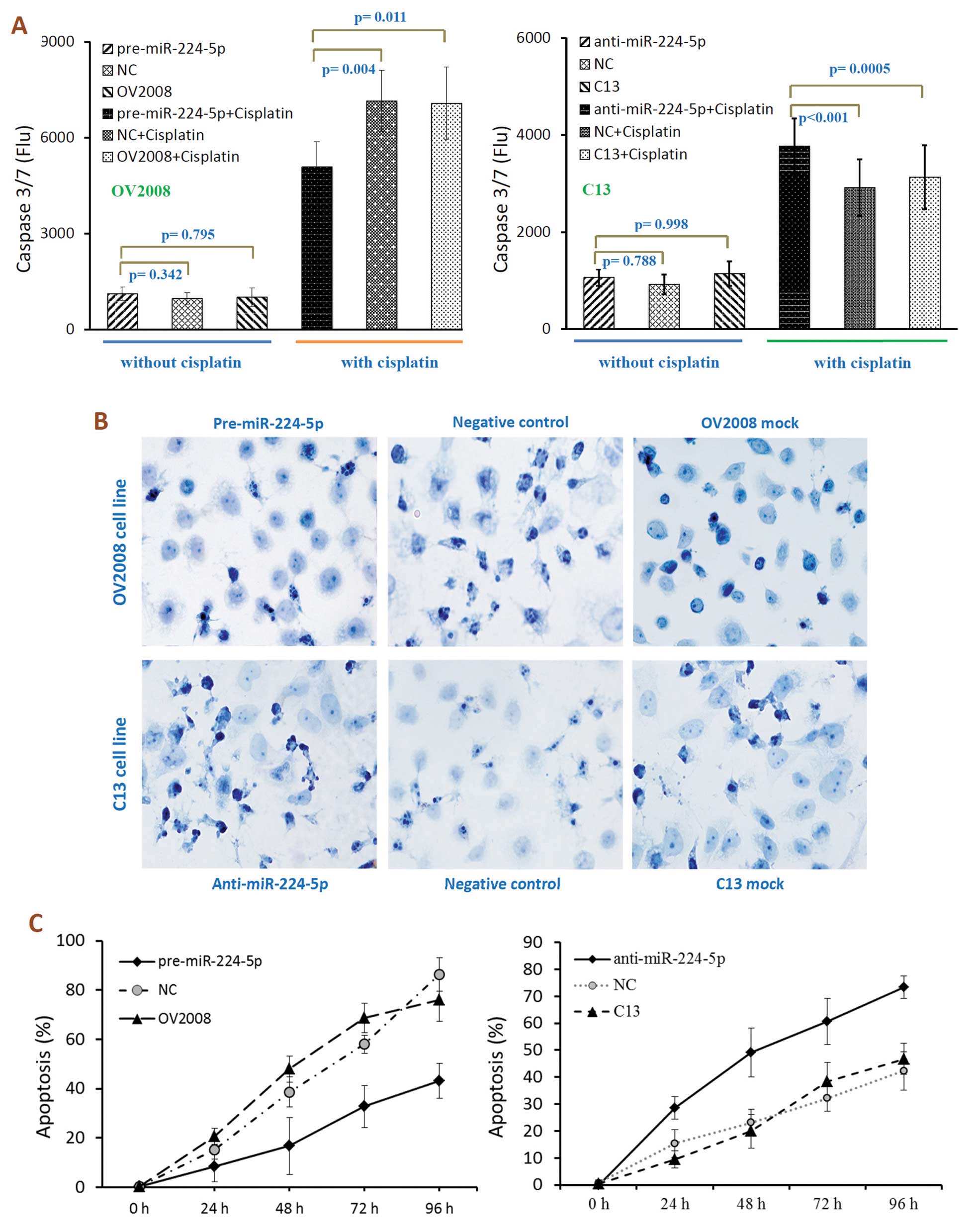
Effects of miR-224-5p on sensitivity of
ovarian cancer cells to cisplatin-induced apoptosis
Cisplatin and other platinum-based cancer drugs
destroy tumor cells by binding to DNA strands and forming
cisplatin-DNA adducts. Vast DNA damage activates apoptosis when
repair is impossible. Thus, we next examined whether miR-224-5p is
involved in cisplatin-induced chemoresistance by affecting
apoptosis. To evaluate the effect of miR-224-5p expression on the
chemosensitivity of ovarian cancer cells, we used
cisplatin-sensitive parental cell line (OV2008 and A2780S) and its
cisplatin-resistant variant (C13 and A2780CP) through transfection
with pre-miR-224-5p (miRNA mimic), anti-miR-224-5p (inhibitor) or
their scrambled negative controls, respectively. After
transfection, cells were treated with cisplatin, followed by
assessment of the activation of caspase-3 and -7 and with TUNEL
assays. As shown in Fig. 3A,
cisplatin-induced apoptosis was reversed by miR-224-5p in tumor
cells, as indicated by the decreased or increased caspase-3/7
activity observed in miR-224-5p upregulated or downregulated tumor
cells, respectively (Fig. 3A).
Furthermore, TUNEL assays revealed that the ratio of
apoptosis in cisplatin-resistant C13 cells after anti-miR-224-5p
treatment was markedly increased compared to the negative control
and mock group (Fig. 3B and C), and
vice versa. These findings suggest that miR-224-5p may protect
ovarian cancer cells from cisplatin-induced damage by preventing
apoptosis.
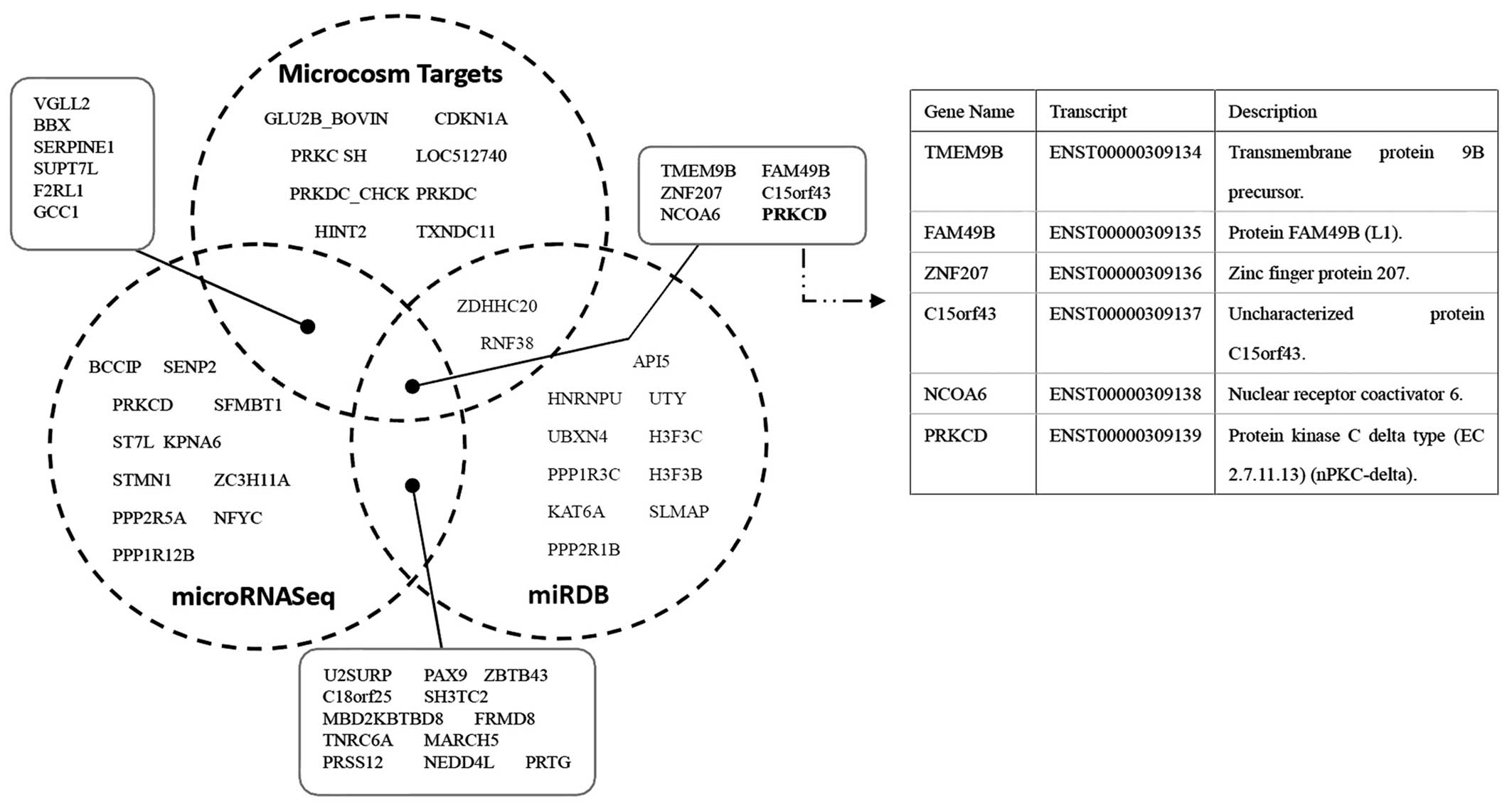
miRNA target prediction and
validations
To further understand the mechanism of miR-224-5p in
regulating chemoresistance of ovarian cancer cells, we chose
several computational approaches, including TargetScan, MicroCosm
Targets and miRNA Target Visualization, to analyze its predicted
targets. In order to retrieve the most relevant targets, we listed
only the top significantly unique miRNAs which were predicted by
these computational approaches, and we found PRKCD, whose gene
product plays a role in both regulating chemoresistance and
promoting apoptosis, is the common target of these approaches
(Fig. 4). These results suggest
that PRKCD may be a potential target gene of miR-224-5p in
regulating cisplatin sensitivity.
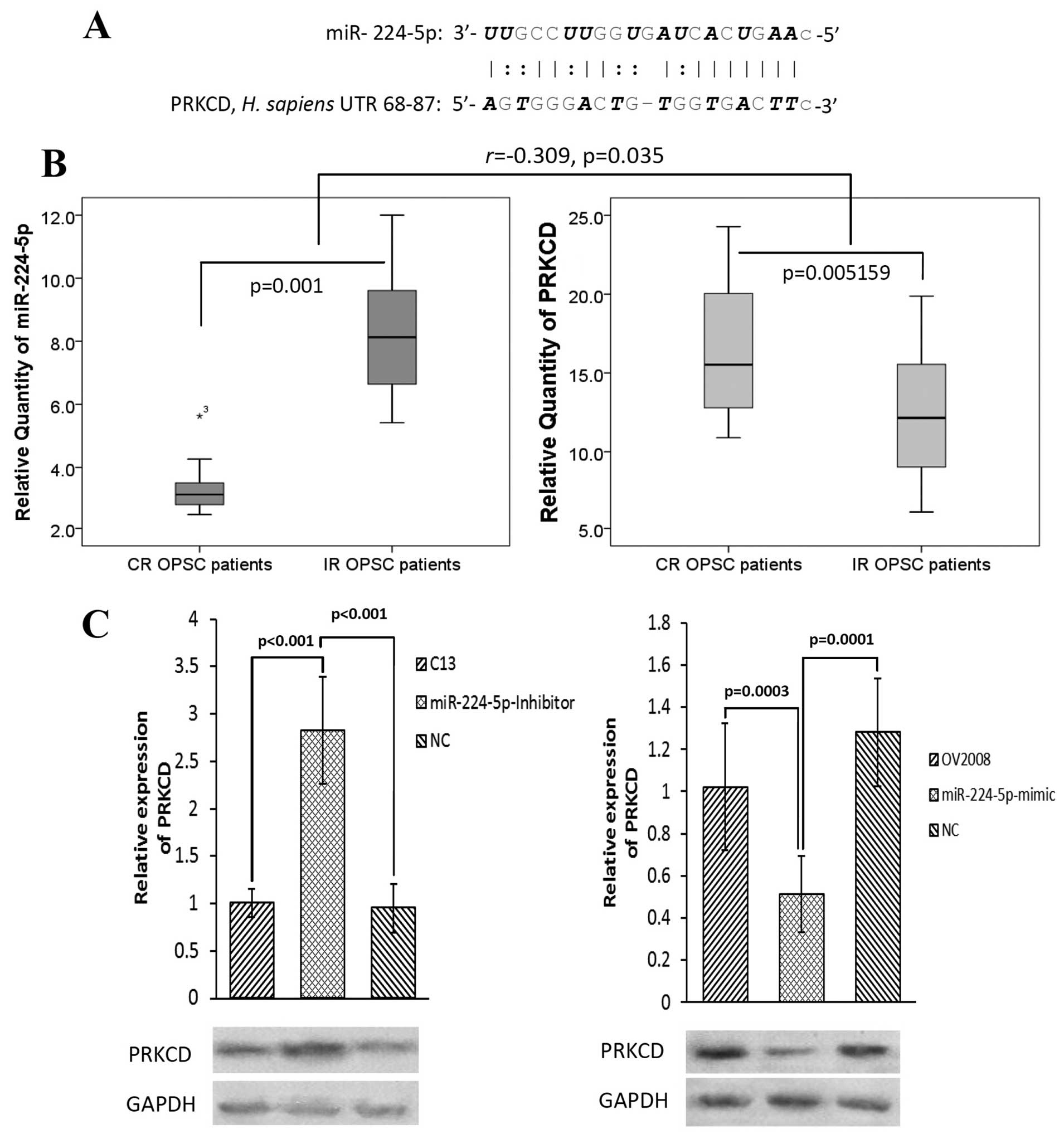
We next validated this prediction with standard
approaches such as PCR and western blot assay (Fig. 5B and C). qPCR analysis for
miR-224-5p expression showed a significant downregulation in CR
patients compared to IR cases (p=0.001, Fig. 5B). However, its candidate target
PRKCD negatively expressed between the same CR and IR patients. The
correlation coefficients (r) and significant levels (p) were −0.309
and 0.035, respectively.
To further examine whether PRKCD is the target of
miR-224-5p at the in vitro level, cisplatin-resistant
parental cell line (C13) and its sensitive variant (OV2008) were
transfected with anti-miR-224-5p and pre-miR-224-5p, respectively.
Western blot analyses revealed that miR-224-5p negatively regulated
PRKCD expression at both cisplatin-resistant parental cell and its
sensitive variant cell levels (Fig.
5C).
Prediction of survival rate in OPSC
patients using miR-224-5p and PRKCD expression patterns
To elucidate the significance of miR-224-5p
expression pattern in EOC chemosensitivity, we performed a
retrospective study to investigate the relationship between
miR-224-5p, PRKCD expression and chemosensitivity in OPSC patients.
A 41-sample training set (CR, 22; IR, 19) was used in analysis of
leave-one-out cross predictions. Using a cut-off of 4.0225 and
8.1578 for miR-224-5p and PRKCD respectively, we found the highest
Youden’s index (Fig. 6A and B).
Mann-Whitney U tests for statistical significance (p<0.001 for
all groups) demonstrated the capacity of the predictor to
distinguish CR patients from IR patients. The mean area under ROC
curve (AUC) values of miR-224-5p and PRKCD were 0.889 and 0.895,
respectively (Fig. 6C and D).
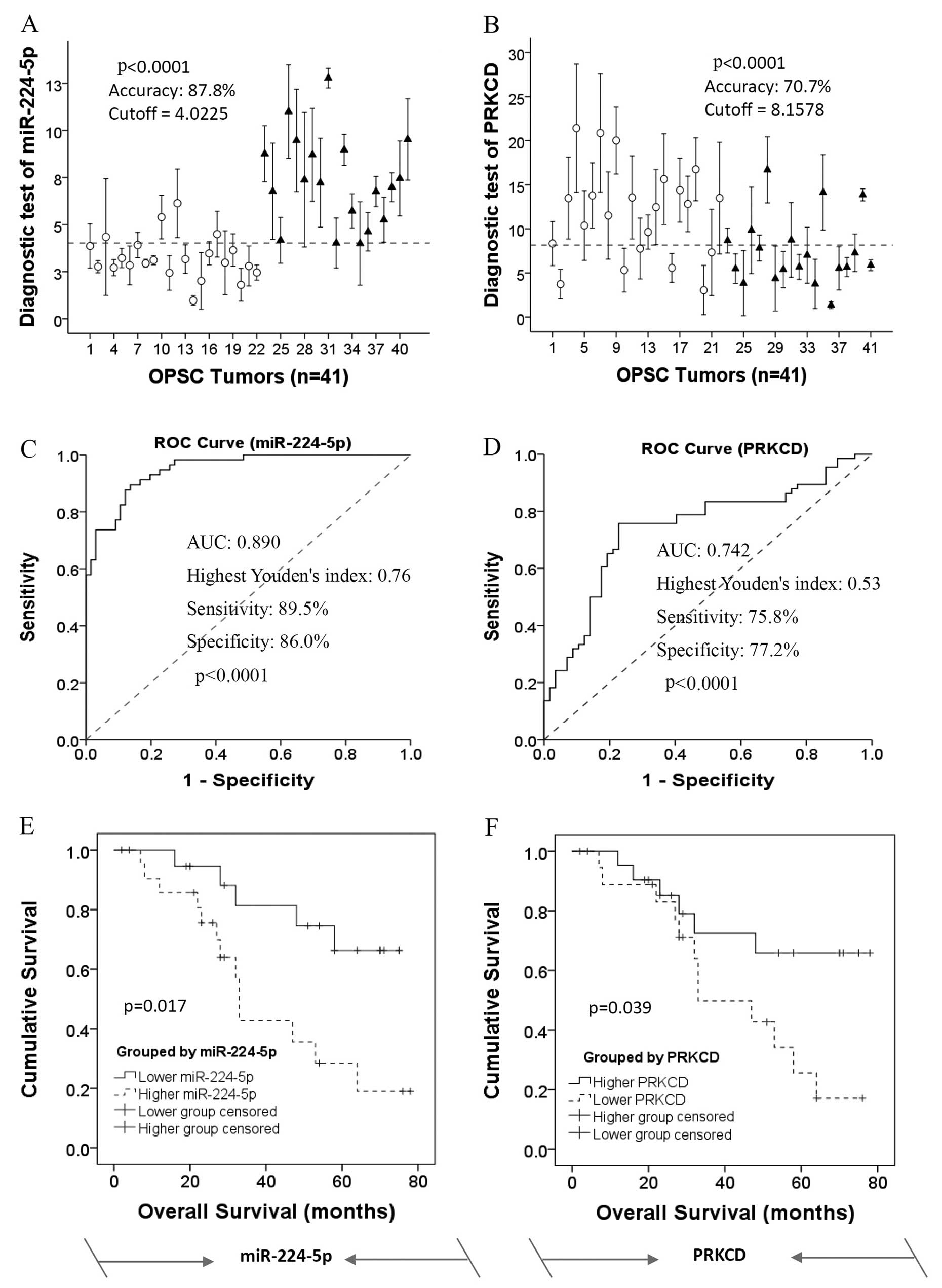 | Figure 6Kaplan-Meier curves showing the
overall survival of OPSC patients stratified by expression levels
of miR-224-5p and PRKCD. (A and B) Leave-one-out cross predictions
of OPSC differentiation (white circle, CR patients; black triangle,
IR patients) with (A) miR-224-5p and (B) PRKCD respectively, n=41
for all; cut-offs were 4.0225 and 8.1578 with the highest Youden’s
index for each of them; accuracy, sensitivity and specificity were
87.8, 89.5 and 86.0% for miR-224-5p and 70.7, 75.8 and 77.2% for
PRKCD, respectively. (C and D) Receiver operating characteristic
(ROC) curves of the predictions of OPSC differentiation according
to (C) miR-224-5p and (D) PRKCD respectively, n=41 for all. AUC
values were 0.878 and 0.707 for miR-224-5p and PRKCD, respectively.
(E and F) miR-224-5p and PRKCD serve as novel predictions and
prognostic biomarkers for the predictive response to survival of
OPSC patients. The overall survival result of a 41-sample training
set divided into two sub-bits according to high and low expression
level of (E) miR-224-5p and (F) PRKCD, as over or under the cut-off
points, respectively. Diagnosis is based on the analysis of the
above ROC research. Kaplan-Meier survival analysis indicated that
the expression levels of miR-224-5p and with PRKCD may serve as
prognostic biomarkers for OPSC patient response to overall
disease-specific survival. CR, complete response; IR, incomplete
response; OPSC, ovarian papillary serous carcinoma. |
Based on the analysis of the above ROC research,
Kaplan-Meier survival analysis indicated that high expression of
miR-224-5p was significantly associated with poor overall survival
of the OPSC patients as compared with the low miR-224-5p expression
group, log-rank p=0.017 (Fig. 6E).
However, the result of PRKCD expression in Kaplan-Meier survival
analysis was contrary to miR-224-5p (Fig. 6F).
These results indicate that the expression levels of
miR-224-5p and with PRKCD attribute to revealing the
chemoresistance of OPSC patients, and may serve as prognostic
biomarkers for OPSC patient response to overall disease-specific
survival.
Discussion
Platinum-based chemotherapy is the standard,
first-line treatment for advanced-stage EOC. Almost all patients
with EOC at the advanced stages receive the same treatment
strategy, which is a platinum-based regimen, usually with cisplatin
(15). However, the major
impediment to a successful treatment is the drug resistance to
chemotherapy. The purpose of the present study was to enhance our
understanding of the biological mechanisms underlying the
chemoresistance of human ovarian cancer and to identify possible
therapies that can induce chemoresistance reversion. By comparing
the miRNA microarray profiles of patients with CR and IR ovarian
cancers, we found that miR-224-5p was significantly upregulated in
IR patients. To examine the association of miR-224-5p with
chemosensitivity of ovarian cancer cells in vitro, we used
transient transfection of miR-224-5p (mimic or inhibitor) in
ovarian cancer cell lines. We found that high level of miR-224-5p
has potent promoting effects on ovarian cancer cell survival and
inhibitory effects on apoptosis, and vice versa. Therefore, we
identified miR-224-5p as a critical contributor to chemoresistance
in epithelial ovarian cancer. Our conclusion is supported by both
in vitro and in vivo experiments.
Recent evidence indicates that miRNAs may play
important roles by affecting various pathways related to anticancer
drug resistance, such as influencing the response to the
conventional chemoagents, cisplatin or microtubule-targeting drugs
(16–18). Therefore, identification of
cancer-specific miRNAs as well as their targets is critical for
understanding the roles of miRNAs in tumor genesis and may be
important for defining novel chemotherapy (4).
To further understand the mechanism of miR-224-5p in
regulating chemoresistance of ovarian cancer cells, we selected
several computational approaches to analyze its predicted targets.
In the present study, we focused on PRKCD, which plays a role in
both regulating chemoresistance and promoting apoptosis. Previous
studies revealed that the protein kinase C (PKC) signal pathway is
a critical regulator of the chemosensitivity in cancers such as
ovarian cancer, non-small cell lung cancer and prostate cancer
(19–21).
PRKCD, known as protein kinase C δ, is a PKC isozyme
that acts as a substrate for caspase-3 (4). Its activity is believed to be required
for apoptosis induced by DNA damaging agents such as cisplatin,
mitomycin C and doxorubicin (22–24).
Additionally, PRKCD has been reported to be a possible positive
regulator of cisplatin-induced cell death in a gastric cancer cell
line, leading to a modest increase in cisplatin uptake, and
associated with the inhibition of cell cycle and tumor progression
(25–28). Collectively, PRKCD has a negative
effect on cell survival. In the present study, we demonstrated that
miR-224-5p could negatively regulate the expression of PRKCD, and
together with PRKCD, they can serve as novel predictors and
prognostic biomarkers for OPSC patient response to overall
disease-specific survival (Fig. 6).
Therefore, the PRKCD pathway may be a molecular mechanism through
which miR-224-5p exerts its functions as an oncogene and enhancer
of chemoresistance to cisplatin in OPSC patients.
In conclusion, we report that alterations in
miR-224-5p expression in ovarian cancer patients and cell lines
confer differential chemosensitivity and that miR-224-5p may
function as an enhancer of chemoresistance at least in part by
targeting PRKCD. Therefore, the miR-224-5p-PRKCD interaction may
become a biomarker for predicting chemosensitivity to cisplatin in
patients with ovarian papillary serous carcinoma.
References
|
1
|
Zhao H, Ding Y, Tie B, et al: miRNA
expression pattern associated with prognosis in elderly patients
with advanced OPSC and OCC. Int J Oncol. 43:839–849.
2013.PubMed/NCBI
|
|
2
|
Saldivar JS, Wu X, Follen M and Gershenson
D: Nucleotide excision repair pathway review I: implications in
ovarian cancer and platinum sensitivity. Gynecol Oncol. 107(Suppl
1): S56–S71. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNAs in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen Y, Ke G, Han D, Liang S, Yang G and
Wu X: MicroRNA-181a enhances the chemoresistance of human cervical
squamous cell carcinoma to cisplatin by targeting PRKCD. Exp Cell
Res. 320:12–20. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang W, Dahlberg JE and Tam W: MicroRNAs
in tumorigenesis: a primer. Am J Pathol. 171:728–738. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: microRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Z, Hu S, Wang J, et al: MiR-27a
modulates MDR1/P-glycoprotein expression by targeting HIPK2 in
human ovarian cancer cells. Gynecol Oncol. 119:125–130. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Duttagupta R, DiRienzo S, Jiang R, et al:
Genome-wide maps of circulating miRNA biomarkers for ulcerative
colitis. PLoS One. 7:e312412012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Metzeler KH, Maharry K, Radmacher MD, et
al: TET2 mutations improve the new European LeukemiaNet risk
classification of acute myeloid leukemia: a cancer and leukemia
group B study. J Clin Oncol. 29:1373–1381. 2011. View Article : Google Scholar
|
|
10
|
Enerly E, Steinfeld I, Kleivi K, et al:
miRNA-mRNA integrated analysis reveals roles for miRNAs in primary
breast tumors. PLoS One. 6:e169152011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Powers MP, Alvarez K, Kim HJ and Monzon
FA: Molecular classification of adult renal epithelial neoplasms
using microRNA expression and virtual karyotyping. Diagn Mol
Pathol. 20:63–70. 2011. View Article : Google Scholar
|
|
12
|
Bansal N, Marchion DC, Bicaku E, et al:
BCL2 antagonist of cell death kinases, phosphatases, and ovarian
cancer sensitivity to cisplatin. J Gynecol Oncol. 23:35–42. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Miller AB, Hoogstraten B, Staquet M and
Winkler A: Reporting results of cancer treatment. Cancer.
47:207–214. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rustin GJ, Nelstrop AE, Bentzen SM,
Piccart MJ and Bertelsen K: Use of tumour markers in monitoring the
course of ovarian cancer. Ann Oncol. 1:21–27. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dressman HK, Berchuck A, Chan G, et al: An
integrated genomic-based approach to individualized treatment of
patients with advanced-stage ovarian cancer. J Clin Oncol.
25:517–525. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Giovannetti E, Erozenci A, Smit J, Danesi
R and Peters GJ: Molecular mechanisms underlying the role of
microRNAs (miRNAs) in anticancer drug resistance and implications
for clinical practice. Crit Rev Oncol Hematol. 81:103–122. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tian W, Chen J, He H and Deng Y: MicroRNAs
and drug resistance of breast cancer: basic evidence and clinical
applications. Clin Transl Oncol. 15:335–342. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kanakkanthara A and Miller JH: MicroRNAs:
novel mediators of resistance to microtubule-targeting agents.
Cancer Treat Rev. 39:161–170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qamar L, Davis R, Anwar A and Behbakht K:
Protein kinase C inhibitor Gö6976 augments caffeine-induced
reversal of chemoresistance to
cis-diamminedichloroplatinum-II (CDDP) in a human ovarian
cancer model. Gynecol Oncol. 110:425–431. 2008.
|
|
20
|
Clark AS, West KA, Blumberg PM and Dennis
PA: Altered protein kinase C (PKC) isoforms in non-small cell lung
cancer cells: PKCδ promotes cellular survival and chemotherapeutic
resistance. Cancer Res. 63:780–786. 2003.
|
|
21
|
Sumitomo M, Asano T, Asakuma J, Asano T,
Nanus DM and Hayakawa M: Chemosensitization of androgen-independent
prostate cancer with neutral endopeptidase. Clin Cancer Res.
10:260–266. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Basu A and Akkaraju GR: Regulation of
caspase activation and
cis-diamminedichloroplatinum(II)-induced cell death by
protein kinase C. Biochemistry. 38:4245–4251. 1999.
|
|
23
|
Blass M, Kronfeld I, Kazimirsky G,
Blumberg PM and Brodie C: Tyrosine phosphorylation of protein
kinase Cδ is essential for its apoptotic effect in response to
etoposide. Mol Cell Biol. 22:182–195. 2002.
|
|
24
|
Basu A, Woolard MD and Johnson CL:
Involvement of protein kinase C-δ in DNA damage-induced apoptosis.
Cell Death Differ. 8:899–908. 2001.
|
|
25
|
Iioka Y, Mishima K, Azuma N, et al:
Overexpression of protein kinase Cδ enhances cisplatin-induced
cytotoxicity correlated with p53 in gastric cancer cell line.
Pathobiology. 72:152–159. 2005.
|
|
26
|
Basu A and Evans RW: Comparison of effects
of growth factors and protein kinase C activators on cellular
sensitivity to cis-diamminedichloroplatinum(II). Int J
Cancer. 58:587–591. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gentilin E, Tagliati F, Filieri C, et al:
miR-26a plays an important role in cell cycle regulation in
ACTH-secreting pituitary adenomas by modulating protein kinase Cδ.
Endocrinology. 154:1690–1700. 2013.PubMed/NCBI
|
|
28
|
Hernández-Maqueda JG, Luna-Ulloa LB,
Santoyo-Ramos P, Castañeda-Patlán MC and Robles-Flores M: Protein
kinase C delta negatively modulates canonical Wnt pathway and cell
proliferation in colon tumor cell lines. PLoS One.
8:e585402013.PubMed/NCBI
|