Introduction
Esophageal cancer is the 5th most common cause of
cancer-related mortality in males worldwide and the 8th most common
in females (1). Esophageal squamous
cell carcinoma (ESCC) is a major histopathologic subtype of
esophageal cancer. The prognosis of patients with ESCC is poor, and
the 5-year survival rate is <30% (2). Identifying new markers and therapeutic
targets may help in the early detection of ESCC and may reverse the
malignant phenotype, thus improving the prognosis.
Karyopherin α 2 (KPNA2) is one of the seven family
members of karyopherin α (3). These
proteins mediate the nucleocytoplasmic transport through large
nuclear pore complexes. With the help of KPNA2, macromolecules
>40 kDa can be shuttled between the cytoplasm and nucleus
(4). During nucleocytoplasmic
transport, karyopherin α can form heterodimers with karyopherin β-1
to generate the nuclear transport complex. As part of the complex,
KPNA2 can recognize cargo proteins via their nuclear localization
signal and act as an adaptor. Karyopherin β-1 then facilitates
protein docking to and translocation through the nuclear pore
complexes. After entering the nucleus, karyopherin α and
karyopherin β-1 can dissociate from the complex and bind RanGTP,
respectively. They are then recycled back into the cytoplasm
(4,5).
Deregulation of the cellular transport machinery
often occurs in tumors. In accordance with this phenomenon, KPNA2
expression has been shown to be elevated in a variety of tumor
tissues, including breast (6–8) and
ovarian cancer (9), melanoma
(10), cervical (11), lung (12), brain (13), prostate (14), liver (15), bladder (16) and esophageal cancer (17). Notably, it has been shown that KPNA2
expression is higher in both tissue and serum in lung cancer
(12), suggesting that KPNA2 can be
secreted into the serum. Since the overexpression of KPNA2 is a
common phenomenon in different types of cancer, it is possible that
KPNA2 serum levels may also be upregulated in other types of
cancer. However, with the exception of lung cancer, similar results
have not been reported until now.
In ESCC, KPNA2 has been shown to be upregulated in
the tumor tissue (17). The high
expression of KPNA2 correlates with a poor prognosis. However, the
expression of KPNA2 in serum and the function of KPNA2 in ESCC
cells remains unclear. In the present study, we observed high
expression of KPNA2 in tissues, cell lines and serum from patients
with ESCC. Knockdown of KPNA2 in an ESCC cell line inhibited cell
proliferation by inducing G2/M phase cell cycle arrest and
attenuated E2F1 nuclear translocation. These results indicate that
KPNA2 may be a potent marker and therapeutic target of ESCC.
Materials and methods
Chemicals
Dulbecco’s modified Eagle’s medium (DMEM) and
RPMI-1640 culture media were purchased from Invitrogen (Carlsbad,
CA, USA). Fetal bovine serum (FBS) was obtained from HyClone
(Logan, UT, USA). Goat anti-KPNA2 (sc-6917) and rabbit anti-E2F1
primary antibody (sc-193) were obtained from Santa Cruz
Biotechnology (Santa Cruz, CA, USA). Goat anti-laminB (ab16048)
primary antibody was purchased from Abcam (Cambridge, UK). Mouse
anti-β-actin primary antibody (A-5316) was obtained from Sigma
(Santa Clara, CA, USA).
Clinical specimens
Tissue specimens were purchased as a microarray
(Outdo Biotech Co., Shanghai, China). The median age of the
patients (71 males, 22 females) from whom the ESCC tissues were
collected for immunochemistry was 64.2 years (range, 49–78 years).
Surgical tissue specimens were collected from ESCC patients with
their informed consent and approval from the Institutional Review
Board of the Cancer Institute and Hospital of Chinese Academy of
Medical Sciences (Beijing, China). Serum samples were collected
from 86 ESCC patients (70 males and 16 females; mean age, 59.5
years; range, 39–76 years) and 60 healthy controls (43 males and 17
females; mean age, 53 years; range, 26–76 years), who all provided
informed consent. Pathological diagnosis was conducted
independently by two senior pathologists. Biochemistry tests and
routine blood tests were performed for the healthy controls, and
the results were within the normal ranges. Sample preparation was
previously described (18).
Cell culture
Kyse30, Kyse140, Kyse150, Kyse170, Kyse410 and
Kyse510 cells were donated by Dr Y. Shimada. These cells were
cultured in RPMI-1640 medium supplemented with 10% FBS, 1% 100 U/ml
penicillin and 100 mg/ml streptomycin. WHCO1 cells were grown in
DMEM supplemented with 10% FBS. All cells were maintained at 37°C
in 5% CO2.
Immunohistochemistry
For immunohistochemical staining, multiple tissue
arrays (MTAs) were incubated with goat anti-KPNA2 polyclonal
antibody. After washing with 1X phosphate-buffered saline (PBS),
slides were reacted with the biotin-labeled second antibody and
then visualized using an ultrasensitive streptavidin-peroxidase
system (ZSGB Biotechnology, Beijing, China). Immunostaining was
scored as follows: 0, negative; 1, weak; 2, moderate; 3, strong
staining. The percentage of KPNA2 staining area was graded as 0, no
positive staining; 1, <5%; 2, 5–25%; 3, 50–75%; or 4, >75%.
The staining index was calculated as the product of staining
intensity and staining area, ranging from 0 to 12.
Enzyme linked immunosorbent assay
(ELISA)
The KPNA2 levels in human serum were measured using
a commercially available ELISA kit (USCN Life Science Inc., Wuhan,
Hubei, China). The ELISA was performed according to the
manufacturer’s instructions. Briefly, 100 μl of diluted
serum samples (at 1:500 dilution) was added to each well and
incubated at 37°C for 2 h. Then, 100 μl reagent A was added
for an additional hour. After 5 washes, reagent B was added for 30
min. The amount of proteins was determined by adding TMB substrate,
and the plate was incubated at 37°C to allow for color development.
The absorbance was measured at 450 nm using a Model 680 microplate
reader (Bio-Rad Laboratory Inc., Hercules, CA, USA).
Western blotting
Approximately 15 μg protein lysate was
separated on SDS-PAGE gels and transferred to PVDF membranes (GE
Healthcare, Piscataway, NJ, USA). After blocking, the membranes
were incubated with primary antibodies and developed using an ECL
system (Applygen Technologies Inc., Beijing, China). LaminB and
β-actin were used as loading controls.
Small interfering RNA (siRNA)
transfection
siRNAs were synthesized by Invitrogen Co. (Shanghai,
China), and the target sequences were: siRNA-1, 5′-GCUCCUGCAUCAUGAU
GAU-3′; siRNA-2, 5′-GUGGCUACUUACGUAAUCU-3′; negative control,
5′-UUCUCCGAACGUGUCACGU-3′. Transient transfection was performed
using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA),
according to the manufacturer’s protocol. Briefly, the cells were
seeded in 6-well plates and allowed to grow for at least 20 h.
Then, 100 nmol siRNA was added to each well. After a 6-h
transfection, the siRNAs were replaced by adding fresh medium.
Western blotting was used to confirm the knockdown efficiency.
Cell proliferation analysis
Cell proliferation was assessed using the WST-8
method based on the metabolic reduction of the monosodium salt
2-(2-methoxy-4-nitrophenyl)-3-(4-
nitrophenyl)-5-(2,4-sulfophenyl)-2H tetrazolium. Three thousand
cells were seeded in 96-well plates and cultured overnight. The
cells were transfected with mock or siRNAs by Lipofectamine 2000.
Then, 10 μl WST-8 solution was added to each well and
incubated for 2 h. The plates were read on a Model 680 microplate
reader at 450 nm. Colony formation assays were performed as
previously described (19).
Flow cytometry
Cells were trypsinized, fixed with ethanol and
analyzed by flow cytometry (LSR II; BD Biosciences, Lexington, KY,
USA). Briefly, fixed cells were washed twice with ice-cold PBS.
RNAse (20 mg/ml) and PI (50 mg/ml) were added and incubated for 30
min at 37°C in the dark. Ten thousand cells per sample were
subjected to flow cytometry. The results were analyzed using the
FlowJo software (Tree Star Inc., Ashland, OR, USA).
Subcellular protein fractionation
Subcellular protein fractionation was performed
using a ProteoExtract Subcellular Proteome Extraction kit
(Calbiochem, La Jolla, CA, USA), according to the manufacturer’s
instructions. Briefly, adherent cells were digested with trypsin
and washed twice with PBS. Extraction buffers I, II and III were
sequentially added to generate different fractions. Fraction 1
includes the cytosolic fraction, fraction 2 includes the membrane
fraction, and fraction 3 includes the nuclear fraction.
Statistical analysis
All statistical comparisons were performed using the
SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). Student’s t-test
was used to determine the statistical significance of the results.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of KPNA2 in ESCC tissues and
cell lines
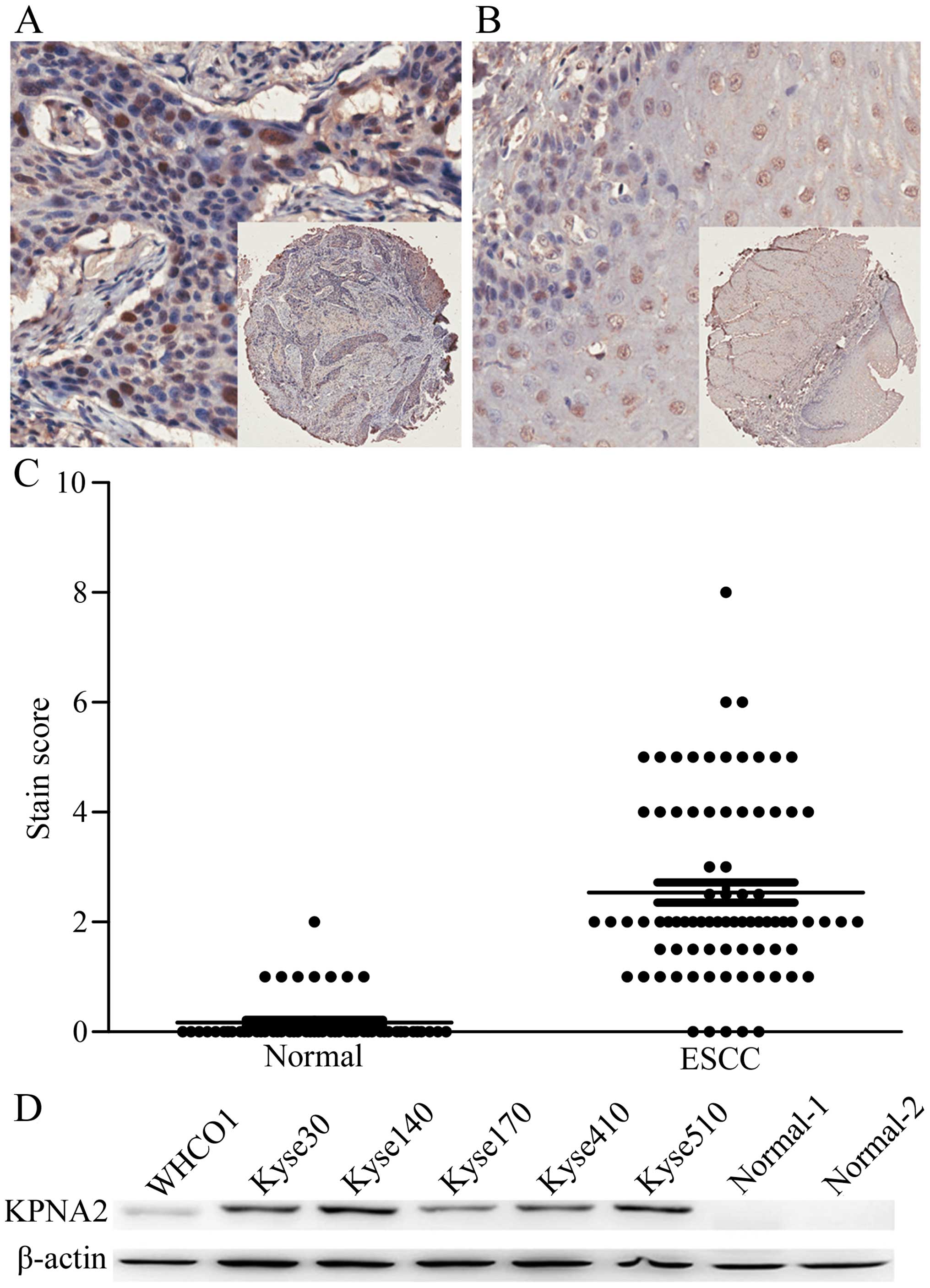
The expression of KPNA2 in ESCC tissues was examined
by immunohistochemistry using tissue microarray. The results showed
that KPNA2 was predominantly localized in the nuclei. Strong
staining was observed in ESCC tissues, while only weak staining was
observed in normal esophageal tissues (Fig. 1A and B). We semi-quantitated the
staining pattern of KPNA2 in ESCC and paired normal esophageal
tissues. The immunohistochemistry scores of KPNA2 ranged from 0 to
8 (median=2) in ESCC patients and from 0 to 2 (median=0) in normal
esophageal tissues. KPNA2 staining was significantly higher in ESCC
patients than in healthy controls (Fig.
1C, Student’s t-test, P<0.0001).
We next examined the KPNA2 expression in the ESCC
cell lines. All 6 ESCC cell lines expressed KPNA2 at different
levels (Fig. 1D). KPNA2 was
detected with a strong signal in Kyse140 and Kyse510 cells, a
moderate signal in Kyse30 and Kyse410 cells and a weak signal in
WHCO1 and Kyse170 cells. In normal human adjacent esophageal
tissues, no obvious KPNA2 signal was detected. These results show
that KPNA2 is also upregulated in ESCC cell lines.
KPNA2 serum levels are higher in ESCC
patients
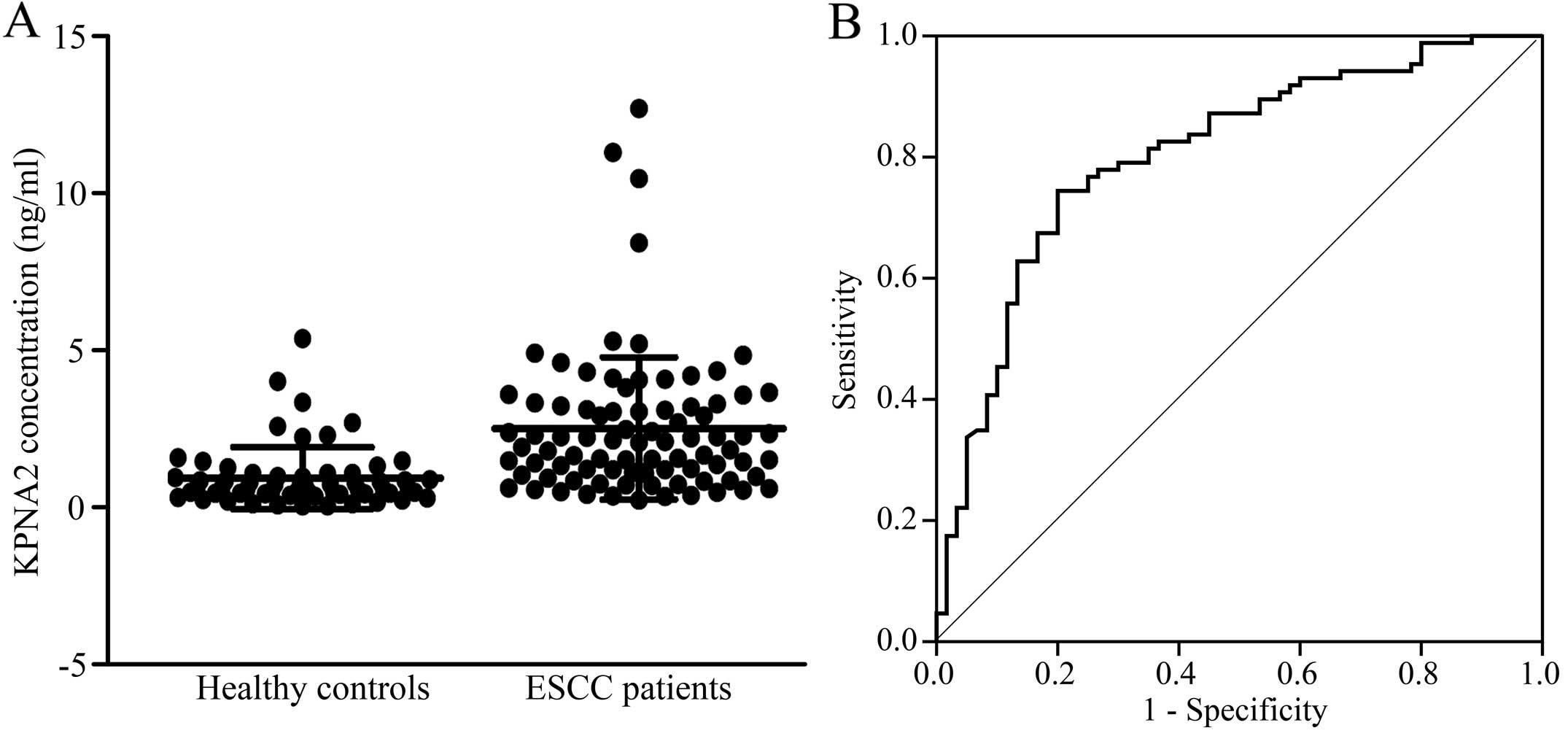
We assessed the serum levels of KPNA2 in ESCC
patients (n=86) and healthy controls (n=60) by ELISA. The serum
concentrations of KPNA2 ranged from 0.19 μg/ml to 12.67
μg/ml (median=1.95 μg/ml) in ESCC patients and from 0
μg/ml to 5.34 μg/ml (median=0.58 μg/ml) in
healthy controls (Fig. 2A). The
KPNA2 levels were significantly higher in ESCC patients compared to
healthy controls (Mann-Whitney U test, P<0.0001). The area under
curve (AUC) was determined to be 0.804 [95% confidence interval
(CI), 0.731–0.877] for KPNA2 (Fig.
2B). When 0.96 μg/ml was used as the cut-off value, the
sensitivity and specificity of serum KPNA2 was 76.7 and 75.0%,
respectively. These results suggest that KPNA2 may be a useful
serum marker for ESCC.
Knockdown of KPNA2 inhibits ESCC cell
proliferation
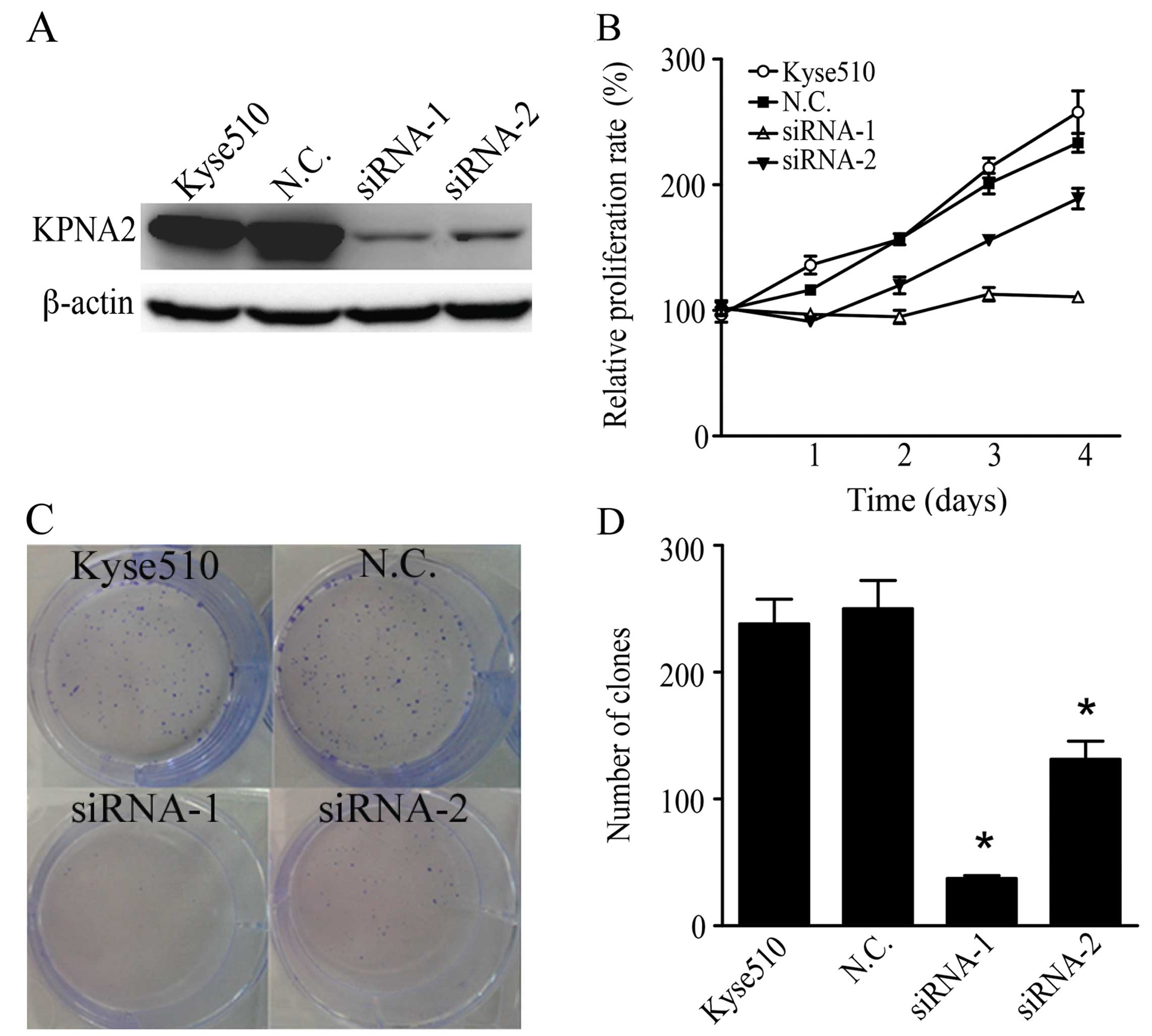
To investigate the role of KPNA2 in human ESCC
cells, we knocked down the KPNA2 expression with siRNA in Kyse510
cells. We used two siRNA sequences targeting different regions of
KPNA2 mRNA and determined the efficiency of knockdown by western
blotting. Both siRNAs significantly suppressed KPNA2 expression,
and siRNA-1 showed higher knockdown efficiency than did siRNA-2
(Fig. 3A). A WST-8 assay showed
that siRNA-1 almost completely abrogated Kyse510 cell growth and
that siRNA-2 showed a moderate effect, whereas scramble siRNA
exhibited no effect on ESCC cell proliferation (Fig. 3B). The colony formation assays
showed that both siRNAs significantly reduced the clonogenicity of
Kyse510 cells (Fig. 3C and D).
Knockdown of KPNA2 induces G2/M phase
arrest
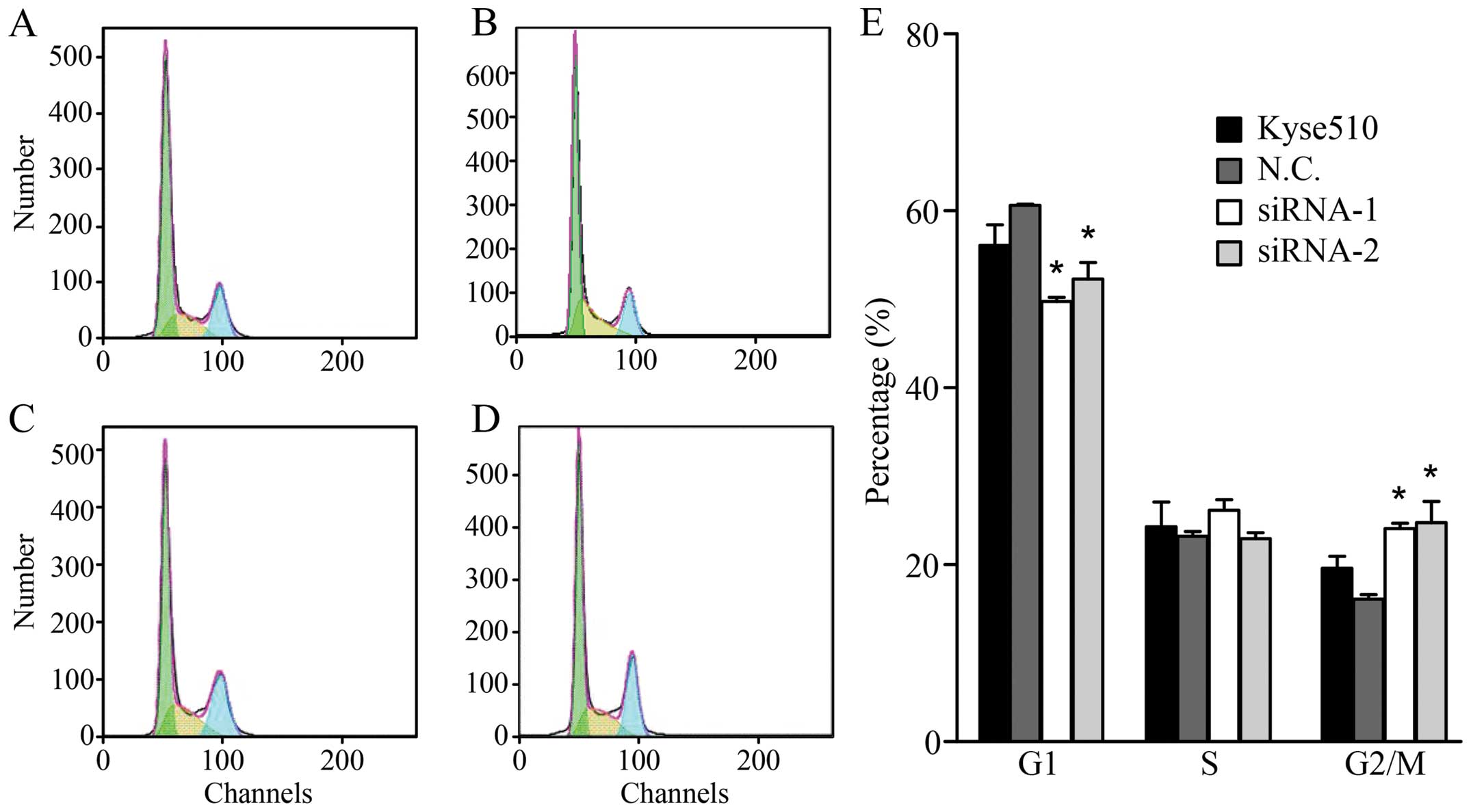
We assessed the effect of KPNA2 knockdown on cell
cycle progression by flow cytometry. The results showed that the
silencing of KPNA2 significantly decreased the percentage of cells
in G0/G1 and increased the percentage of cells in the G2/M phase of
the cell cycle (Fig. 4). The
percentage of cells at S phase did not change. These results
clearly show that the knockdown of KPNA2 induces a G2/M phase
arrest in ESCC cells.
Knockdown of KPNA2 attenuates E2F1
nuclear translocation
E2F1 is a central regulator of proliferation and
cell cycle progression and has been shown to be a target protein of
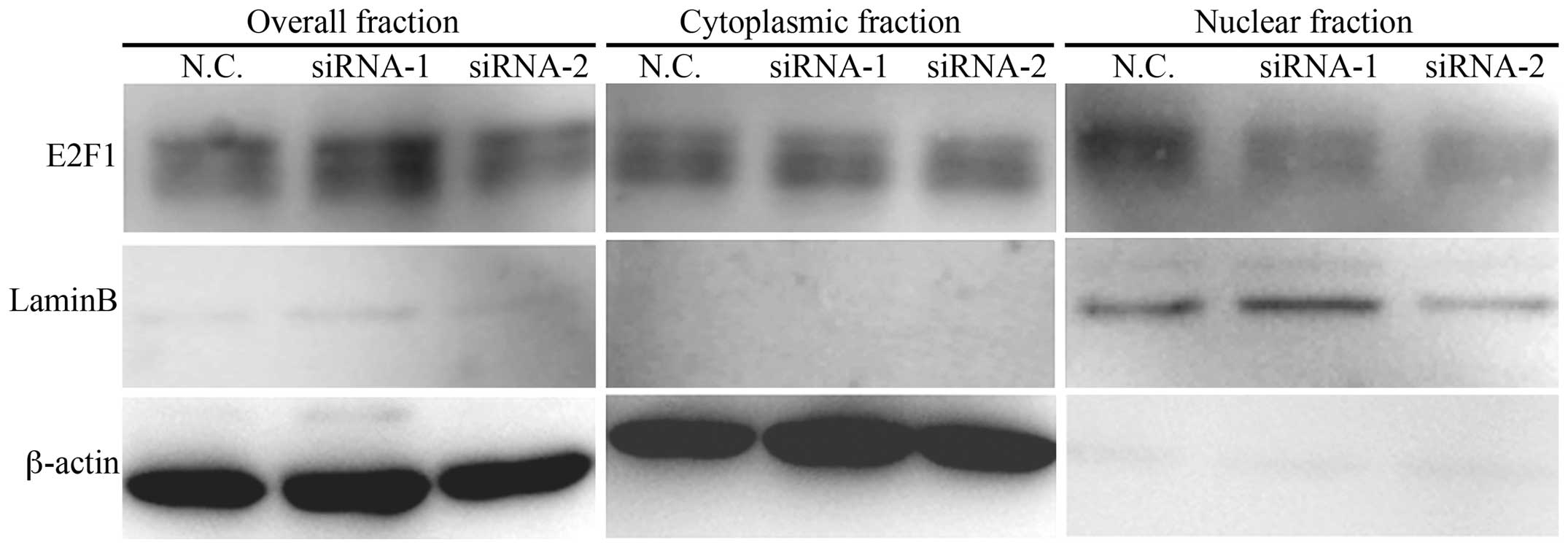
KPNA2. We examined the expression of E2F1 in KPNA2 knockdown cells
by subcellular fractionation. The fractionation efficiency was
verified by a western blot analysis of laminB and β-actin in the
cytoplasmic and nuclear fractions. The expression level of E2F1 was
reduced in the nuclear fraction of KPNA2 knockdown ESCC cells
compared with the scramble siRNA-transfected cells (Fig. 5). The results show that E2F1 nuclear
translocation is inhibited in ESCC cells following KPNA2
knockdown.
Discussion
In the present study, we reported that KPNA2 is
upregulated in ESCC tissues, cell lines and sera. The knockdown of
KPNA2 in ESCC cells inhibited cell proliferation and survival and
induced G2/M cell cycle arrest. These effects may be partially
mediated by reducing E2F1 nuclear translocation.
KPNA2 has been shown to be upregulated in various
types of cancer. Our results showed that KPNA2 was higher in ESCC
tissues, cell lines and serum. The increased expression of KPNA2
may be due to the altered transcriptional activity of tumors.
Indeed, the upregulation of KPNA2 at the transcriptional level has
been detected in cervical cancer (20). Additionally, its promoter activity
has been studied extensively. The promoter region is located −80 to
−24 bp upstream of the KPNA2 gene locus. In this region, there is a
highly conserved binding site for E2F transcription factors. It was
previously suggested that KPNA2 expression is initiated by E2F
transcription factors. Notably, the nuclear import of E2F1 is also
mediated by KPNA2 (21). This
finding raised the possibility that a positive feedback loop may
exist. In that feedback loop, E2F1 could increase the expression of
KPNA2 and, in turn, KPNA2 increases the nuclear accumulation of
E2F1. The expression of KPNA2 is also regulated by the cell cycle.
The highest expression of KPNA2 occurs during the G2/M phase
(22,23).
Currently, ESCC diagnosis is predominantly made
using cytological methods. The identification of useful ESCC serum
markers remains limited. Our laboratory has demonstrated that the
serum levels of GROβ are elevated in ESCC patients (24). However, the use of only a single
marker may be inefficient due to low sensitivity or specificity.
Therefore, combining several markers as a panel could potentially
provide better results. Here, we showed that KPNA2 is higher in
serum from ESCC patients. We propose that KPNA2 could be used as a
single or combined biomarker for the diagnosis of ESCC.
The effect of KPNA2 in tumor cell survival is
controversial. KPNA2 has been shown to be involved in the
proliferation of lung (12), liver
(15) and prostate cancer (14), while no effect on proliferation has
been observed in cervical cancer (11,25).
Our results showed that the knockdown of KPNA2 expression
attenuated tumor cell proliferation in ESCC cells. This finding is
consistent with phenomena observed in lung, liver and prostate
cancer. These previous studies indicated that KPNA2 was a potential
oncogene that functioned in sustaining cell proliferation. Since
KPNA2 was upregulated in ESCC tissues and since reducing KPNA2
levels could attenuate cell proliferation, KPNA2 may be a potential
therapeutic target of ESCC.
Either G1/S or G2/M cell cycle arrest can be induced
by KPNA2 knockdown, depending on the cell context (21,26).
Our results showed that KPNA2 knockdown induced G2/M cell cycle
arrest in ESCC cells. The synthesis of the proteins required for
mitosis occurs during the G2/M phase. Notably, E2F1 or its homolog
protein E2F4 has been shown to modulate the G2/M checkpoint by
regulating target gene expression (27). E2F target genes include genes that
are involved in chromosome segregation, chromatin
assembly/condensation and the mitotic spindle checkpoint. All of
these proteins play important roles in G2/M phase progression. In
ESCC cells, the knockdown of KPNA2 attenuated E2F1 nuclear
translocation and induced G2/M cell cycle arrest. These results
emphasize the possibility that the primary function of KPNA2 is
mediated by E2F1 in ESCC cells.
As a protein functioning in nuclear transport, the
deregulation of KPNA2 may affect cargo protein translocation in
cancer, thus promoting carcinogenesis. Many cancer-associated
proteins have been identified as cargo proteins of KPNA2, including
Chk2 (28), BRCA1 (29), NBS1 (30), RAC-1 (31), c-myc and P53 (21). A proteomic approach was previously
adopted to find new cargo proteins of KPNA2 in lung cancer. Of
particular interest in that investigation was E2F1, since both E2F1
nuclear translocation and its target gene expression are inhibited
by KPNA2 knockdown. The E2F1-mediated signaling pathways were the
most disrupted after KPNA2 knockdown. E2F1 is also regulated by
KPNA2 in ESCC cells. The expression of E2F1 was higher and
correlated with a poor prognosis in ESCC (32). Further analysis of the relationship
between E2F1 and KPNA2 in ESCC tissues will provide definitive
insight into the mechanism of KPNA2-mediated carcinogenesis and may
suggest a new strategy for ESCC therapy.
In summary, the expression of KPNA2 was higher in
tissues, cell lines and sera in ESCC. The knockdown of KPNA2
expression in ESCC cells attenuated cell proliferation and survival
by inducing G2/M phase cell cycle arrest through the E2F1 signaling
pathway. KPNA2 may be a potent marker and therapeutic target for
ESCC.
Acknowledgements
We thank Dr Yulin Sun, Dr Ming Liu and Miss Fang Liu
for great support. This study was supported by the National
High-Tech R&D Program (no. 2012AA020206), NSFC (nos. 31070673,
31170780, 81372591, 81321091) and BJNSFC (no. 5112012) of
China.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Wang GQ, Abnet CC, Shen Q, Lewin KJ, Sun
XD, Roth MJ, Qiao YL, Mark SD, Dong ZW, Taylor PR and Dawsey SM:
Histological precursors of oesophageal squamous cell carcinoma:
results from a 13 year prospective follow up study in a high risk
population. Gut. 54:187–192. 2005.PubMed/NCBI
|
|
3
|
Kelley JB, Talley AM, Spencer A, Gioeli D
and Paschal BM: Karyopherin α7 (KPNA7), a divergent member of the
importin α family of nuclear import receptors. BMC Cell Biol.
11:632010.
|
|
4
|
Stewart M: Molecular mechanism of the
nuclear protein import cycle. Nat Rev Mol Cell Biol. 8:195–208.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goldfarb DS, Corbett AH, Mason DA,
Harreman MT and Adam SA: Importin α: a multipurpose
nuclear-transport receptor. Trends Cell Biol. 14:505–514. 2004.
|
|
6
|
Dahl E, Kristiansen G, Gottlob K, Klaman
I, Ebner E, Hinzmann B, Hermann K, Pilarsky C, Dürst M,
Klinkhammer-Schalke M, Blaszyk H, Knuechel R, Hartmann A, Rosenthal
A and Wild PJ: Molecular profiling of laser-microdissected matched
tumor and normal breast tissue identifies karyopherin α2 as a
potential novel prognostic marker in breast cancer. Clin Cancer
Res. 12:3950–3960. 2006.PubMed/NCBI
|
|
7
|
Dankof A, Fritzsche FR, Dahl E, Pahl S,
Wild P, Dietel M, Hartmann A and Kristiansen G: KPNA2 protein
expression in invasive breast carcinoma and matched peritumoral
ductal carcinoma in situ. Virchows Arch. 451:877–881. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gluz O, Wild P, Meiler R, Diallo-Danebrock
R, Ting E, Mohrmann S, Schuett G, Dahl E, Fuchs T, Herr A, Gaumann
A, Frick M, Poremba C, Nitz UA and Hartmann A: Nuclear karyopherin
α2 expression predicts poor survival in patients with advanced
breast cancer irrespective of treatment intensity. Int J Cancer.
123:1433–1438. 2008.
|
|
9
|
Zheng M, Tang L, Huang L, Ding H, Liao WT,
Zeng MS and Wang HY: Overexpression of karyopherin-2 in epithelial
ovarian cancer and correlation with poor prognosis. Obstet Gynecol.
116:884–891. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Winnepenninckx V1, Lazar V, Michiels S,
Dessen P, Stas M, Alonso SR, Avril MF, Ortiz Romero PL, Robert T,
Balacescu O, Eggermont AM, Lenoir G, Sarasin A, Tursz T, van den
Oord JJ and Spatz A: Melanoma Group of the European Organization
for Research and Treatment of Cancer: Gene expression profiling of
primary cutaneous melanoma and clinical outcome. J Natl Cancer
Inst. 98:472–482. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van der Watt PJ, Maske CP, Hendricks DT,
Parker MI, Denny L, Govender D, Birrer MJ and Leaner VD: The
Karyopherin proteins, Crm1 and Karyopherin β1, are overexpressed in
cervical cancer and are critical for cancer cell survival and
proliferation. Int J Cancer. 124:1829–1840. 2009.
|
|
12
|
Wang CI, Wang CL, Wang CW, Chen CD, Wu CC,
Liang Y, Tsai YH, Chang YS, Yu JS and Yu CJ: Importin subunit
alpha-2 is identified as a potential biomarker for non-small cell
lung cancer by integration of the cancer cell secretome and tissue
transcriptome. Int J Cancer. 128:2364–2372. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gousias K, Becker AJ, Simon M and
Niehusmann P: Nuclear karyopherin a2: a novel biomarker for
infiltrative astrocytomas. J Neurooncol. 109:545–553. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mortezavi A, Hermanns T, Seifert HH,
Baumgartner MK, Provenzano M, Sulser T, Burger M, Montani M,
Ikenberg K, Hofstädter F, Hartmann A, Jaggi R, Moch H, Kristiansen
G and Wild PJ: KPNA2 expression is an independent adverse predictor
of biochemical recurrence after radical prostatectomy. Clin Cancer
Res. 17:1111–1121. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yoshitake K, Tanaka S, Mogushi K, Aihara
A, Murakata A, Matsumura S, Mitsunori Y, Yasen M, Ban D, Noguchi N,
Irie T, Kudo A, Nakamura N, Tanaka H and Arii S: Importin-α1 as a
novel prognostic target for hepatocellular carcinoma. Ann Surg
Oncol. 18:2093–2103. 2011.
|
|
16
|
Jensen JB1, Munksgaard PP, Sørensen CM,
Fristrup N, Birkenkamp-Demtroder K, Ulhøi BP, Jensen KM, Ørntoft TF
and Dyrskjøt L: High expression of karyopherin-α2 defines poor
prognosis in non-muscle-invasive bladder cancer and in patients
with invasive bladder cancer undergoing radical cystectomy. Eur
Urol. 59:841–848. 2011.
|
|
17
|
Sakai M, Sohda M, Miyazaki T, Suzuki S,
Sano A, Tanaka N, Inose T, Nakajima M, Kato H and Kuwano H:
Significance of karyopherin-α 2 (KPNA2) expression in esophageal
squamous cell carcinoma. Anticancer Res. 30:851–856. 2010.
|
|
18
|
Sun Y, Mi W, Cai J, Ying W, Liu F, Lu H,
Qiao Y, Jia W, Bi X, Lu N, Liu S, Qian X and Zhao X: Quantitative
proteomic signature of liver cancer cells: tissue transglutaminase
2 could be a novel protein candidate of human hepatocellular
carcinoma. J Proteome Res. 7:3847–3859. 2008. View Article : Google Scholar
|
|
19
|
Xu Y, Zhou L, Huang J, Liu F, Yu J, Zhan
Q, Zhang L and Zhao X: Role of Smac in determining the
chemotherapeutic response of esophageal squamous cell carcinoma.
Clin Cancer Res. 17:5412–5422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
van der Watt PJ, Ngarande E and Leaner VD:
Overexpression of Kpnβ1 and Kpnα2 importin proteins in cancer
derives from deregulated E2F activity. PLoS One. 6:e277232011.
|
|
21
|
Wang CI, Chien KY, Wang CL, Liu HP, Cheng
CC, Chang YS, Yu JS and Yu CJ: Quantitative proteomics reveals
regulation of karyopherin subunit alpha-2 (KPNA2) and its potential
novel cargo proteins in nonsmall cell lung cancer. Mol Cell
Proteomics. 11:1105–1122. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ishida S, Huang E, Zuzan H, Spang R, Leone
G, West M and Nevins JR: Role for E2F in control of both DNA
replication and mitotic functions as revealed from DNA microarray
analysis. Mol Cell Biol. 21:4684–4699. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu W, Giangrande PH and Nevins JR: E2Fs
link the control of G1/S and G2/M transcription. EMBO J.
23:4615–4626. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dong QM, Zhang JQ, Li Q, Bracher JC,
Hendricks DT and Zhao XH: Clinical significance of serum expression
of GROβ in esophageal squamous cell carcinoma. World J
Gastroenterol. 17:2658–2662. 2011.
|
|
25
|
Quensel C, Friedrich B, Sommer T, Hartmann
E and Kohler M: In vivo analysis of importin α proteins reveals
cellular proliferation inhibition and substrate specificity. Mol
Cell Biol. 24:10246–10255. 2004.
|
|
26
|
Huang L, Wang HY, Li JD, Wang JH, Zhou Y,
Luo RZ, Yun JP, Zhang Y, Jia WH and Zheng M: KPNA2 promotes cell
proliferation and tumorigenicity in epithelial ovarian carcinoma
through upregulation of c-Myc and downregulation of FOXO3a. Cell
Death Dis. 4:e7452013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ren B, Cam H, Takahashi Y, Volkert T,
Terragni J, Young RA and Dynlacht BD: E2F integrates cell cycle
progression with DNA repair, replication, and G2/M
checkpoints. Genes Dev. 16:245–256. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zannini L, Lecis D, Lisanti S, Benetti R,
Buscemi G, Schneider C and Delia D: Karyopherin-α2 protein
interacts with Chk2 and contributes to its nuclear import. J Biol
Chem. 278:42346–42351. 2003.
|
|
29
|
Narod SA and Foulkes WD: BRCA1 and
BRCA2: 1994 and beyond. Nat Rev Cancer. 4:665–676. 2004.
View Article : Google Scholar
|
|
30
|
Tseng SF, Chang CY, Wu KJ and Teng SC:
Importin KPNA2 is required for proper nuclear localization and
multiple functions of NBS1. J Biol Chem. 280:39594–39600. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sandrock K, Bielek H, Schradi K, Schmidt G
and Klugbauer N: The nuclear import of the small GTPase Rac1 is
mediated by the direct interaction with karyopherin α2. Traffic.
11:198–209. 2010.PubMed/NCBI
|
|
32
|
Ebihara Y, Miyamoto M, Shichinohe T,
Kawarada Y, Cho Y, Fukunaga A, Murakami S, Uehara H, Kaneko H,
Hashimoto H, Murakami Y, Itoh T, Okushiba S, Kondo S and Katoh H:
Overexpression of E2F-1 in esophageal squamous cell carcinoma
correlates with tumor progression. Dis Esophagus. 17:150–154. 2004.
View Article : Google Scholar : PubMed/NCBI
|