Introduction
Osteosarcoma (OS), originating from bone as an
aggressive bone tumor, is an infrequent but the most common and
destructive primary bone tumor in children and adolescents. It is
the second highest cause of cancer-related mortality, mainly due to
the development of often fatal metastasis (1). In the past few decades, surgical
resection therapy has resulted in the poor prognosis of OS
patients. With the application of neoadjuvant chemotherapy in OS,
the five-year survival rate has significantly increased (2,3). To
date, however, the molecular pathogenesis and etiology of OS are
still not clearly elucidated.
Members of the RASSF family (RASSF1-10) have been
reported to participate in a variety of important biological
processes, and in particular have been identified as candidate
tumor suppressors in cancers (4),
of which RASSF5 (also called NORE1) is expressed in most normal
tissues but is downregulated in several cancer cell lines (5). The protein expression of RASSF5 is
frequently downregulated in lung tumor cell lines and primary lung
tumors, impairs cell growth and mediates Ras-dependent apoptosis
(6). RASSF5 has also been
identified as a breakpoint-spanning gene downregulated in clear
cell renal cell carcinoma (CCRCC), and inhibits cell proliferation,
representing a new candidate tumor suppressor for CCRCC (7). Some studies show that no inactivating
somatic mutation for RASSF5 is found in lung tumor lines, but the
RASSF5 promotor region is hypermethylated in primary tumors and
tumor cell lines (8). It is known
that methylation of promotor CpG islands is a common mechanism
inactivating tumor-suppressor genes in cancer. RASSF family genes
including RASSF5 to various degrees are methylated in neuroblastoma
cell lines and primary tumors (9).
Methylation of RASSF5 frequently occurs in squamous cell carcinomas
of the head and neck (10), and
even has the potential for serving as a recurrence biomarker for
bladder cancer (11).
However, few studies indicate that no promotor
methylation of RASSF5 is detected in thyroid tumor (12) and methylation is an uncommon event
in primary thyroid tumors (13).
Moreover, promotor methylation is not the molecular mechanism
responsible for RASSF5 suppression in neuroblastic tumors (14). Thus, to further clarify the function
and molecular mechanism of RASSF5 in cancer, we examined the
expression of RASSF5 in OS tissues by immunohistochemical (IHC)
assay, and investigated the effects of RASSF5 overexpression on
cell growth and invasion in vitro. We hypothesized that
RASSF5 may function as a tumor suppressor implicated in the
development of OS.
Materials and methods
Materials
The OS cell lines (MG-63 and U-2 OS) used for the
experiments were obtained from the Institute of Biochemistry and
Cell Biology (Shanghai, China). Lentiviral-mediated RASSF5 vector
(Lv-RASSF5), negative control vector, and virion-packaging elements
were purchased from GeneChem (Shanghai, China). Human OS tissues
and the corresponding ANCT were collected from the Department of
Orthopedic Surgery, Changzheng Hospital. OS tissue microarray was
constructed by Shanghai Outdo Biotech Co., Ltd. (Shanghai, China).
All the antibodies were purchased from Cell Signaling Technology
(Boston, MA, USA). RASSF5 primer was synthesized by ABI
(Framingham, MA, USA).
Drugs and reagents
Dulbecco’s modified Eagle’s medium (DMEM) and fetal
bovine serum (FBS) were purchased from Thermo Fisher Scientific
Inc. (Waltham, MA, USA); TRIzol reagent and Lipofectamine 2000 were
obtained from Invitrogen (Carlsbad, CA, USA); M-MLV reverse
transcriptase was purchased from Promega (Madison, WI, USA);
SYBR-Green Master Mix was obtained from Takara (Otsu, Japan); and
the ECL Plus kit was obtained from GE Healthcare (Piscataway, NJ,
USA). Cell apoptosis kit [propidium iodide (PI), RNase A, Annexin
V-FITC] was from KeyGen Biology (Nanjing, China).
Clinical samples and data
The tissue microarray was prepared for IHC test.
Human OS tissues and the corresponding ANCT were obtained from
biopsies in a total of 45 consecutive OS cases admitted to our
hospital from January 2005 to December 2011. The baseline
characteristics of the patients before neo-adjuvant chemotherapy
are summarized (Table II). The
study was approved by the Medical Ethics Committee of the Second
Military Medical University, and written informed consent was
obtained from the patients or their parents before sample
collection. Two pathologists respectively reviewed all of the
cases.
 | Table IIAssociation of RASSF5 expression with
clinicopathological factors of the OS patients. |
Table II
Association of RASSF5 expression with
clinicopathological factors of the OS patients.
| | RASSF5
expression | |
|---|
| |
| |
|---|
| Variables | Cases (n) | − | + | P-value |
|---|
| Total | 45 | 27 | 18 | |
| Age (years) | | | | 0.901 |
| <20 | 28 | 17 | 11 | |
| ≥20 | 17 | 10 | 7 | |
| Gender | | | | 0.330 |
| Male | 26 | 14 | 12 | |
| Female | 19 | 13 | 6 | |
| Histology | | | | 0.805 |
| Osteoblastic | 18 | 10 | 8 | |
|
Chondroblastic | 15 | 9 | 6 | |
| Fibroblastic | 7 | 4 | 3 | |
| Others | 5 | 4 | 1 | |
| Ennecking
staging | | | | 0.868 |
| I | 13 | 7 | 6 | |
| II | 24 | 15 | 9 | |
| III | 8 | 5 | 3 | |
| Distant
metastases | | | | 0.010 |
| No | 27 | 12 | 15 | |
| Yes | 18 | 15 | 3 | |
Tissue microarray
The Advanced Tissue Arrayer (ATA-100; Chemicon
International, Temecula, CA, USA) was used to create holes in a
recipient paraffin block and to acquire cylindrical core tissue
biopsies with a diameter of 1 mm from the specific areas of the
‘donor’ block. The tissue core biopsies were transferred to the
recipient paraffin block at defined array positions. The tissue
microarrays contained tissue samples from 45 formalin-fixed
paraffin-embedded cancer specimens with known diagnosis, and
corresponding ANCT from these patients. The block was incubated in
an oven at 45°C for 20 min to allow complete embedding of the
grafted tissue cylinders in the paraffin of the recipient block,
and then stored at 4°C until microtome sectioning.
IHC staining
Tissue microarray sections were processed for IHC
analysis of RASSF5 protein as follows. Immunohistochemical
examinations were carried out on 3-mm sections. For anti-RASSF5
IHC, unmasking was performed with 10 mM sodium citrate buffer, pH
6.0, at 90°C for 30 min. For anti-RASSF5 IHC, antigen unmasking was
not necessary. Sections were incubated in 0.03% hydrogen peroxide
for 10 min at room temperature, to remove endogenous peroxidase
activity, and then in blocking serum [0.04% bovine serum albumin,
A2153; Sigma-Aldrich, Shanghai, China and 0.5% normal goat serum
X0907; Dako Corporation, Carpinteria, CA, USA, in
phosphate-buffered saline (PBS)] for 30 min at room temperature.
Anti-RASSF5 antibody was used at a dilution of 1:200. The antibody
was incubated overnight at 4°C. Sections were then washed three
times for 5 min in PBS. Non-specific staining was blocked with 0.5%
casein and 5% normal serum for 30 min at room temperature. Finally,
staining was developed using diaminobenzidine substrate, and
sections were counterstained with hematoxylin. Normal serum or PBS
was used to replace the anti-RASSF5 antibody in negative
controls.
Quantification of protein expression
The expression of RASSF5 was semi-quantitatively
estimated as total immunostaining scores, which were calculated as
the product of a proportion score and an intensity score. The
proportion and intensity of the staining were evaluated
independently by two observers. The proportion score reflected the
fraction of positive staining cells (0, none; 1, <10%; 2, 10 to
≥25%; 3, >25 to 50%; and 4, >50%), and the intensity score
represented the staining intensity (0, no staining; 1, weak; 2,
intermediate; and 3, strong). Finally, a total expression score was
obtained ranging from 0 to 12. Based on the analysis in advance,
RASSF5 expression was categorized into two groups: low-level RASSF5
expression (score 0–3) and high-level RASSF5 expression (score
4–12). The scoring was independently assessed by two
pathologists.
Cell culture and transfection
OS cells were cultured in DMEM supplemented with 10%
heat-inactivated FBS, 100 U/ml of penicillin, and 100 μg/ml of
streptomycin. Cells in this medium were placed in a humidified
atmosphere containing 5% CO2 at 37°C. Cells were
subcultured at a 1:5 dilution in medium containing 300 μg/ml G418
(an aminoglycoside antibody; a commonly used stable transfection
reagent in molecular genetic testing). On the day of transduction,
OS cells were replated at 5×104 cells/well in 24-well
plates containing serum-free growth medium with Polybrene (5
mg/ml). When reaching 50% confluency, the cells were transfected
with recombinant experimental virus or control virus at the optimal
multiplicity of infection (MOI) of 50, and cultured at 37°C and 5%
CO2 for 4 h. Then the supernatant was discarded and
serum containing growth medium was added. At 4 days of
post-transduction, transfection efficiency was measured by the
frequency of green fluorescent protein (GFP)-positive cells.
Positive and stable transfectants were selected and expanded for
further study. The Lv-RASSF5 vector-infected clone, the negative
control vector-infected and untreated OS cells were respectively
named as the Lv-RASSF5, NC and control (CON) group.
Quantitative real-time PCR
To quantitatively determine the mRNA expression
level of RASSF5 in OS cells, real-time PCR was performed. Total RNA
was extracted from each clone using TRIzol according to the
manufacturer’s protocol. Reverse transcription was carried out
using M-MLV and cDNA amplification was performed using the
SYBR-Green Master Mix kit according to the manufacturer’s
guidelines. The RASSF5 gene was amplified using a specific
oligonucleotide primer, and the β-actin gene was used as an
endogenous control. The PCR primer sequences were as follows:
RASSF5, 5′-TTAGGAAAGAGGAATATTTTAT-3′ and 5′-TAAACCTT
CAACCCTACCTCTTTC-3′; β-actin, 5′-CAACGAATTTGG CTACAGCA-3′ and
5′-AGGGGTCTACATGGCAACTG-3′. Data were analyzed using the
comparative Ct method (2−ΔΔCt). Three separate
experiments were performed for each clone.
Western blot assay
OS cells were harvested and extracted using lysis
buffer (Tris-HCl, SDS, mercaptoethanol and glycerol). Cell extracts
were boiled for 5 min in loading buffer, and then an equal amount
of cell extracts was separated on 15% SDS-PAGE gels. Separated
protein bands were transferred onto polyvinylidene fluoride (PVDF)
membranes, which were subsequently blocked in 5% skim milk powder.
Primary antibodies against RASSF5, p-MST1, p-LATS1, PCNA, MMP-2 and
p53 were diluted according to the manufacturer’s instructions and
incubated overnight at 4°C. Subsequently, horseradish
peroxidase-linked secondary antibodies were added at a dilution of
1:1,000 and incubated at room temperature for 2 h. The membranes
were washed 3 times with PBS, and the immunoreactive bands were
visualized using the ECL Plus kit according to the manufacturer’s
instructions. The relative protein levels in the different cell
lines were normalized to the concentration of β-actin. Three
separate experiments were performed for each clone.
Cell proliferation assay
Cell proliferation was analyzed using the MTT assay.
Briefly, cells infected with Lv-RASSF5 viruses were incubated in
96-well plates at a density of 1×105 cells/well with
DMEM supplemented with 10% FBS. Cells were treated with 20 μl of
MTT dye at 0, 24, 48 and 72 h, and subsequently incubated with 150
μl of DMSO for 5 min. The color reaction was measured at 570 nm
using an enzyme immunoassay analyzer (Bio-Rad, Hercules, CA, USA).
The proliferation activity was calculated for each clone.
Transwell invasion assay
Transwell filters were coated with Matrigel (3.9
μg/μl; 60–80 μl) on the upper surface of a polycarbonate membrane
(diameter, 6.5 mm; pore size, 8 μm). After incubating at 37°C for
30 min, the Matrigel solidified and served as the extracellular
matrix for analysis of tumor cell invasion. Harvested cells
(1×105) in 100 μl of serum-free DMEM were added into the
upper compartment of the chamber. A total of 200 μl of conditioned
medium derived from NIH3T3 cells was used as a source of
chemoattractant, which was placed in the bottom compartment of the
chamber. After 24 h of incubation at 37°C with 5% CO2,
the medium was removed from the upper chamber. The non-invaded
cells on the upper side of the chamber were scraped off with a
cotton swab. Cells that had migrated from the Matrigel into the
pores of the inserted filter were fixed with 100% methanol, stained
with hematoxylin, then mounted and dried at 80°C for 30 min. The
number of cells invading through the Matrigel was counted in 3
randomly selected visual fields from the central and peripheral
portion of the filter by using an inverted microscope (x200
magnification). Each assay was repeated 3 times.
Cell apoptosis analysis
To detect cell apoptosis, OS cells treated with
Lv-RASSF5 were trypsinized, washed with cold PBS and resuspended in
binding buffer according to the instructions of the apoptosis kit.
FITC-Annexin V and PI were added to the fixed cells for 20 min in
darkness at room temperature. Then, Annexin V binding buffer was
added to the mixture before the fluorescence was measured on a
FACsort flow cytometer. The cell apoptosis was analyzed using
CellQuest software (Becton-Dickinson, USA). Three separate
experiments were performed for each clone.
Statistical analysis
SPSS 20.0 was used for the statistical analysis.
Kruskal-Wallis H and Chi-square tests were used to analyze the
expression rate in all groups. One-way analysis of variance (ANOVA)
was used to analyze the differences between groups. The LSD method
of multiple comparisons was used when the probability for ANOVA was
statistically significant. Statistical significance was set at
P<0.05.
Results
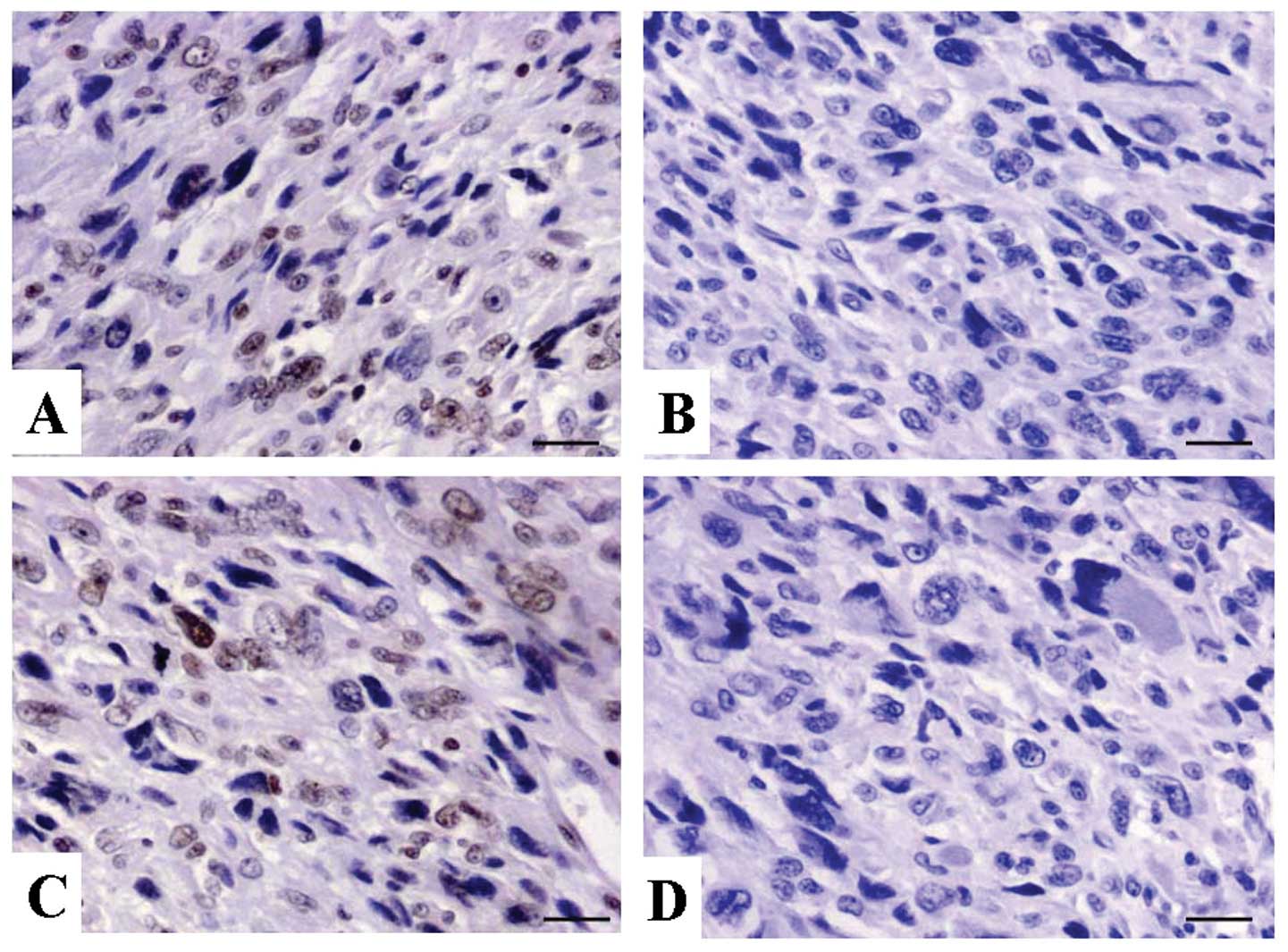
Expression of RASSF5 in OS tissues
The expression of RASSF5 protein was examined using
IHC staining in OS tissues. As shown in Fig. 1, the level of positive expression of
RASSF5 protein was detected in OS and ANCT tissues. Positive-RASSF5
immunostaining was mainly localized in the nucleus of OS tissue
cells. According to the RASSF5 immunoreactive intensity, the
positive expression of RASSF5 in OS tissues was significantly
downregulated compared with that in ANCT (P=0.002) (Table I).
 | Table IExpression of RASSF5 protein in the OS
tissues. |
Table I
Expression of RASSF5 protein in the OS
tissues.
| | RASSF5 protein
(n) | | | | |
|---|
| |
| | | | |
|---|
| Target | Sample | − | + | ++ | +++ | Total | Positive rate
(%) | χ2 | P-value |
|---|
| RASSF5 | OS | 27 | 11 | 5 | 2 | 45 | 40.0 | | |
| ANCT | 12 | 17 | 11 | 5 | 45 | 73.3 | 9.965 | 0.002 |
Association between RASSF5 expression and
clinicopathological characteristics
The association of RASSF5 expression with various
clinicopathological factors was analyzed. As shown in Table II, decreased expression of RASSF5
was closely correlated with distant metastasis of OS patients
(P=0.01). However, no significant correlation was found between
RASSF5 expression and other factors including age, gender of the
patients, and histology and Ennecking staging of the tumor
(P>0.05, respectively).
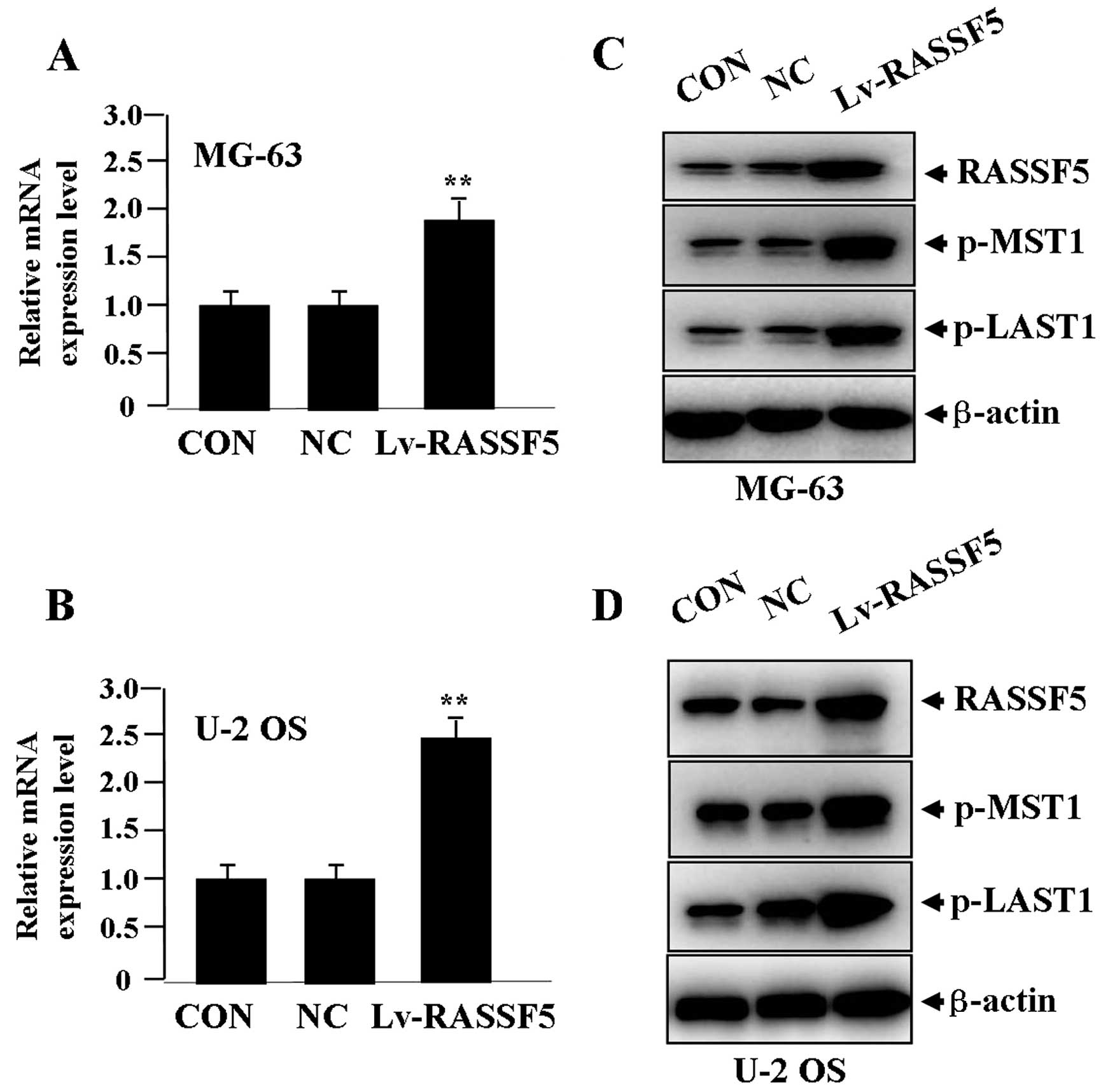
Effect of RASSF5 overexpression on the
expression of MST1 and LATS1
After Lv-RASSF5 (MOI=50) was transfected into OS
cells for 24 h, the expression levels of RASSF5 mRNA (Fig. 2A and B) and protein (Fig. 2C and D), and p-MST1 and p-LATS1
proteins (Fig. 2C and D) were
detected by real-time PCR and western blot assays, indicating that,
when RASSF5 expression was significantly upregulated, p-MST1 and
p-LATS1 expression was also increased in the Lv-RASSF5 group when
compared with the NC and CON groups in both cell lines.
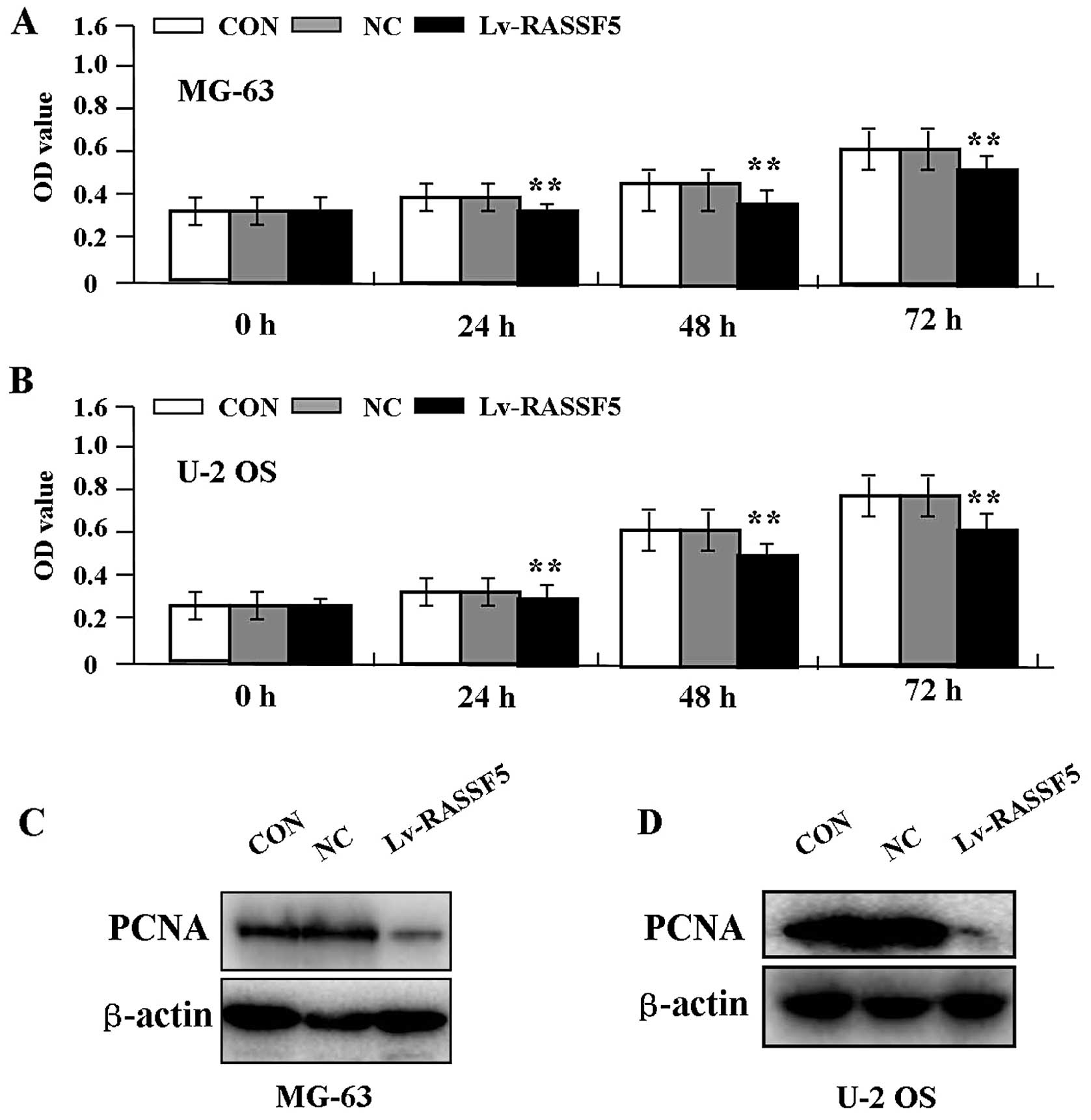
Effect of RASSF5 overexpression on cell
proliferation
Deregulated cell proliferation is a hallmark of
cancer. To verify the effect of RASSF5 overexpression on tumor
growth in OS cells, we examined cell proliferative activities by
MTT assay. The results showed that RASSF5 overexpression markedly
diminished the proliferative activities of OS cells in a
time-dependent manner compared to the NC and CON groups (Fig. 3A and B, P<0.01). In addition, the
expression level of PCNA protein, examined by western blot assay
(Fig. 3C and D), was found to be
significantly downregulated in the Lv-RASSF5 group when compared
with the NC and CON groups in both cell lines.
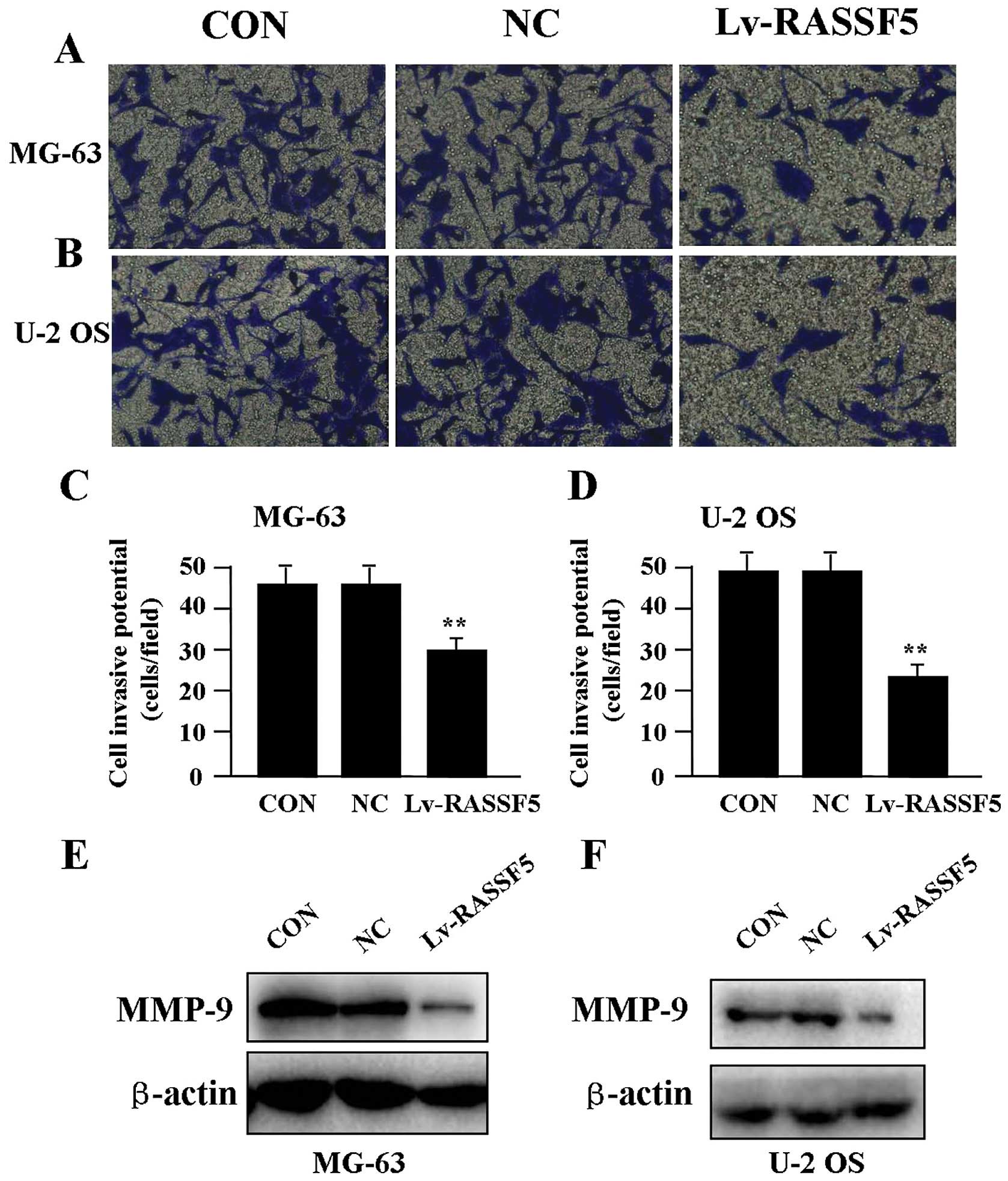
Effect of RASSF5 overexpression on cell
invasion
To determine the effect of RASSF5 overexpression on
cell invasion, a Transwell assay was performed. The invasive
potential of OS cells in the Transwell assay was determined by the
ability of cells to invade a matrix barrier containing laminin and
type IV collagen, major components of the basement membrane.
Representative micrographs of Transwell filters are shown in
Fig. 4A and B. It was found that
the invasive potential of OS cells was apparently weakened in the
Lv-RASSF5 groups compared to that in the NC and CON groups in both
cell lines (P<0.01) (Fig. 4C and
D). In addition, the expression level of MMP-9 protein,
examined by western blot assay (Fig. 4E
and F), was found significantly downregulated in Lv-RASSF5
compared with the NC group.
Effect of RASSF5 overexpression on cell
apoptosis
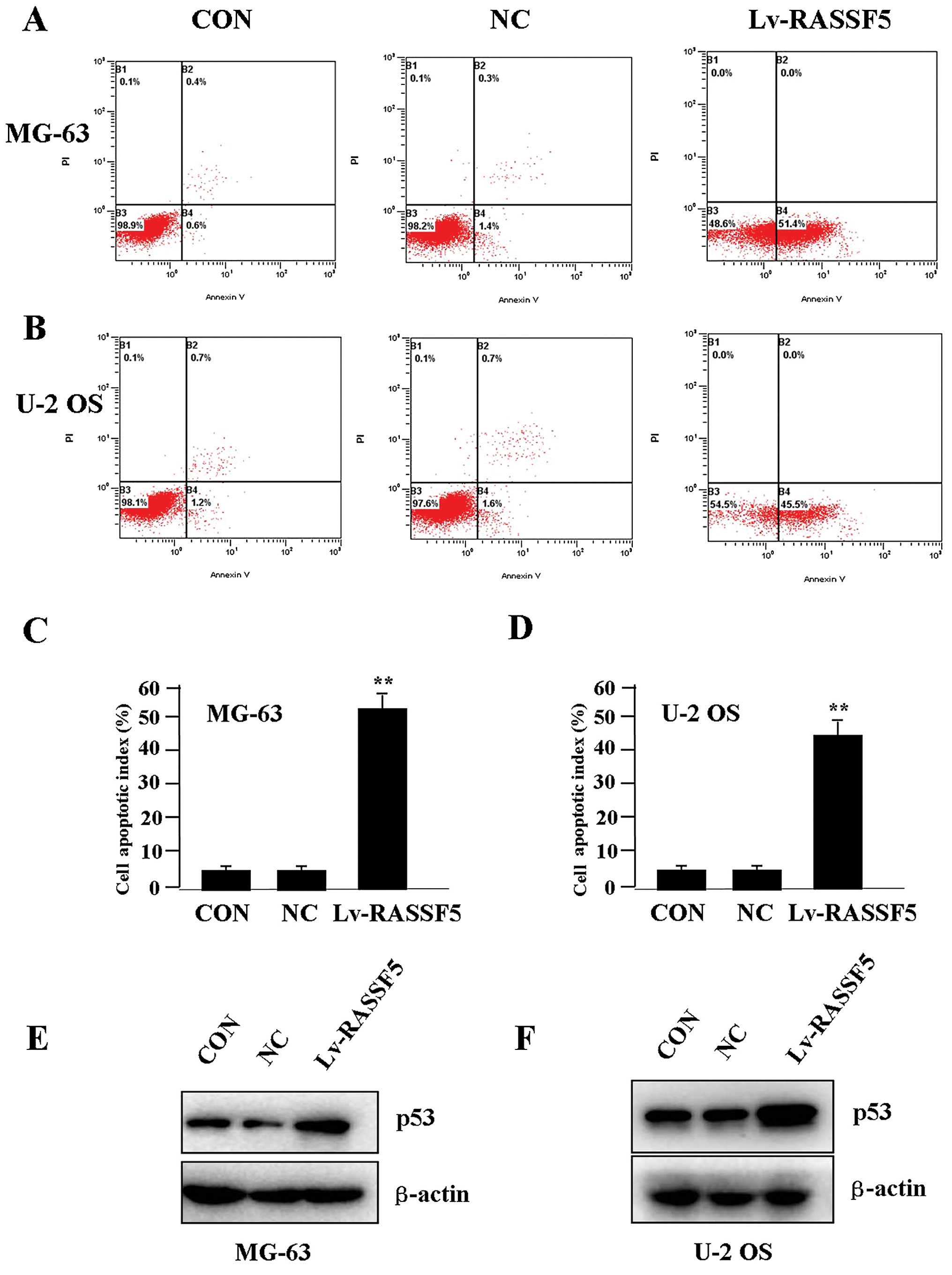
To determine whether RASSF5 overexpression
influences OS cell apoptosis, flow cytometric analysis with
PI/FITC-Annexin V staining was performed. It was shown that the
apoptosis indices of the OS cells in the Lv-RASSF5 group were
markedly higher than indices in the NC group in both cell lines
(Fig. 5A–D). Additionally, p53
pathway initiates DNA repair, cell-cycle arrest, senescence and
importantly, apoptosis (16). To
determine whether RASSF5 overexpression regulates the expression of
p53 protein, western blotting was carried out. It was found that
the protein level of p53 was increased in the Lv-RASSF5 group in
comparison with the NC group in both cell lines (Fig. 5E and F).
Discussion
The RASSFs comprise 10 members, RASSF1 to RASSF10,
and are implicated in various cellular mechanisms including cell
apoptosis, cell cycle distribution and metastases, of which RASSF5
has been reported to function as a tumor suppressor (17). Studies have shown that the candidate
tumor-suppressor gene RASSF5 is epigenetically downregulated and
plays a role in pathogenetic and prognostic significance in
hepatocellular carcinoma (HCC) (18,19).
It is lowly expressed in non-small cell lung carcinoma (NSCLC)
which implies a key preventive role of RASSF5 against the
carcinogenesis of NSCLC (20).
RASSF5 is frequently silenced mainly by aberrant promoter
methylation, and is associated with tumor metastasis in CRC,
implicating RASSF5 as a potential biomarker for the development of
CRC (21). Yet, a limited amount of
data have been reported concerning the expression of RASSF5 in OS
tissues. Our present study demonstrated that RASSF5, mainly
localized in the nucleus, was markedly downregulated in OS tissues
when compared to the adjacent non-tumor tissues, and is negatively
associated with distant metastases of the tumor, suggesting that
loss of RASSF5 may represent a new biomarker involved in the
development of OS.
Although RASSF5 may serve as a tumor-suppressor
gene, the functions of RASSF5 are largely unknown. A
loss-of-function experiment revealed that loss of RASSF5 leads to
uncontrolled growth and transformation of HCC, but overexpression
of RASSF5 suppresses cell replication and transformation (22). RASSF5 is suppressed in
pheochromocytoma and abdominal paraganglioma, resulting in enhanced
apoptosis and impaired colony formation (23). It also decreased cellular growth and
induced cell apoptosis, implicated in the blockade of malignant
progression of colorectal tumors (24). Thus, to further clarify the role of
RASSF5 in cancer, we assessed the function of RASSF5 in the
biological behaviors of OS cells, and found that overexpression of
RASSF5 suppressed growth and invasion, and induced cell apoptosis
in OS cells, indicating that RASSF5 may serve as a promising
therapeutic target for the treatment of OS.
Furthermore, RASSF1A interacts with the
pro-apoptotic kinase MST1, through activation of MST1 by promoting
MST1 autophosphorylation and LATS2 phosphorylation (25), suggesting RASSF1A as part of the
MST1/LATS1 signaling pathway, which is a major conserved mechanism
governing cell contact inhibition, organ size control, and cancer
development (26,27). RASSF binds to MST1, regulates
mammalian cell proliferation (28),
and triggers cell apoptosis via activation of mammalian MST1
(29). However, a few studies have
demonstrated that MST1 exhibits a growth promoting activity in HCC
cells (30), and LASSF5 suppresses
the growth of lung cancer cells independent of MST1/2 kinase
(31). To demonstrate the
regulation of RASSF5 on MST1/LATS1 signaling in cancer, we found
that RASSF5 inhibited the growth and invasion of OS cells with
increased expression of p-MST1 and p-LATS1, suggesting that RASSF5
may function as a tumor suppressor in OS through activation of
MST1/LATS1 signaling.
Evidence indicates that a number of biomarkers
including PCNA (32), MMP-9
(33) and p53 (34) are responsible for tumor development
and prognosis in a variety of cancers. MTS1 can enhance
p53-dependent apoptosis of tumor cells, leading to the decreased
growth of tumor cells (35). The
present study revealed that overexpression of RASSF5 downregulated
the expression of PCNA and MMP-9 and upregulated the expression of
p-MST1, p-LATS1 and p53 in OS cells, suggesting that RASSF5 may
inhibit growth and invasion via downregulation of PCNA and MMP-9
expression, and enhance pro-apoptotic effects through MST1-mediated
p53 expression in OS cells.
In conclusion, our findings demonstrate that
downregulation of RASSF5 expression is correlated with distant
metastasis of OS tissues, and overexpression of RASSF5 may function
as a tumor suppressor in OS cells through activation of the
MST1/LATS1 pathway, suggesting that RASSF5 may serve as a potential
therapeutic target for the treatment of OS.
References
|
1
|
Ta HT, Dass CR, Choong PF and Dunstan DE:
Osteosarcoma treatment: state of the art. Cancer Metastasis Rev.
28:247–263. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tan ML, Choong PF and Dass CR:
Osteosarcoma: conventional treatment vs. gene therapy. Cancer Biol
Ther. 8:106–117. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar
|
|
4
|
Chan JJ, Flatters D, Rodrigues-Lima F, et
al: Comparative analysis of interactions of RASSF1-10. Adv Biol
Regul. 53:190–201. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tommasi S, Dammann R, Jin SG, et al:
RASSF3 and NORE1: identification and cloning of two
human homologues of the putative tumor suppressor gene
RASSF1. Oncogene. 21:2713–2720. 2002. View Article : Google Scholar
|
|
6
|
Vos MD, Martinez A, Ellis CA, et al: The
pro-apoptotic Ras effector Nore1 may serve as a Ras-regulated tumor
suppressor in the lung. J Biol Chem. 278:21938–21943. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen J, Lui WO, Vos MD, et al: The t(1;3)
breakpoint-spanning genes LSAMP and NORE1 are
involved in clear cell renal cell carcinomas. Cancer Cell.
4:405–413. 2003.PubMed/NCBI
|
|
8
|
Hesson L, Dallol A, Minna JD, et al:
NORE1A, a homologue of RASSF1A tumour suppressor gene
is inactivated in human cancers. Oncogene. 22:947–954. 2003.
View Article : Google Scholar
|
|
9
|
Djos A, Martinsson T, Kogner P and Carén
H: The RASSF gene family members RASSF5, RASSF6 and RASSF7 show
frequent DNA methylation in neuroblastoma. Mol Cancer. 11:402012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Steinmann K, Sandner A, Schagdarsurengin U
and Dammann RH: Frequent promoter hypermethylation of tumor-related
genes in head and neck squamous cell carcinoma. Oncol Rep.
22:1519–1526. 2009.PubMed/NCBI
|
|
11
|
Meng W, Huebner A, Shabsigh A, et al:
Combined RASSF1A and RASSF2A promoter methylation
analysis as diagnostic biomarker for bladder cancer. Mol Biol Int.
2012:701812012.
|
|
12
|
Foukakis T, Au AY, Wallin G, et al: The
Ras effector NORE1A is suppressed in follicular thyroid
carcinomas with a PAX8-PPARγ fusion. J Clin Endocrinol
Metab. 91:1143–1149. 2006.
|
|
13
|
Nakamura N, Carney JA, Jin L, et al:
RASSF1A and NORE1A methylation and
BRAFV600E mutations in thyroid tumors. Lab
Invest. 85:1065–1075. 2005. View Article : Google Scholar
|
|
14
|
Geli J, Kogner P, Lanner F, et al:
Assessment of NORE1A as a putative tumor suppressor in human
neuroblastoma. Int J Cancer. 123:389–394. 2008.
|
|
15
|
Hanahan D and Weinberg RA: The hallmarks
of cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vazquez A, Bond EE, Levine AJ and Bond G:
The genetics of the p53 pathway, apoptosis and cancer therapy. Nat
Rev Drug Discov. 7:979–987. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Richter AM, Pfeifer GP and Dammann RH: The
RASSF proteins in cancer; from epigenetic silencing to functional
characterization. Biochim Biophys Acta. 1796:114–128.
2009.PubMed/NCBI
|
|
18
|
Macheiner D, Heller G, Kappel S, et al:
NORE1B, a candidate tumor suppressor, is epigenetically silenced in
human hepatocellular carcinoma. J Hepatol. 45:81–89. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Calvisi DF, Evert M and Dombrowski F:
Pathogenetic and prognostic significance of inactivation of RASSF
proteins in human hepatocellular carcinoma. Mol Biol Int.
2012:8498742012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shinmura K, Tao H, Nagura K, et al:
Suppression of hydroxyurea-induced centrosome amplification by
NORE1A and down-regulation of NORE1A mRNA expression in non-small
cell lung carcinoma. Lung Cancer. 71:19–27. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fernandes MS, Carneiro F, Oliveira C and
Seruca R: Colorectal cancer and RASSF family - a special emphasis
on RASSF1A. Int J Cancer. 132:251–258. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Macheiner D, Gauglhofer C, Rodgarkia-Dara
C, et al: NORE1B is a putative tumor suppressor in
hepatocarcinogenesis and may act via RASSF1A. Cancer Res.
69:235–242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Geli J, Kiss N, Lanner F, et al: The Ras
effectors NORE1A and RASSF1A are frequently
inactivated in pheochromocytoma and abdominal paraganglioma. Endocr
Relat Cancer. 14:125–134. 2007.
|
|
24
|
Lee CK, Lee JH, Lee MG, et al: Epigenetic
inactivation of the NORE1 gene correlates with malignant
progression of colorectal tumors. BMC Cancer. 10:5772010.PubMed/NCBI
|
|
25
|
Guo C, Tommasi S, Liu L, et al: RASSF1A is
part of a complex similar to the Drosophila
Hippo/Salvador/Latstumor-suppressor network. Curr Biol. 17:700–705.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zeng Q and Hong W: The emerging role of
the hippo pathway in cell contact inhibition, organ size control,
and cancer development in mammals. Cancer Cell. 13:188–192. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chan SW, Lim CJ, Chen L, et al: The Hippo
pathway in biological control and cancer development. J Cell
Physiol. 226:928–939. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Avruch J, Praskova M, Ortiz-Vega S, et al:
Nore1 and RASSF1 regulation of cell proliferation and of the MST1/2
kinases. Methods Enzymol. 407:290–310. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ehrkamp A, Herrmann C, Stoll R and Heumann
R: Ras and rheb signaling in survival and cell death. Cancers.
5:639–661. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ng YK, Lau WS, Lui VW, et al: Full-length
Mst1 exhibits growth promoting function in human hepatocellular
carcinoma cells. FEBS Lett. 587:496–503. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Aoyama Y, Avruch J and Zhang XF: Nore1
inhibits tumor cell growth independent of Ras or the MST1/2
kinases. Oncogene. 23:3426–3433. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Naryzhny SN: Proliferating cell nuclear
antigen: a proteomics view. Cell Mol Life Sci. 65:3789–3808. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brinckerhoff CE, Rutter JL and Benbow U:
Interstitial collagenases as markers of tumor progression. Clin
Cancer Res. 6:4823–4830. 2000.PubMed/NCBI
|
|
34
|
Trieb K and Kotz R: Proteins expressed in
osteosarcoma and serum levels as prognostic factors. Int J Biochem
Cell Biol. 33:11–17. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Grigorian M and Lukanidin E: Activator of
metastasis in cancer cells, Mst1/S100A4 protein binds to tumor
suppressor protein p53. Genetika. 39:900–908. 2003.(In
Russian).
|