Introduction
Cytosine methylation is an epigenetic function
involved in the control of the gene expression of eukaryotic cells.
In mammalian cells, ~3–5% of the cytosine residues in genomic DNA
are methylated, 70–80% of which are found in CpG dinucleotide-rich
regions known as CpG islands (1–5). In
normal cells, CpG methylation patterns in genomes are established
and maintained precisely after DNA replication via the action of
the DNA methyltransferases (DNMT) (4–7).
However, cancer cells have two contrasting features: genome-wide
hypomethylation and specific CpG island hypermethylation (7–9). It is
also known that hypermethylation of tumor-suppressor genes causes
inactivation and facilitates gene mutation associated with allelic
loss (1,7,10).
The enzymatic methylation machinery is composed of
catalytically active DNMTs, including DNMT1, DNMT3a and DNMT3b.
DNMT1 is the most abundant DNMT and has preference for
hemimethylated DNA substrates. It is responsible for copying the
methylation pattern after each round of DNA replication (6,11).
DNMT3a and DNMT3b (also known as de novo methytransferases)
target unmethylated DNA and are essential for embryonic development
(6,11,12).
Increased mRNA and protein expression levels of the three DNMTs
have been reported in the majority of human cancers (7,11,13–15).
It is commonly described that the hypermethylation of
tumor-suppressor genes correlates with a higher expressiono of
DNMTs (1,9,12). We
hypothesized that the overexpression of the enzymes is an early
event that precedes the appearance of preneoplastic lesions and
tumors. Deleterious mutations in DNMT1 or DNMT3b have not been
described thus far in human tumors (16). Although deleterious mutations in
DNMT3a are not common in cancer, they have been associated with
poor prognosis (17). To gain
insight into the specific contribution of each DNMT to
carcinogenesis, several animal models have been studied in the
absence of one of the methyltransferases with contrasting results.
In murine models, it has been shown that heterozygous mutations for
DNMT1 caused a global reduction in DNA methylation and a decrease
in the development of intestinal tumors, while simultaneously
enhancing lymphomagenesis (18). It
has also been reported that deletion of DNMT3a in a K-ras-dependent
murine lung cancer model, promoted tumor progression (16), whereas the loss of DNMT3b
accelerated the lymphomagenesis in a murine model of MYC-induced
cancer (18,19).
Animal models also allow the analysis of stages
prior to tumor formation. The early stages of carcinogenesis are
characterized by the presence of altered cells foci or
preneoplastic lesions (20,21). Although the analysis of these
lesions are potentially useful for the identification of early
genetic and epigenetic events leading to cancer in human beings,
such dysplastic nodules are difficult to detect in biopsies and
commonly coexist with other tissue pathologies (20). Thus, animal models for
carcinogenesis have proven to be important tools for the analysis
of nodules and cancer progression (22). The resistant hepatocyte model (RHM)
consistently reproduces the development of hepatocellular carcinoma
(HCC) in male rats (23–25). The hepatic chemocarcinogenesis
induced in the rat share several morphological, biochemical and
molecular characteristics with human HCC (21,26).
Since neoplastic pathology is a complex process, it has been
divided into three phases: initiation (exposure to a carcinogen),
promotion (initiated cell growth and the formation of altered cell
foci) and progression (establishment of preneoplastic lesions and
futher dedifferentiation to tumors). These phases can be
distinguished in the RHM (20,24,25). A
necrogenic dose of diethylnitrosamine induces DNA damage and
oxidative stress in hepatocytes during cancer initiation. The
initiated cells can be stimulated to develop into altered
hepatocyte foci (AHF) and nodules by a proliferative stimulus that
consists of the administration of carcinogen 2-acetylaminofluorene
in combination with partial hepatectomy (promotion). The AHF
accumulates genomic damage and altered genetic expression
ultimately progressing into HCC without any additional treatment
with the carcinogen (progression) (20,24,25).
In the present study, we investigated the time course expression of
the three DNMTs and the methylation patterns of three tumor-related
genes exploiting the synchronous development of preneoplastic
lesions and tumors in the RHM. We analyzed the methylation of
timp3, rassf1a and p16. It has been suggested
that the downregulation of these genes enhances cancer development.
Timp3 irreversibly inhibits metalloproteinases and is thought to be
inactivated in cancer to facilitate metastasis (8). Rassf1a inhibits cell proliferation by
negatively regulating the G1/S-phase transition and enhances murine
double minute 2 (MDM2) self-ubiquitination in response to DNA
damage (27,28). p16 is an inhibitor of cyclin
D-dependent protein kinases, and its inactivation results in the
cell being unable to halt the cell cycle in the G1 phase (28–30).
These genes were also selected as they have been found to be
frequently hypermethylated in several human and rat cancers,
including HCC (7,8), and thus may be useful to determine
whether methylation changes occurred in our model. The RHM
exhibited a transient expression of the de novo DNMTs as
part of the carcinogenic process. Our results also demonstrated
that the RHM is a suitable model for the analysis of dynamic
methylation changes.
Materials and methods
Animals and treatments
All the experiments followed the Institutional
Animal Care and Use Committee Guidelines and the protocols were
performed in accordance with and approved by the Comité Interno
para el Cuidado y Uso de Animales de Laboratorio (CICUAL) of
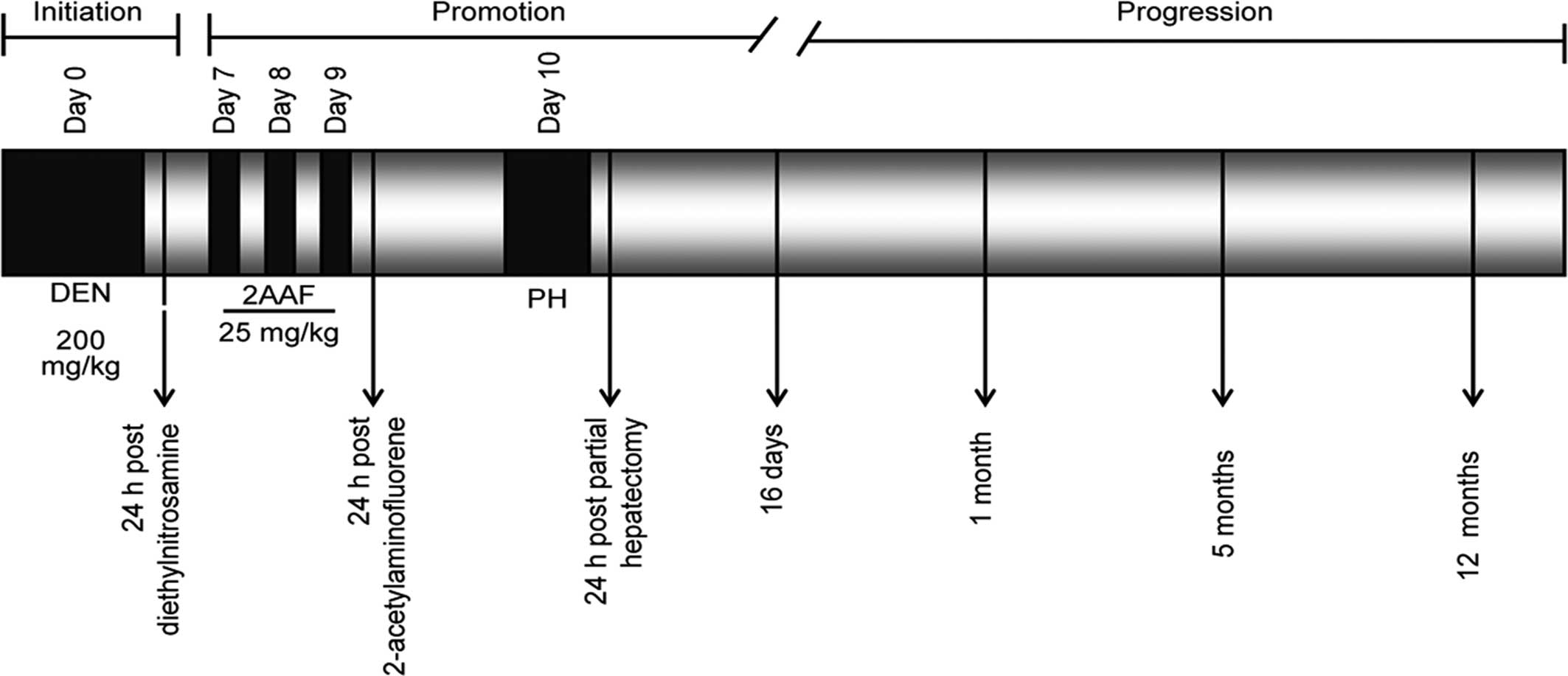
Cinvestav (Permit Number: 0001-02). Male F344 rats weighing 180–200
g (UPEAL-Cinvestav, Mexico, DF, Mexico) were subjected to a 10-day
carcinogen treatment. The rats were initiated with an
intraperitoneal dose of diethylnitrosamine (200 mg/kg).
2-Acetylaminofluorene was administered by gavage at a dose of 25
mg/kg during three consecutive days, beginning on day 7 after
initiation. On day 10, the rats were subjected to partial
hepatectomy. The rats were fed with a germ-free diet and water
ad libitum in a pathogen-free environment. The animals were
housed in an air-ventilated room whose temperature was maintained
at 24°C and a 12-h light/dark cycle. Groups of 4–5 animals were
sacrificed by exsanguination under ether anesthesia at different
time periods from 24 h after initiation up to 12 months as
described in Fig. 1. Livers were
excised, washed in physiological saline solution, frozen in
2-methylbutane with liquid nitrogen and stored at −70°C.
Histological analysis
The putative preneoplastic lesions and tumors in rat
livers were identified with the histoenzymatic staining of the
γ-glutamil transpeptidase (GGT). Briefly, 4 μm sections were cut
from frozen liver tissue and mounted on poly-L-lysine-coated glass
slides. The tissue sections were fixed in acetone for 5 min at 4°C,
followed by the addition of 0.3 mg/ml
α-glutamyl-4-methoxy-2-naphthylamine, 2 mg/ml glycyl-glycine and
0.5 mg/ml Fast Blue BB salt in Tris base (100 mM) solution for 10
min at room temperature. GGT-positive tissue was used as a guide to
dissect preneoplastic lesions and tumors with a stainless steel
cork borer (internal diameter, 1 mm) from the frozen tissues.
Separate slides were fixed and counterstained with hematoxylin and
eosin (H&E). Representative images were captured by optical
microscopy (Olympus 1 70; Olympus Europa GmbH, Hamburg,
Germany).
Methylation-specific PCR (MSP) and
bisulfite sequencing PCR (BSP)
Genomic DNA purified from the liver of control and
experimental animals underwent bisulfite modification according to
the instructions for the EZ DNA Methylation kit (Zymo Research).
MSP primers (M, methylated; U, unmethylated primers) were designed
as previously reported (8).
Products were amplified as follows: M-timp3 and
U-timp3: 40 cycles at 94°C for 30 sec, 57.5°C for 45 sec and
72°C for 45 sec; M-rassf1a: 40 cycles at 94°C for 30 sec,
55°C for 45 sec and 72°C for 45 sec; U-rassf1a: 30 cycles at
94°C for 30 sec, 50°C for 30 sec and 72°C for 45 sec; M-p16
and U-p16: 40 cycles at 94°C for 30 sec, 52.5°C for 30 sec
and 72°C for 45 sec; in all the cases, an initial denaturation step
at 94°C for 10 min and final extension at 72°C for 10 min were
included. Fully methylated genomes (positive controls) were
obtained by the in vitro methylation of genomic DNA derived
from non-treated rats with Sss1 methylase according to the
manufacturer’s instructions (New England Biolabs) and subsequent
bisulfite conversion. BSP of timp3 was performed in a 20-μl
reaction mixture under the conditions: initial denaturation at 94°C
for 10 min; 40 cycles at 94°C for 30 sec, 60°C for 45 sec, and 72°C
for 45 sec; and a final extension at 72°C for 10 min. The primer
sequence for timp3 (GenBank: AC136645) BSP was: sense:
5′-AAGGGGAAATTTTTTTTGAGGTTTT-3′ and antisense:
5′-CCCCTCTAACCAATAACAACCC-3′. Amplified products were visualized by
2% agarose gel electrophoresis. PCR amplicons were purified using
isopropanol precipitation and then sequenced in reverse direction,
from at least three independent amplification products. Purified
DNA was diluted and cycle-sequenced using the ABI BigDye Terminator
kit v3.1 (Applied Biosystems, Foster City, CA, USA) according to
the manufacturer’s instructions. Sequencing reactions were
electrophoresed on an ABI 3100 genetic analyzer. Electropherograms
were analyzed for the presence of methylated or unmethylated CpG
islands. The set of primers were designed by using Methyl Primer
Express v1.0 software (Life Technologies Corp.). The CpG islands
were verified using the Methyl Primer Express v1.0 software (GC
percentage <55%, observed-to-expected CpG ratio <65%).
Semi-quantitative RT-PCR
RNA was extracted from frozen liver tissue using
TriPure Isolation reagent according to the manufacturer’s
instructions (Roche). cDNA synthesis and PCR were performed in One
Step (SuperScript One-Step RT-PCR; Invitrogen). Appropriate primers
(0.2 mM each) and 150 ng of total RNA per 20 μl of reaction were
used. The absence of DNA contamination was verified by PCR assay.
The cycling parameters used were: 45 min at 45°C for cDNA
synthesis, followed by 2 min at 94°C; denaturing for 15 sec at
94°C, annealing for 30 sec between 50°C and 55°C, followed by 30
sec at 72°C; and final elongation for 10 min at 72°C. An adequate
number of cycles corresponding to exponential amplification were
performed to avoid saturated products with a kinetic analysis of
20–35 cycles for each gene. Primer set sequences were: for
timp3 (GenBank: NM_012886): sense,
5′-GCGTGTATGAAGGCAAGATG-3′ and antisense,
5′-GGTCACAAAGCAAGGCAAGT-3′; for rassf1a (GenBank:
NM_001037555): sense, 5′-CCGCACCTCTTTTTACTTGC-3′ and antisense,
5′-GGCGTTCAGTTCGTTCAAA-3′; for p16 (GenBank: NM_031550):
sense, 5′-TACCCCGATACAGGTGATGA-3′ and antisense,
5′-TCGTGATGTCCCCGCTCTA-3′; for dnmt1 (GenBank: NM_053354)
sense, 5′-CGGATTGGTCGGATAAAAGA-3′ and antisense,
5′-GCTTCCTCATCGCTCCAGTA-3′; for dnmt3a (GenBank:
NM_001003958): sense, 5′-GGAGAGGAAAGGGAGAGAGG-3′ and antisense,
5′-AGGGATGGTGCTGTTGAGAC-3′; for dnmt3b (GenBank: NM_031144):
sense, 5′-AAACCCAACAACAAGCAACC-3′ and antisense,
5′-ACATCAGAAGCCATCCGTTC-3′; and for β-actin (GenBank: NM_031144):
sense, 5′-CCTCTATGCCAACACAGTGC-3′ and antisense,
5′-CATCGTACTCCTGCTTGCTG-3′.
Immunohistochemical staining
Tissue sections (4 μm) mounted on
poly-L-lysine-coated glass slides were obtained as previously
described. Briefly, the frozen tissue sections were fixed in a 0.2%
glutaraldehyde solution (31).
Endogeneous peroxidase activity was blocked by incubating sections
with 3% H2O2. Following treatment with 1%
bovine albumin for 10 min to block non-specific protein binding
sites, rabbit polyclonal antibodies for DNMT1 (sc-20701, dilution
1:25; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), DNMT3a
(sc-20703, dilution 1:25; Santa Cruz Biotechnology, Inc.) and
DNMT3b (sc-20704, dilution 1:25; Santa Cruz Biotechnology, Inc.)
were incubated overnight at 4°C. Antigen-antibody complexes were
visualized using standard staining protocol (LSAB Plus-Kit; Dako).
Slides were counterstained with methyl green for 1 h to visualize
nuclei. Representative images were captured by optical microscopy
(Olympus 170; Olympus Europa GmbH).
Statistical analysis
The experiments were repeated at least three times.
Results are reported as means ± SEM. Statistical significance was
determined using t-test with P<0.05 as the level of
significance. Statistical analysis was performed using Prism 4
(GraphPad Software, La Jolla, CA, USA). In order to quantify the
methylation changes in the BSP, values of 1, 0.5 and 0 were
assigned to a methylated cytosine, hemimethylated and unmethylated
cytosine, respectively, and a multinomial distribution analysis was
performed. Briefly, the sum (according to the values assigned) and
the distribution of three possible states (methylated,
hemimethylated and unmethylated) in the treated groups were
compared against the non-treated samples. Statistical significance
was determined when the P-value was <0.05 (Computer Software
Microsoft Excel; Microsoft Redmond, WA, USA).
Results
Preneoplastic lesions and tumor
detection
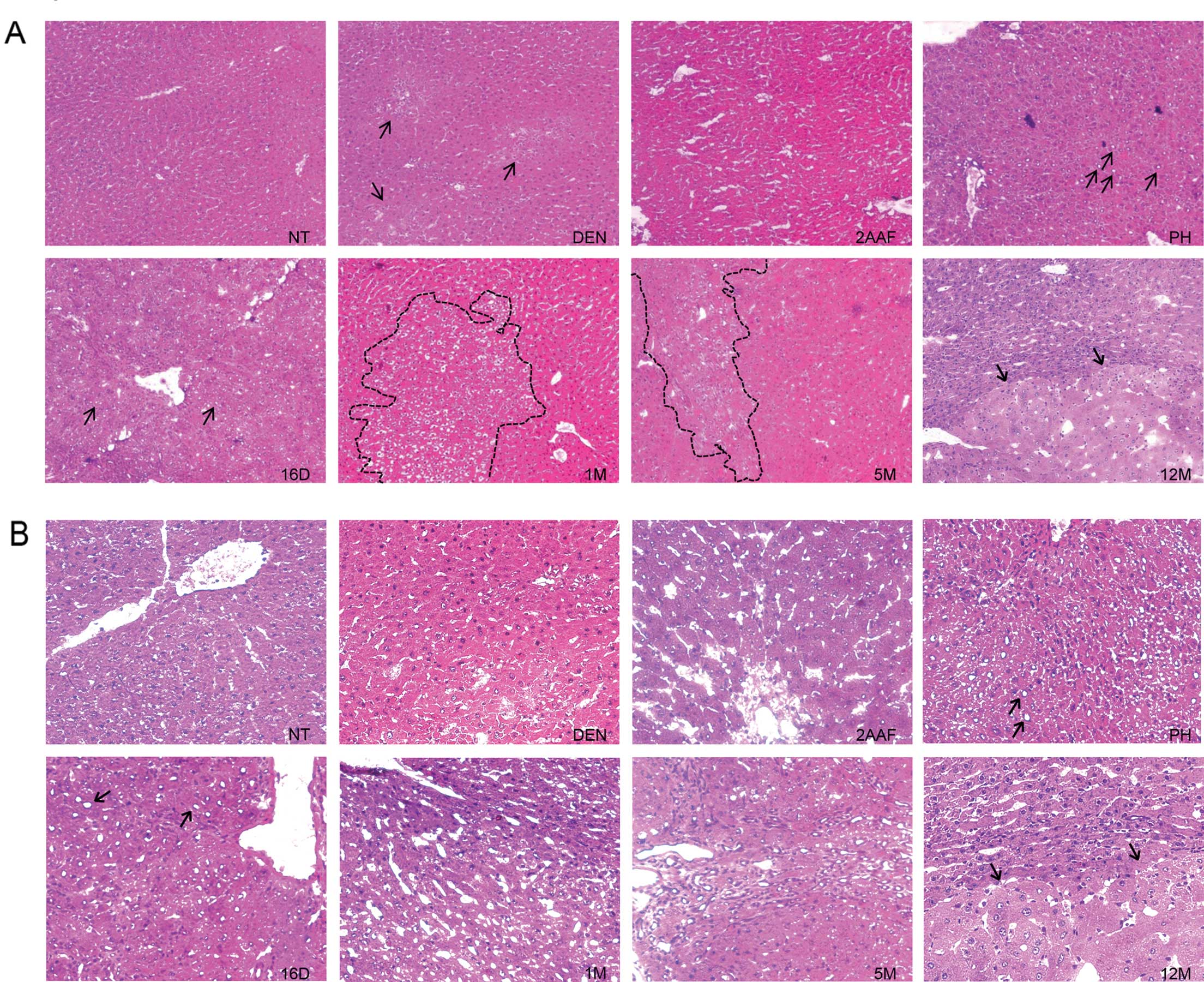
The hepatic tissue throughout the RHM was analyzed.
Changes in the morphology of the liver due to the carcinogenic
treatment, including necrosis, was detected 24 h after
diethylnitrosamine administration by H&E staining. Hepatocytes
with nuclear atypia were initially observed 24 h after
2-acetylaminofluorene administration, and persisted 24 h after
partial hepatectomy and 16 days after complete treatment, but
disappeared at 1 month. Hepatocyte nodules and tumor lesions were
identified by H&E staining (Fig.
2) and by determining the presence of the GGT marker (Fig. 3). Numerous nodules were uniformly
stained for GGT activity at 1 month. These nodules contained
hepatocytes with minimal nuclear atypia. The number of nodules
decreased from 1 to 5 months, which is associated with the presence
of remodeling and persistent nodules, as described by Enomoto and
Farber (32). HCC were found after
12 months. These lesions exhibited increased cytoplasmic size of
hepatocytes, nuclear atypia, and anisonucleosis (Fig. 2).
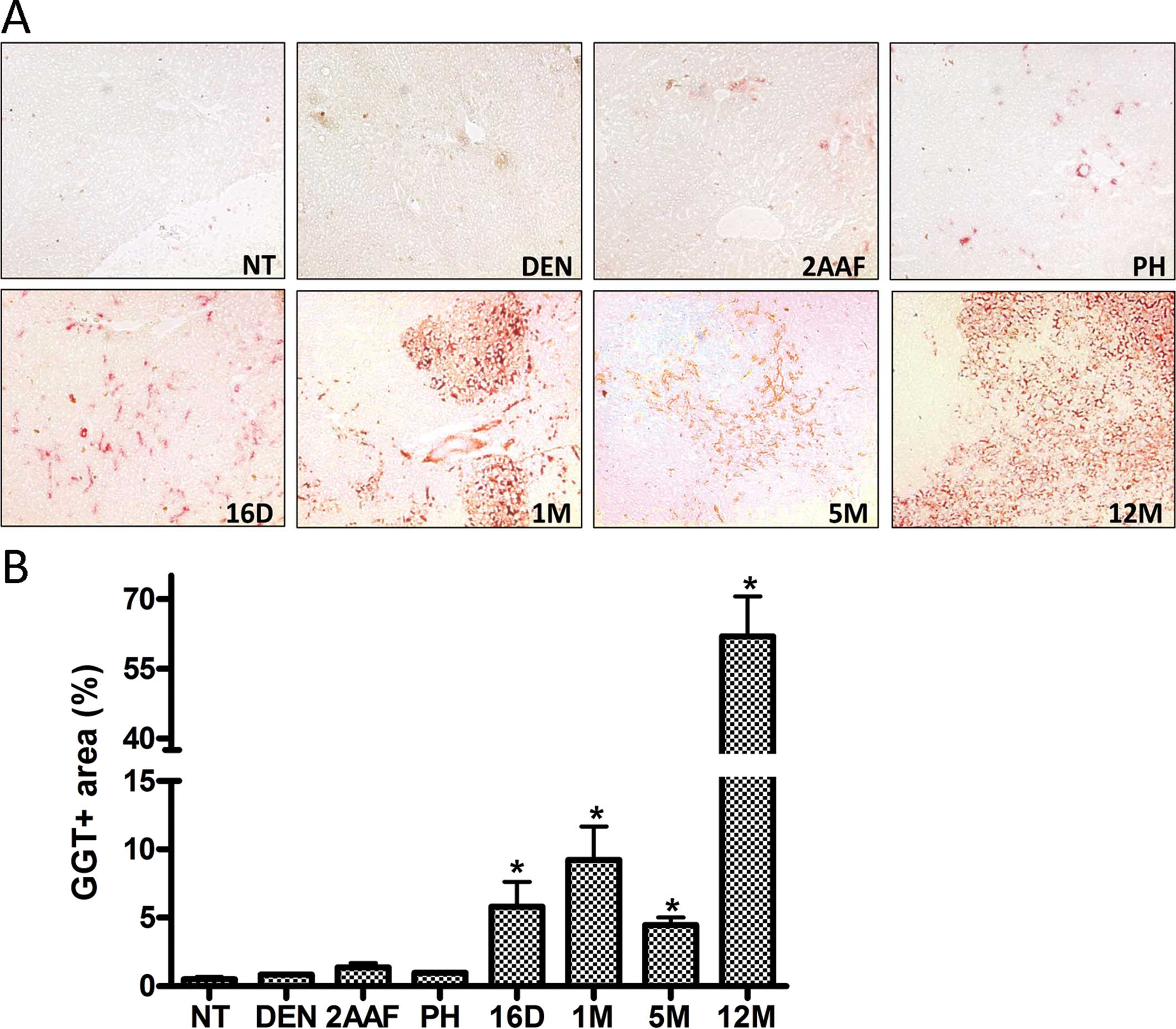 | Figure 3Preneoplastic lesions and tumors
identified by GGT activity. (A) Representative histological
sections from the livers of non-treated rats (NT) and rats
sacrificed 24 h after administration of DEN (DEN), 24 h after
administration of 2-AAF (2AAF), 24 h after partial hepatectomy
(PH); and 16 days (16D), 1 month (1M), 5 months (5M) and 12 months
(12M) after complete carcinogenic treatment. Images are shown at a
magnification of ×10. (B) Percentage of positive GGT area in NT-non
treated rats, DEN, 24 h after administration of DEN; 2AAF, 24 h
after the last administration of 2-AAF (2AAF); PH, 24 h after
partial hepatectomy; and 16D, 16 days; 1M, 1 month; 5M, 5 months;
and 12M, 12 months after complete carcinogenic treatment,
respectively. Data were obtained from three independent experiments
with the quantification of 5 fields at ×10 each, and are expressed
as the means ± SEM. Samples were statistically significant when
*P<0.05. |
DNA methylation changes of timp3, rassf1a
and p16
To assess whether our rat model had an aberrant DNA
methylation pattern, the methylation status of the tumor-related
genes was analyzed by MSP. Products from non-treated samples (NT)
were only amplified with the unmethylated primers (U). timp3
and rassf1a were amplified with the methylated (M) primers
in the majority of the samples, while p16 showed consistent
amplification only at 12 months (Fig.
4B and Table I). The expression
level of the three genes was investigated for differential mRNA
expression. As expected, the mRNA of the three tumor suppressors
decreased as the carcinogenic process was enhanced. timp3
and p16 expression showed a decrease in rats sacrificed 16
days after complete treatment and remained underexpressed in later
stages. The subexpression of rassf1a was statistically
significant in 12M (Fig. 4C and D).
To gain a quantitative understanding of the methylation changes
seen in the MSP we performed BSP of timp3. The methylation
level observed in treated rats gradually decreased when compared to
NT (Fig. 4E). Few CpGs remained
methylated at 5 months. However, the methylation increased at 12
months. Of note, the first, second, fourth and sixth CpGs appeared
to be consistently methylated in tumors. The methylation of CpGs
9–13 also occurred more frequently in tumors than in normal samples
(Fig. 4E).
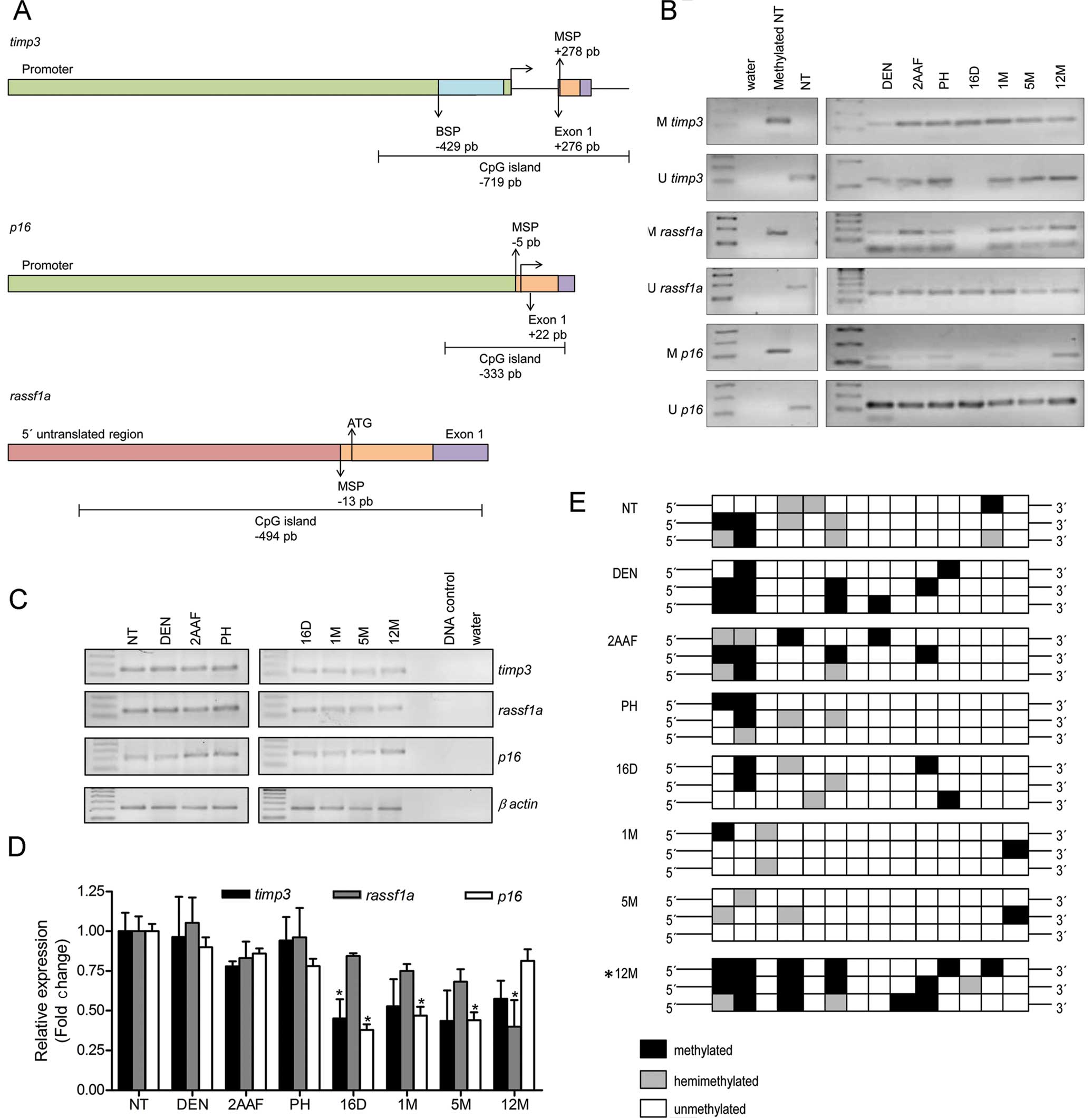 | Figure 4Methylation status of
tumor-suppressor genes. (A) Schematic diagram of the fragments
analyzed by MSP and BSP. The horizontal arrow shows the
transcription start site. In timp3, the green box shows the
promoter encompassing 2832 bp, the purple box is exon 1 and
includes 178 bp and the orange box is the MSP amplified region (120
bp). The fragment analyzed in the BSP is shown as the blue box (373
bp). In p16 (GenBank: AC_000073), the green box shows the
promoter comprising 1408 bp, the purple box is exon 1 and spans 126
bp and the orange box represents the MSP amplified region (123 bp).
In rassf1a (GenBank: NW_047801), the first translated codon
is shown as ATG. The pink box shows the 5′ untranslated region
comprising 629 bp, the purple box is exon 1 and spans 250 bp and
the orange box is the MSP amplified region (169 bp). (B) MSP
results of tumor-related genes timp3, rassf1a and
p16 24 h after administration of diethylnitrosamine (DEN),
2-acetylaminofluorene (2AAF), partial hepatectomy (PH) and 16 days
(16D), 1 month (1M), 5 and 12 months (5M and 12M, respectively)
after complete carcinogenic treatment are shown. Non-treated rat
genomic DNA (NT), water and genomic DNA in vitro methylated
by Sss1 were used as controls. (C) Expression of timp3,
rassf1a and p16 as determined by semi-quantitative
RT-PCR. The expression level of β-actin was used to confirm the
quality and quantity of total RNA from each sample. (D)
Quantification of the expression of three tumor suppressor genes
relative to NT. Data were obtained from three independent
experiments and are expressed as the means ± SEM. Results were
statistically significant when *P<0.05. (E) Overview
of bisulfite sequencing. Each box represents one sequenced CpG
whose methylation status is indicated by coloration. Results were
statistically significant when *P<0.05, as described
in Materials and methods. |
 | Table IM and U amplification frequency in
three independent experiments. |
Table I
M and U amplification frequency in
three independent experiments.
| Gene names | NT | DEN | 2AAF | PH | 16D | 1M | 5M | 12M |
|---|
| M-timp3 | 0/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| U-timp3 | 3/3 | 3/3 | 3/3 | 2/3 | 2/3 | 3/3 | 3/3 | 3/3 |
|
M-rassf1a | 0/3 | 2/3 | 3/3 | 3/3 | 2/3 | 2/3 | 3/3 | 3/3 |
|
U-rassf1a | 3/3 | 3/3 | 3/3 | 3/3 | 2/3 | 3/3 | 3/3 | 3/3 |
| M-p16 | 0/3 | 1/3 | 1/3 | 1/3 | 1/3 | 2/3 | 0/3 | 3/3 |
| U-p16 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
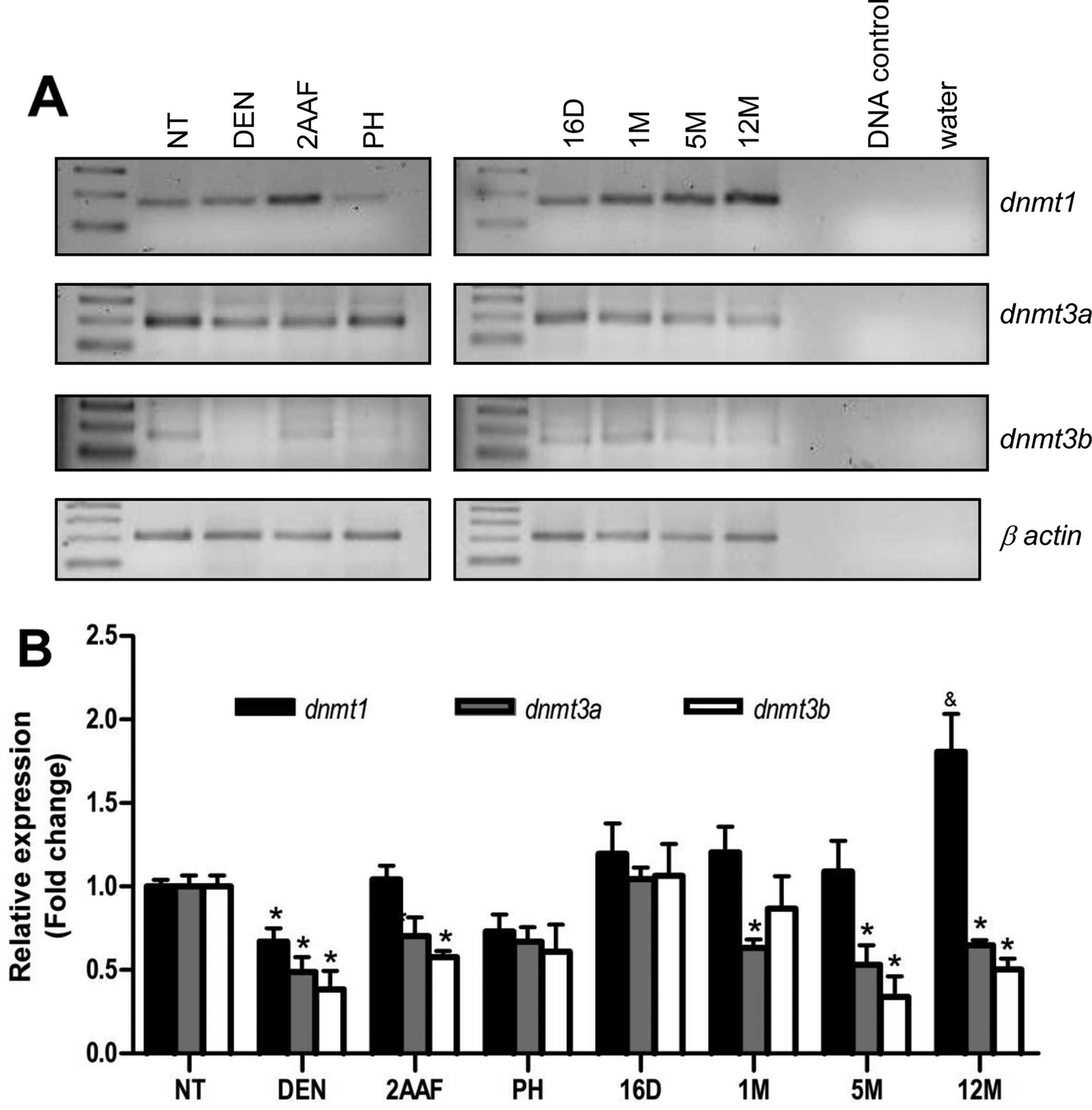
Expression of DNMT1, DNMT3a and DNMT3b
during the RHM model
After confirming that the DNA methylation is
considerably modified in our rat model, the RNA and protein
expression of the three DNMTs was analyzed. Fig. 5 shows the representative mRNA
expression of the DNMTs across different stages of the RHM.
dnmt1 was mildly subexpressed during carcinogen
administration, but significantly overexpressed in the 12-month
samples. dnmt3a and dnmt3b were also subexpressed
during carcinogen administration and peaked at 16 days
post-treatment. The two de novo methyltransferases were
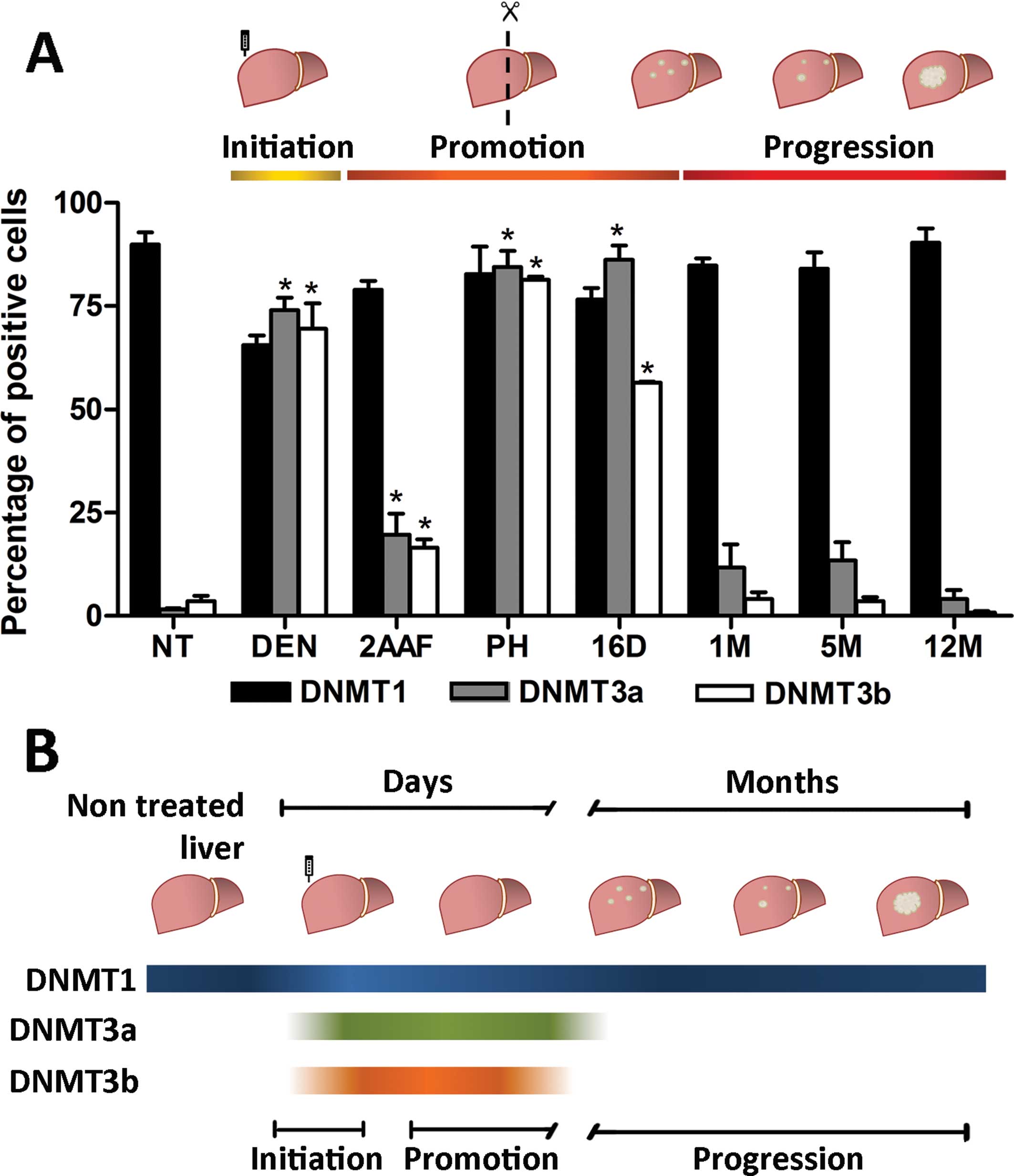
subexpressed in preneoplastic lesions and tumors (Fig. 5). Immunohistochemical staining
patterns of the DNMTs were obtained to support the results of the
semi-quantitative RT-PCR. Only DNMT1 nuclear immunostaining was
positive and evenly distributed in normal tissue (Fig. 6A). A slight decrease in the DNMT1
nuclear intensity was observed 24 h after diethylnitrosamine and
2-acetylaminofluorene administration, whereas DNMT3a and DNMT3b
were positive 24 h after diethylnitrosamine exposure. While the
nuclear signal of DNMT1 was sustained between 24 h after partial
hepatectomy, 16 days post-treatment, and the first and fifth months
(Figs. 6A and 7A), DNMT3a and DNMT3b expression was
decreased and could not be detected in the preneoplastic lesions
(first and fifth months) (Fig. 6B and
C, respectively). Tumor cells at 12 months exhibited an
increased nuclear size with DNMT1-positive expression in the
periphery of the nucleus. Nevertheless, no significant
overexpression was evident at 12 months (Fig. 6A and 7A). In accordance with the
semi-quantitative RT-PCR, DNMT3a and DNMT3b were again negative in
the tumor samples.
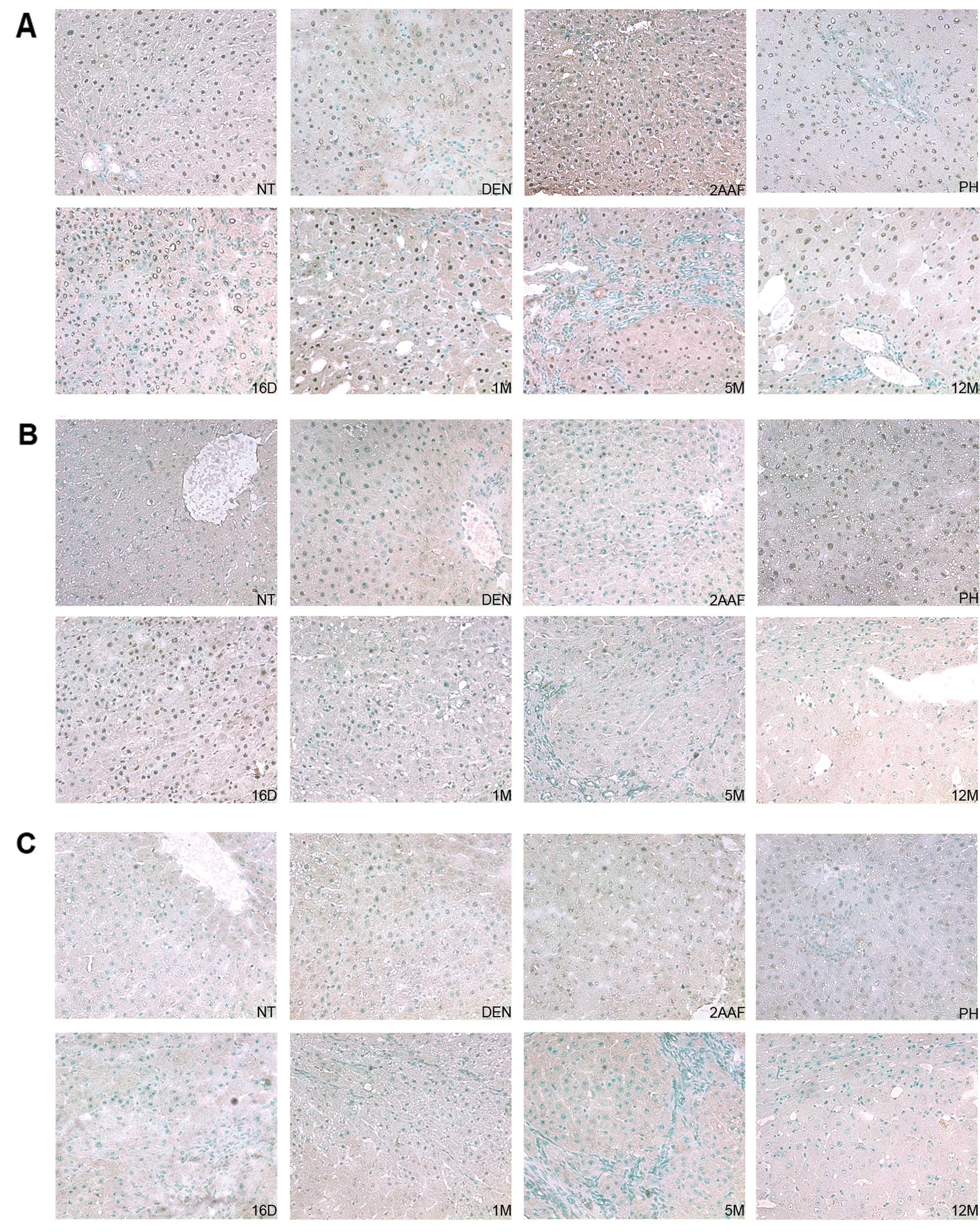 | Figure 6DNMTs nuclear localization by
immunohistochemistry in the RHM model. (A) Representative
histological sections from the livers of non-treated rats (NT) and
rats sacrificed 24 h after administration of DEN (DEN), 24 h after
administration of 2-AAF (2AAF), 24 h after partial hepatectomy
(PH); 16 days (16D), 1 month (1M), 5 months (5M) and 12 months
(12M) after complete carcinogenic treatment. Normal hepatocytes
were positive for nuclear protein expression of DNMT1 (NT). DNMT1
showed an intense nuclear signal in the majority of the samples and
was notably absent within the necrotic areas (DEN). (B)
Representative histological sections from the livers of non-treated
rats (NT) and rats sacrificed 24 h after administration of DEN
(DEN), 24 h after administration of 2-AAF (2AAF), 24 h after
partial hepatectomy (PH); and 16 days (16D), 1 month (1M), 5 months
(5M) and 12 months (12M) after complete carcinogenic treatment.
DNMT3a was detected at the early stages of the RHM (DEN, 2AAF, PH
and 16D) before its nuclear expression was decreased at 1M, 5M and
12M. (C) A) Representative histological sections from the livers of
non-treated rats (NT) and rats sacrificed 24 h after administration
of DEN (DEN), 24 h after administration of 2-AAF (2AAF), 24 h after
partial hepatectomy (PH); and 16 days (16D), 1 month (1M), 5 months
(5M) and 12 months (12M) after complete carcinogenic treatment.
Positive nuclear signal for DNMT3b was weakly detected at the early
stages of the RHM: DEN, 2AAF and PH prior to its nuclear expression
being decreased at 16D, 1M, 5M and 12M. All the sections were
counterstained with methyl green. Magnification, ×20. |
Discussion
Animal models of hepatocarcinogenesis have provided
reliable data for understanding the cellular development of human
HCC. The RHM is characterized by the evolution from nodules to HCC
without additional carcinogen treatment, mirroring the three main
stages of carcinogenesis: initiation, promotion and progression
(20,23–26).
It may be hypothesized that any epigenetic changes detected
following carcinogen administration can be related to the
development of cancer and not to the carcinogen itself.
Aberrant DNA methylation is a frequent epigenetic
event in cancer and it has been established that it depends upon
the overexpression of the DNMTs. However, it has been difficult to
track the methylation changes and the expression of DNMTs during
the different stages of carcinogenesis. Therefore, we examined the
expression of the DNMTs in the RHM. First, we confirmed that the
methylation status of tumor-suppressor genes was affected as
described previously (7,8). The MSP indicated that DNA methylation
for timp3 and rassf1a occurred after carcinogen
administration, while p16 only appeared consistently
methylated in tumors. It was also found that the expression of the
three tumor suppressors declined after the complete carcinogenic
treatment. Thus, future studies should focus on other mechanisms
that may co-operate with DNA methylation to inactivate these tumor
suppressors. BSP of timp3 was performed to gain a better
understanding of the methylation changes observed in the MSP. The
basal methylation level of the timp3 promoter was reduced
during the formation of preneoplastic lesions but was regained in
tumors (Fig. 4). In the MSP, the
amplified region was designed to analyze CpGs that were included in
the exon 1 of these genes (Fig. 4A)
(8). We found that the analysis of
the CpGs within the promoter had a greater impact on gene
transcription (1,5,16).
Thus, the amplified region in the BSP incorporated CpGs within the
promoter and not near exon 1. Therefore, the discrepancies between
the methylation status of timp3 in the MSP and the BSP were
the result of differential methylation patterns of CpGs across the
promoter and exon 1. Taking into consideration that the non-treated
samples were not amplified with the ‘M’ primers, the results show
that timp3, rassf1a and p16 were methylated
due to the carcinogenic process. Furthermore, the CpGs within the
promoter of timp3 exhibited a gradual demethylation that
spanned months prior to being hypermethylated in tumors (Fig. 4E and Table II). The results supported the
subsequent analysis of the DNMTs.
 | Table IIMultinomial distribution analysis of
the timp3 promoter methylation. |
Table II
Multinomial distribution analysis of
the timp3 promoter methylation.
| Variables | Accumulated
probability |
|---|
| NT | 0.49428037 |
| DEN | 0.39282388 |
| 2AAF | 0.25557738 |
| PH | 0.8489242 |
| 16D | 0.8489242 |
| 1M | 0.94073454 |
| 5M | 0.9857591 |
| 12M |
0.0192686a |
The overexpression of the DNMTs has been reported in
various types of cancer (14,15).
We found DNMT1 overexpressed at mRNA level in tumors. However,
dnmt3a and dnmt3b were decreased in preneoplastic
lesions and tumors and these results were congruent with the
immunohistochemical analysis. Although DNMT1 protein overexpression
was not evident between normal hepatic tissue and preneoplastic
lesions or tumors (Fig. 7A), we
observed different nuclear immunoreactivity associated with the
morphology of the nucleus (Fig.
6A). Notably, DNMT1 exhibited a diffuse nuclear signal in DEN
samples which was also less intense than in NT. DNMT3a and DNMT3b,
which were absent in non-treated samples, were localized similarly
after diethylnitrosamine exposure. It was previously mentioned that
diethylnitrosamine damages DNA and exacerbates hepatocyte oxidative
stress in the RHM (22). Oxidative
stress involves modifications in DNA bases, strand breaks, cross
linkages, all of which increase mutational rates (22,33)
and it has been described that oxygen reactive species inhibit the
methionine adenosyltransferases, responsible for supplying the
DNMTs with the methyl donor, the s-adenosyl methionine (SAM)
(34,35). Thus, oxidative stress at the
initiation of the model used in this study may hinder the
availability of methyl donors. Thus, the diffuse nuclear
localization following diethylnitrosamine administration may be
associated with a low abundance of SAM. After partial hepatectomy
and 16 days post-treatment, a positive expression of the DNMTs was
detected, which peaked for DNMT3a and DNMT3b expression (Fig. 6B and C, respectively). It has been
suggested that de novo methyltransferases are required for
proper tissue regeneration in other animal models (36,37).
Thus, it is possible that DNMT3 was recruited as part of the
hepatic regeneration between DEN and 16D. In spite of the
downregulation of DNMT3a and DNMT3b, DNMT1 was still visible in
preneoplastic lesions and tumors. Notably, at 12M DNMT1 was
localized in the periphery of the nucleus. Hypermethylated regions
have been found to co-localize with the nuclear lamina in human
cell lines, colorectal cancer (38)
and murine lung cancer models (16). Thus, the presence of DNMT1 at the
edge of the nucleus corresponds to the hypermethylated regions near
the nuclear lamina. During the time lapse between the fifth and
twelfth months, the preneoplastic lesions dedifferentiated into
tumors. At these stages, DNMT1 was the only enzyme present.
However, if DNMT1 is important in the late dedifferentiation of
tumors, the performance of the enzyme, when it is not overexpressed
as described in other types of cancer remains to be determined. It
is possible that specific protein interactions along
post-translational modifications regulate DNMT1 performance and
localization during the RHM. It has been reported that sumoylation
of DNMT1 increases its catalytic activity (10). The phosphorylation of DNMT1 may be
involved in conformational changes, enzyme stability and abundance
of the enzyme in the replication bubbles and DNA repair sites
(39,40). However, a more or less active enzyme
cannot account for the hypermethylation of specific genes in a
hypomethylated background. We hypothesize that protein interactions
allow the enzyme to identify its targets. DNMT1 is known to
interact with proteins that assist its recruitment to replication
sites (PCNA-proliferating cell nuclear antigen,
URHF1-ubiquitin-like with PHD and RING finger domains 1) and with
those that cooperate in gene silencing (pRB-retinoblastoma protein,
HDAC1-histone deacetylase 1, HDAC2-histone deacetylase 2,
EZH2-enhancer of zeste homologue 2) (41–43).
As discussed earlier, the RHM allows the analysis of
the initiation, promotion and progression stages of carcinogenesis
(20,24,25).
Although the de novo methyltransferases are downregulated in
preneoplastic lesions and tumors, it is noteworthy that the two
enzymes were detected in specific stages, i.e., after initiation
and during promotion. Considering the literature available, the
expression profile of the DNMT3s poses two scenarios: i) the
enzymes may play an early role establishing methylation patterns
that ‘drive’ cancer initiation and progression (1). When DNMT3s fulfill this role, they are
downregulated as the DNMT1 copies the methylation patterns during
subsequent cell replication (44).
ii) The DNMT3s function as tumor supressors protecting cells from
oncogene and transposable elements activation following
demethylation (16,45). As discussed earlier, after
carcinogen administration a wave of oxidative stress ensues and
oxygen reactive species enhance DNA hypomethylation (22,34,35).
Furthermore, Takasugi and colleagues (37) showed that DNA hypomethylation is
induced by partial hepatectomy. Therefore, DNMT3a and DNMT3b may be
required to prevent hypomethylation.
In summary, our MSP results indicate that
timp3, rassf1a and p16 became methylated
during and after complete carcinogenic treatment. Through BSP we
found that a CpG-rich region within the promoter of timp3
first became hypomethylated in preneoplastic lesions and then
hypermethylated in tumors. Although the RHM did not exhibit a
statistically significant overexpression of DNMT1, it was the only
enzyme detectable during the late dedifferentiation of
preneoplastic lesions to tumors. By contrast, DNMT3a and DNMT3b
were recruited specifically during initiation and promotion. To the
best of our knowledge, this is the first report concerning the
activation and subsequent downregulation of the de novo
methyltransferases as part of the natural course of carcinogenesis
in an animal model (Fig. 7B). The
synchronous events evident in the RHM make it a suitable model to
identify more specific functions and interactions of the DNMTs in
shorter time windows in future studies; for example, to study the
mechanisms involved in the genetic regulation of the DNMT3s by the
end of the promotion stage and the protein interactions of the
DNMT1 during the progression stage.
Acknowledgements
The authors would like to thank Samia Fattel,
Leobardo García-Molina, Sergio Hernández-García, Julia Torres-Mena
and Ruth Pacheco-Rivera for their technical support. They also
thank Dr José de Jesús Serrano-Luna at CINVESTAV and Dr Julio Isael
Pérez-Carreón at the Instituto Nacional de Medicina Genómica, for
useful comments in the preparation of the manuscript. The present
study was supported by the Multidisciplinary Project, CINVESTAV, by
CONACyT, grant no. CB12-178558 and scholarship: 207267 and COMECYT
scholarship: 13BTD0531.
Abbreviations:
|
DNMT(s)
|
DNA methyltransferase(s)
|
|
RHM
|
resistant hepatocyte model
|
|
HCC
|
hepatocellular carcinoma
|
|
MSP
|
methylation-specific PCR
|
|
BSP
|
bisulphite sequencing PCR
|
|
AHF
|
altered hepatocyte foci
|
|
GGT
|
g-glutamil transpeptidase
|
|
NT
|
non-treated sample
|
|
DEN
|
24 h after diethylnitrosamine
administration
|
|
2AAF
|
24 h after 2-acetylaminofluorene
administration
|
|
PH
|
24 h after partial hepatectomy
|
|
16D
|
16 days after complete carcinogenic
treatment
|
|
1M
|
1 month after complete carcinogenic
treatment
|
|
5M
|
5 months after complete carcinogenic
treatment
|
|
12M
|
12 months after complete carcinogenic
treatment
|
References
|
1
|
Aryee MJ, Liu W, Engelmann JC, Nuhn P,
Gurel M, Haffner MC, Esopi D, Irizarry RA, Getzenberg RH, Nelson
WG, Luo J, Xu J, Isaacs WB, Bova GS and Yegnasubramanian S: DNA
methylation alterations exhibit intra-individual stability and
inter-individual heterogeneity in prostate cancer metastases. Sci
Transl Med. View Article : Google Scholar : 2013.
|
|
2
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:85–93.
2002.
|
|
3
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rishi V, Bhattacharya P, Chatterjee R,
Rozenberg J, Zhao J, Glass K, Fitzgerald P and Vinson C: CpG
methylation of half-CRE sequences creates C/EBPα binding sites that
activate some tissue-specific genes. Proc Natl Acad Sci.
107:20311–20316. 2010.PubMed/NCBI
|
|
5
|
Ziller MJ, Gu H, Müller F, Donaghey J,
Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein
BE, Gnirke A and Meissner A: Charting a dynamic DNA methylation
landscape of the human genome. Nature. 500:477–481. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheng X and Blumenthal RM: Mammalian DNA
methyltransferases: a structural perspective. Structure. 3:341–350.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sigalotti L, Fratta E, Coral S, Cortini E,
Covre A, Nicolay HJ, Anzalone L, Pezzani L, Di Giacomo A, Fonsatti
E, Colizzi F, Altomonte M, Calabrò L and Maio M: Epigenetic drugs
as pleiotropic agents in cancer treatment: biomolecular aspects and
clinical applications. J Cell Physiol. 212:330–344. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Asada K, Asada R, Yoshiji H, Fukui H,
Floyd RA and Kotake Y: DNA cytosine methylation profile in various
cancer-related genes is altered in cultured rat hepatocyte cell
lines as compared with primary hepatocytes. Oncol Rep.
15:1241–1248. 2006.
|
|
9
|
Feinberg AP and Vogelstein B:
Hypomethylation distinguishes genes of some human cancers from
their normal counterparts. Nature. 301:89–92. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee B and Muller MT: SUMOylation enhances
DNA methyltransferase 1 activity. Biochem J. 421:449–461. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ling Y, Sankpal UT, Robertson AK, McNally
JG, Karpova T and Robertson KD: Modification of de novo DNA
methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction
with histone deacetylases (HDACs) and its capacity to repress
transcription. Nucleic Acids Res. 32:598–610. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kanai Y and Hiroshida S: Alterations of
DNA methylation associated with abnormalities of DNA
methyltransferases in human cancers during transition from a
precancerous to a malignant state. Carcinogenesis. 28:2434–2442.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oh BK, Kim H, Park HJ, Shim YH, Choi J,
Park C and Park YN: DNA methyltransferase expression and DNA
methylation in human hepatocellular carcinoma and their
clinicopathological correlation. Int J Mol Med. 20:65–73.
2007.PubMed/NCBI
|
|
14
|
Choi MS, Shim YH, Hwa JY, Lee SK, Ro JY,
Kim JS and Yu E: Expression of DNA methyltransferases in multistep
hepatocarcinogenesis. Hum Pathol. 34:11–18. 2013. View Article : Google Scholar
|
|
15
|
Yoshimasa S, Yae K, Tohru N, Michiie S,
Hidetsugu S, Hiromasa I and Setsuo H: Increased protein expression
of DNA methyltransferase (DNMT) 1 is significantly correlated with
the malignant potential and poor prognosis of human hepatocellular
carcinomas. Int J Cancer. 105:527–532. 2003. View Article : Google Scholar
|
|
16
|
Raddatz G, Gao Q, Bender S, Jaenisch R and
Lyko F: Dnmt3a protects active chromosome domains against
cancer-associated hypomethylation. PLoS Genet. 8:e10031462012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y,
Shi JY, Zhu YM, Tang L, Zhang XW, Liang WX, Mi JQ, Song HD, Li KQ,
Chen Z and Chen SJ: Exome sequencing identifies somatic mutations
of DNA methyltransferase gene DNMT3A in acute monocytic leukemia.
Nat Genet. 43:309–315. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hlady RA, Novakova S, Opavska J,
Klinkebiel D, Peters LS, Bies J, Hannah J, Iqbal J, Anderson KM,
Siebler HM, Smith LM, Greiner TC, Bastola D, Joshi S, Lockridge O,
Simpson MA, Felsher DW, Wagner KU, Chan WC, Christman JK and
Opavsky R: Loss of Dnmt3b function upregulates the tumor modifier
Ment and accelerates mouse lymphomagenesis. J Clin Invest.
122:163–177. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Trinh BN, Long TI, Nickel AE, Shibata D
and Laird PW: DNA methyltransferase deficiency modifies cancer
susceptibility in mice lacking DNA mismatch repair. Mol Cell Biol.
22:2906–2917. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pérez-Carreón JI, López-García C,
Fattel-Fazenda S, Arce-Popoca E, Alemán-Lazarini L,
Hernández-García S, Le Berrey V, Sokoly S, Francoisy JM and
Villa-Treviño S: Gene expression profile related to the progresión
of preneoplastic nodules toward hepatocellular carcinoma in rats.
Neoplasia. 8:373–383. 2006.
|
|
21
|
Rotstein J, Macdonald PD, Rabes HM and
Farber E: Cell cycle kinetics of rat hepatocytes in early putative
preneoplastic lesions in hepatocarcinogenesis. Cancer Res.
44:2913–2917. 1984.PubMed/NCBI
|
|
22
|
Sánchez-Pérez Y, Carrasco-Legleu C,
García-Cuellar C, Pérez-Carreón JI, Hernández-García S,
Salcido-Neyoy M, Alemán-Lazarini L and Villa-Treviño S: Oxidative
stress in carcinogenesis. Correlation between lipid peroxidation
and induction of preneoplastic lesions in rat hepatocarcinogenesis.
Cancer Lett. 217:25–32. 2005.PubMed/NCBI
|
|
23
|
Solt D and Farber E: New principle for the
analysis of chemical carcinogenesis. Nature. 263:701–703. 1976.
View Article : Google Scholar
|
|
24
|
Solt DB, Medine A and Farber E: Rapid
emergence of carcinogen-induced hyperplastic lesions in a new model
for the sequential analysis of liver carcinogenesis. Am J Pathol.
88:595–618. 1977.PubMed/NCBI
|
|
25
|
Solt DB, Cayama E, Tsuda H, Enomoto K, Lee
G and Farber E: Promotion of liver cancer development by brief
exposure to dietary 2-acetylaminofluorene plus partial hepatectomy
or carbon tetrachloride. Cancer Res. 43:188–191. 1983.PubMed/NCBI
|
|
26
|
Pascale RM, Simile MM, De Miglio MR,
Muroni MR, Calvisi DF, Asara G, Casabona D, Frau M, Seddaiu MA and
Feo F: Cell cycle deregulation in liver lesions of rats with and
without genetic predisposition to hepatocarcinogenesis. Hepatology.
35:1341–1350. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wen BL, Ao L, Zhou ZY, Cui ZH, Zhou YH,
Yuan XY, Xiang YL, Cao J and Liu JY: CpG island hypermethylation of
multiple tumor suppressor genes associated with loss of their
protein expression during rat lung carcinogenesis induced by
3-methylcholanthrene and diethylnitrosamine. Biochem Biophys Res
Commun. 402:507–514. 2010. View Article : Google Scholar
|
|
28
|
Agathanggelou A, Cooper WN and Latif F:
Role of the Ras-association domain family 1 tumor suppressor gene
in human cancers. Cancer Res. 65:3497–3508. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Du YP, Peng JS, Sun A, Tang ZH, Ling WH
and Zhu HL: Assessment of the effect of betaine on p16 and
c-myc DNA methylation and mRNA expression in a chemical
induced rat liver cancer model. BMC Cancer. View Article : Google Scholar : 2009.
|
|
30
|
Li J, Knobloch TJ, Poi MJ, Zhang Z, Davis
AT, Muscarella P and Weghorst CM: Genetic alterations of
RDINK4/ARF enhancer in human cancer cells. Mol Carcinog.
53:211–218. 2014.
|
|
31
|
Gutierrez-Reyes G, del Carmen Garcia de
Leon M, Varela-Fascinetto G, Valencia P, Pérez Tamayo R, Rosado CG,
Labonne BF, Rochilin NM, Garcia RM, Valadez JA, Latour GT, Corona
DL, Diaz GR, Zlotnik A and Kershenobich D: Cellular senescence in
livers from children with end stage liver disease. PLoS One.
5:e102312010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Enomoto K and Farber E: Kinetics of
phenotypic maturation of remodeling of hyperplastic nodules during
liver carcinogenesis. Cancer Res. 42:2330–2335. 1982.PubMed/NCBI
|
|
33
|
Ziech D, Franco R, Pappa A and
Panayiotidis MI: Reactive oxygen species (ROS) - induced genetic
and epigenetic alterations in human carcinogenesis. Mutat Res.
711:167–173. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Torres L, Avila MA, Carretero MV, Latasa
MU, Caballería J, López-Rodas G, Boukaba A, Lu SC, Franco L and
Mato JM: Liver-specific methionine adenosyltransferase MAT1A gene
expression is associated with a specific pattern of promoter
methylation and histone acetylation: implications for MAT1A
silencing during transformation. FASEB J. 14:95–102. 2004.
|
|
35
|
Yang H, Huang ZZ, Zeng Z, Chen C, Selby RR
and Lu SC: Role of promoter methylation in increased methionine
adenosyltransferase 2A expression in human liver cancer. Am J
Physiol Gastrointest Liver Physiol. 280:184–190. 2001.PubMed/NCBI
|
|
36
|
Takayama K, Shimoda N, Takanaga S, Hozumi
S and Kikuchi Y: Expression patterns of dnmt3aa, dnmt3ab and dnmt4
during development and fin regeneration in zebrafish. Gene Expr
Patterns. 14:105–110. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Takasugi M, Hayakawa K, Arai D and Shiota
K: Age- and sex-dependent DNA hypomethylation controlled by growth
hormone in mouse liver. Mech Ageing Dev. 134:331–337. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Berman BP, Weisenberger DJ, Aman JF,
Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CPE, van Dijk MC,
Tollenaar RAEM, van den Berg D and Laird PW: Regions of focal DNA
hypermethylation and long-range hypomethylation in colorectal
cancer coincide with nuclear lamina-associated domains. Nat Genet.
44:40–46. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hervouet E, Lalier L, Debien D, Cheray M,
Geairon A, Rogniaux H, Loussouarn D, Martin SA, Vallette FM and
Cartron PF: Disruption of Dnmt1/PCNA/UHRF1 interactions promotes
tumorigenesis from human and mice glial cells. PLoS One.
5:e113332010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mortusewicz O, Schermelleh L, Walter J,
Cardoso MC and Leonhardt H: Recruitment of DNA methyltransferase I
to DNA repair sites. Proc Natl Acad Sci. 102:8905–8909. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gazin C, Wajapeyee N, Gobeil S, Virbasius
CM and Green MR: An elaborate pathway required for Ras-mediated
epigenetic silencing. Nature. 449:1073–1077. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J,
Koseki H and Wong J: UHRF1 targets DNMT1 for DNA methylation
through cooperative binding of hemi methylated DNA and methylated
H3K9. Nat Commun. View Article : Google Scholar : 2013.PubMed/NCBI
|
|
43
|
Shamma A, Suzuki M, Hayashi N, Kobayashi
M, Sasaki N, Nihiuchi T, Doki Y, Okamoto T, Kohno S, Muranaka H,
Kitajima S, Yamamoto K and Takahashi C: ATM mediates pRB function
to control DNMT1 protein stability and DNA methylation. Mol Cell
Biol. 33:3113–3124. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cartron PF, Blanquart C, Hervouet E,
Gregoire M and Vallette FM: HDAC1-mSin3a-NCOR1, Dnmt3b-HDAC1-Egr1
and Dnmt1-PCNA-UHRF1-G9a regulate the NY-ESO1 gene expression. Mol
Oncol. 7:452–463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xionga Y, Dowdya SC, Xueb A, Shujuana J,
Eberhardtc NL, Podratza KC and Jianga SW: Opposite alterations of
DNA methyltransferase gene expression in endometrioid and serous
endometrial cancers. Gynecol Oncol. 96:601–609. 2005. View Article : Google Scholar : PubMed/NCBI
|