Introduction
Osteosarcoma is a primary bone malignancy with high
rates of metastasis, mortality and disability, and usually occurs
in children and adolescents (1).
The most common treatments for osteosarcoma are surgery,
chemotherapy and biotherapy. However, the prognosis of osteosarcoma
is still poor because of the high degree of malignancy, rapid
disease progression and early metastasis (2).
Gene amplification can be defined as increased copy
numbers of certain regions of the genome. Gene amplification often
results in the upregulation of gene expression by increasing the
number of templates available for transcription. Gene amplification
is commonly found in tumor cells and is considered an important
mechanism whereby tumor cells gain increased levels of expression
of critical genes. Thus, identification and characterization of
these genes should provide important insights into the pathobiology
of cancer.
Culllin 4B (CUL4B) is a component of the
Cullin4B-Ring E3 ligase complex (CRL4B) that functions in
proteolysis. It has been implicated in tumorigenesis since the core
components participate in a broad variety of physiologically and
developmentally controlled processes such as cell progression,
replication and DNA damage response (3). In mammals, there are two Cullin 4
members, CUL4A and CUL4B, which share 82% sequence
identity. Although Cul4a-null mice do not exhibit evident
developmental defects (4–6), CUL4A has been shown to target
p53 and several cyclin-dependent kinase inhibitors, such as p21 and
p27 for proteolysis in cultured cells (7–9) and
was found to be highly expressed in breast and hepatocellular
carcinomas (10,11). Loss-of-function mutation in the
X-linked CUL4B, in contrast, causes mental retardation,
short state, absence of speech and other phenotypes in humans
(12,13), and Cul4B knockout mice are
embryonically lethal, indicating a unique function of CUL4B
that cannot be compensated by CUL4A. Recently, Yang et
al (14) showed that depletion
of CUL4B resulted in loss of not only H2AK119 mono-ubiquitination
but also H3K9 tri-methylation and DNA methylation, leading to
depression of a collection of genes, including the tumor-suppressor
IGFBP3. Further experiments revealed that CUL4B promoted cell
proliferation and invasion, which are consistent with a tumorigenic
phenotype, at least partially by repressing IGFBP3. Further
findings revealed that the expression of CUL4B was markedly
upregulated in samples of human cervical carcinoma and was
negatively correlated with the expression of IGFBP3.
Although experiments have shown that CULB
possesses an intrinsic transcription repressive activity, (3,15) the
exact role of CULB in the process and progression of osteosarcoma
is still poorly understood. Here, we established an osteosarcoma
cell model in vitro, and transfected osteosarcoma SAOS-2
cells using CUL4B RNAi and assessed the apoptosis of SAOS-2 cells.
We demonstrated that CUL4B promotes cell proliferation and inhibits
the apoptosis of osteosarcoma cells. These results add to the
understanding of the role of CUL4B in the proliferation and
apoptosis of osteosarcoma cells and its molecular mechanisms.
Materials and methods
Cell line and culture conditions
Human osteosarcoma cell line SAOS-2 was purchased
from the Shanghai Institute of the Chinese Academy of Sciences
(Shanghai, China) and grown in Dulbecco’s modified Eagle’s medium
(DMEM) with 10% (v/v) fetal calf serum, streptomycin (100 U/ml) and
penicillin (100 U/ml). DMEM, fetal bovine serum (FBS) and Dimethyl
sulfoxide (DMSO) were purchased from Gibco Biotechnology
(Gibco-BRL, MD, USA). Cultures were maintained at 37°C in an
incubator with a humidified atmosphere of 5% CO2.
Reverse transcription-polymerase chain
reaction (RT-PCR) analysis of CUL4B gene expression in tumor
cells
Total-RNA was extracted under RNase-free conditions
(carried out according to the operation manual for TRIzol;
Invitrogen Corp., Carlsbad, CA, USA) with
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal
reference. Primer design was as follows: the CUL4B upstream
sequence was 5′-GGGAAAGGAATGGTGAA-3′; and the downstream sequence
was 5′-TGCATAGAGCCGGTTAG-3′. The GAPDH upstream sequence was
5′-TGACTTCAACAGCGACAC CCA-3′; and its downstream sequence was
5′-CACCCTGTT GCTGTAGCCAAA-3′. The Access RT-PCR kit was used to
perform single-step reverse transcription and PCR amplification. An
aliquot of 5 μl of the amplified products was subjected to
electrophoresis on 2% agarose gel. The gels were examined under a
UV lamp.
Construct design: lentiviral-mediated
small interfering RNA delivery system
We targeted the gene of interest by designing small
interfering RNAs (siRNAs) using the design software developed by
Ambion Corp. (Naugatuck, CT, USA) to select the best parameters for
the RNA interference target. We determined the effective target
sequence: PSCSI2891, AGCAGTGGAAGCTATTCAGAA (CUL4B mRNA). We
designed the DNA oligonucleotides of siRNAs (Shanghai Genechem Co.,
Ltd.): PSCSI2891-1, 5′-CcggAGCAGTGGA
AGCTATTCAGAATTCAAGAGATTCTGAATAGCTTCCA CTGCTTTTTTg-3′ and
PSCSI2891-2, 5′-aattcaaaaaAGCAG
TGGAAGCTATTCAGAATCTCTTGAATTCTGAATAGCT TCCACTGCT-3′. After
annealing, the double-stranded DNA was digested with Agel
and EcoRI (New England Biolabs) to linearize the pGCSIL-GFP
vector. We modified the double-stranded DNA after annealing and
linked it with the pGCSIL-GFP vector following the double
digestion. We used calcium chloride to prepare competent cells of
Escherichia coli afresh and cultured it at 37°C for 16 h. We
used computer-aided high-throughput cloning of bacteria in liquid
medium for sequencing (Shanghai Genechem Co., Ltd.).
Preparation and grouping of cells
The human osteosarcoma cell line SAOS-2 was cultured
in the RPMI-1640 culture solution with fetal calf serum (volume
fraction 10%) and incubated with 5% CO2 at 37°C. The
cells that remained in the logarithmic phase were divided into two
groups: i) negative control, in which the normal target cells were
infected withthe negative control virus and ii) knockdown, in which
the normal target cells were infected with the RNAi target
virus.
Real-time PCR and western blotting to
test knockdown efficiency
The human osteosarcoma SAOS-2 cells grew well on the
day prior to viral introduction was recovered. The cell suspension
was incubated with 5% CO2 at 37°C. When the degree of
cell fusion reached 30%, adequate viral load was introduced in the
different groups, to a multiplicity of infection (MOI) of 100.
Following incubation for three days, the expression level of GFP
was observed under a fluorescence microscope. The culture was
continued if the efficiency of infection exceeded 50%. After
incubation for five days, the cells were collected; the mRNA
expression of the gene of interest was analyzed using real-time PCR
for RNA interference. The upstream primer sequence for the
CUL4B gene was 5′-GGGAAAGGAATGGTGAA-3′ while the downstream
primer sequence was 5′-TGCATAGAGCCGGTTAG-3′. The cell culture
solution was aspirated and the cells were washed twice in
phosphate-buffered saline (PBS). An adequate amount of pre-cooled
2X lysis buffer was added. After deplating, the cells were
transferred to the tube and then lysed on ice for 10–15 min. The
protein concentration was determined and adjusted to 2 μg/μl. Next,
2X loading buffer was added to each sample and subsequently boiled
for 5–10 min. Forty micrograms of total protein was loaded into
each well containing 10% SDS-polyacrylamide gel, subjected to
electrophoresis at 30 mA for 2 min, and then transferred to a
polyvinylidene fluoride (PVDF) membrane at 400 mA for 2 h. The
membrane was blocked with 5% milk in Tris-buffered saline (TBS) for
1 h at room temperature. The primary antibody was added and
incubated with the membrane for 2 h at room temperature, and then
washed three times with TBS/Tween-20. A secondary antibody was
added to the membrane and incubated at room temperature with gentle
agitation. Two hours later, the membrane was washed three times
with TBS/Tween-20 for 10 min per wash. The bands were visualized
using an enhanced chemilluminescence (ECL) kit followed by exposure
to X-ray film.
Cellomics to test inhibition of
osteosarcoma cell proliferation following downregulation of the
CUL4B gene
After trypsinization, the cell suspension was
resuspended in complete medium and density was adjusted to
2×104/ml. We used the blood counting chamber to count
the cells, laying them at 2,000 cells/well. Each group comprised
three to five compound perforations. Each perforation was filled
with 100 μl, and the same quantity of cells. The cells were
incubated with 5% CO2 and cultured at 37°C. Starting the
next day, the plates were tested and read once a day with Cellomics
for five days. After adjusting the input parameters of Cellomics
ArrayScan (Thermo Fisher Scientific Inc., Waltham, MA, USA), the
quantity of cells with green fluorescence was accurately calculated
while scanning the perforations in the plates. The data were
collected and analyzed to create a proliferation curve for the five
days.
Fluorescence-activated cell sorting
(FACS) to assess osteosarcoma cell cycle distribution following
CUL4B gene silencing
The cell culture supernatant was aspirated when the
coverage rate for the 6-cm dish cells in the experimental group
increased to 80%, ensuring that the cells did not enter the plateau
phase. The cells were washed once with D-Hank’s solution and
subjected to trypsinization. The complete medium was removed and
the cells were collected in a 5-ml centrifuge tube. We set three
compound perforations in each group and performed timed cycle tests
ensuring an adequate number of cells for computerized analysis,
with at least 1,000,000 each time. We used PBS (pH, 7.2–7.4) that
was pre-cooled at 4°C to wash and precipitate cells once. The cells
were collected after centrifugation at 1,500 rpm for 5 min. The
cells were fixed with 70% ethanol, which was pre-cooled to 4°C, for
at least 1 h. The stationary liquid was abandoned by centrifugation
at 1,500 rpm for 5 min. We used PBS to wash and precipitate cells
once. Adequate quantity of cell staining fluid (1–1.5 ml) was added
for suspension, based on the volume of cells, ensuring that the
pass rate of cells reached 200–350 cell/sec for computer analysis.
We used a 300-mesh screen cloth to filter within the tube while
streaming onto the computer.
Fluorescence-activated cell sorting
(FACS) analysis of apoptosis following downregulation of the CUL4B
gene
We used D-Hank’s solution to wash cells once after
collecting the culture supernatant from each experimental group
following transfer into the 5-ml centrifuge tube. We used
pancreatic enzymes to digest the cells. The culture supernatants
were then abandoned, and cells were collected into one 5-ml
centrifuge tube, with three compound perforations in each group.
The supernatants were aspirated after centrifugation at 1,500 rpm
for 5 min. We used PBS to wash and precipitate the cells once and
then cells were collected after centrifugation at 1,500 rpm for 5
min. A 1X binding buffer was then used to wash and precipitate
cells once, and centrifuged at 1,500 rpm for 5 min. The collected
cells were resuspended using 1 ml 1X staining buffer; the volume of
staining buffer solution was determined according to the
precipitation capacity of the cells, adjusting the final density of
the cell suspension to 1×106–1×107 cells/ml.
We took a 100-μl cell suspension (1×105–1×106
cells) and added 5 μl Annexin V-APC for dyeing. The mixture was
placed in a dark place at room temperature for 10–15 min, and then
transferred to the tube for streaming onto the computer for further
analysis.
Inhibition of clonability of the
osteosarcoma cells following downregulation of the CUL4B gene
We digested cells that remained in the logarithmic
phase in each experimental group, with pancreatic enzymes. The
cells were resuspended in complete medium. Using a blood counting
chamber, we inoculated the 96-well culture plate at the rate of 500
cells per perforation in each experimental group. We set three
compound perforations in each experimental group, transferred the
cells that were already inoculated into the incubator and they were
cultured for three days or when the number of cells in most single
clones exceeded five. During the process, the solution was changed
and cells were monitored every three days. Using Cellomics
ArrayScan, we scanned and photographed the perforations, analyzed
the number and size of the clones within the perforations and the
number of cells in each clone.
Statistical analysis
All data are expressed as mean ± standard deviation
(SD). We used the statistical software SPSS 12.0 to perform the
relevant analysis. The significance level of statistics was set at
P<0.05.
Results
Expression of the CUL4B gene in SAOS-2
cells, preparation of RNA-interfering lentivirus vector and
assessment of knockdown efficiency
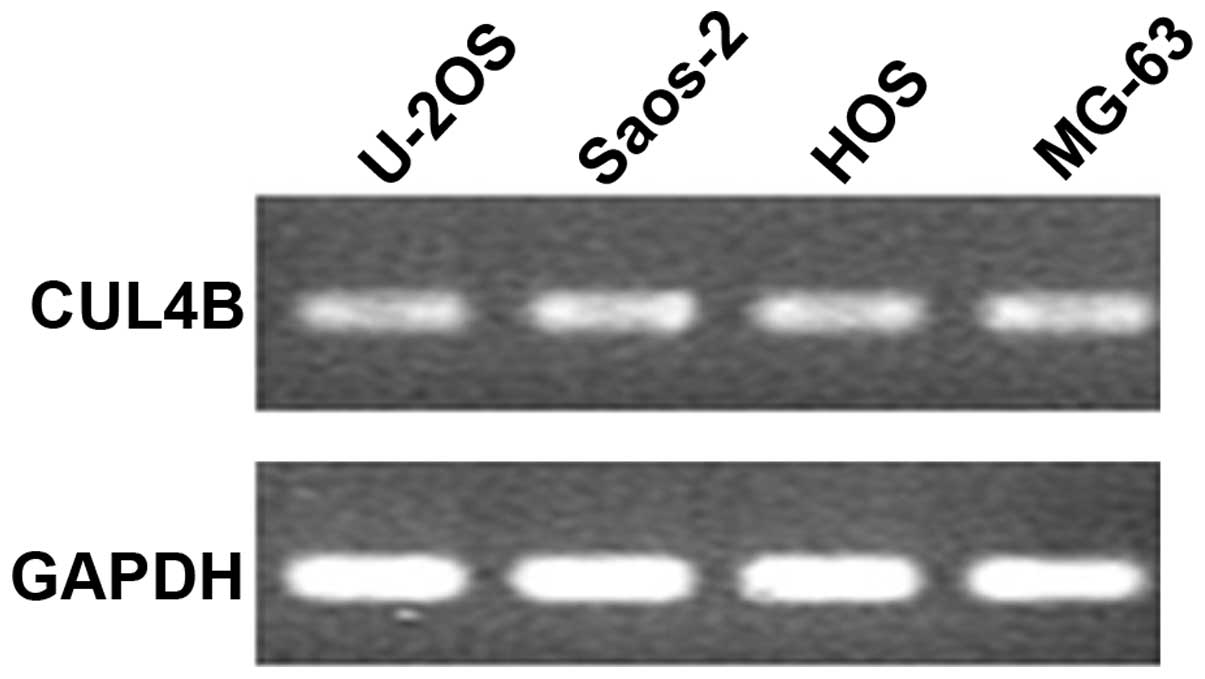
The results of the semi-quantitative PCR, which set
GAPDH as an internal reference, showed that the CUL4B gene
was abundantly expressed in the osteosarcoma SAOS-2 cells (Fig. 1). The length of the PCR fragment in
the positive clone that annealed with the fragment of vshRNA was
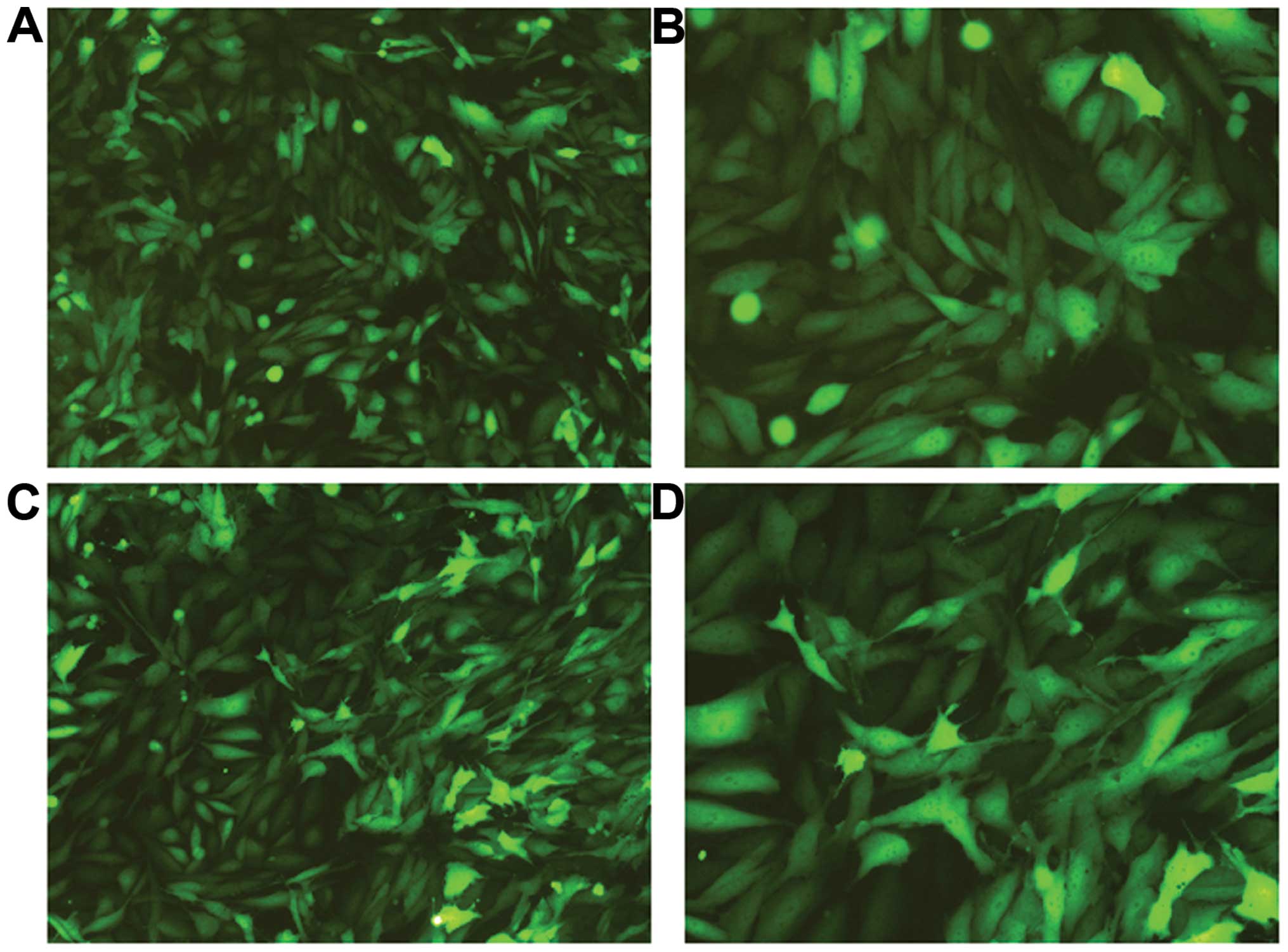
343 bp. After transfection with the siRNA lentivirus for three
days, GFP expression was observed under fluorescence microscopy
(Fig. 2). After five days, it was
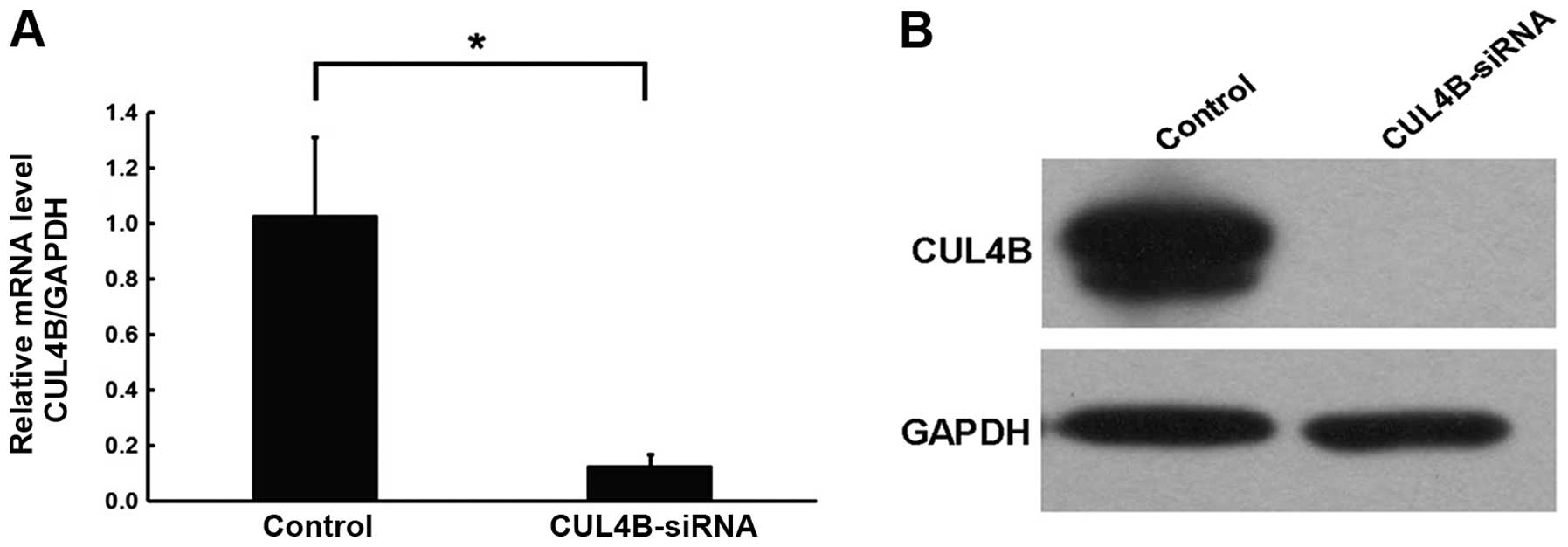
found that the mRNA expression level of the CUL4B gene in
the SAOS-2 cells of the knockdown group was inhibited, which was
significantly different compared with the negative control group.
The western blotting results confirmed inhibition of the protein
level by the silencing of the CUL4B gene in SAOS-2 cells.
The RNA-interfering lentivirus of the CUL4B gene construct
effectively inhibited the expression of the CUL4B gene and
several targets following gene silencing (Fig. 3).
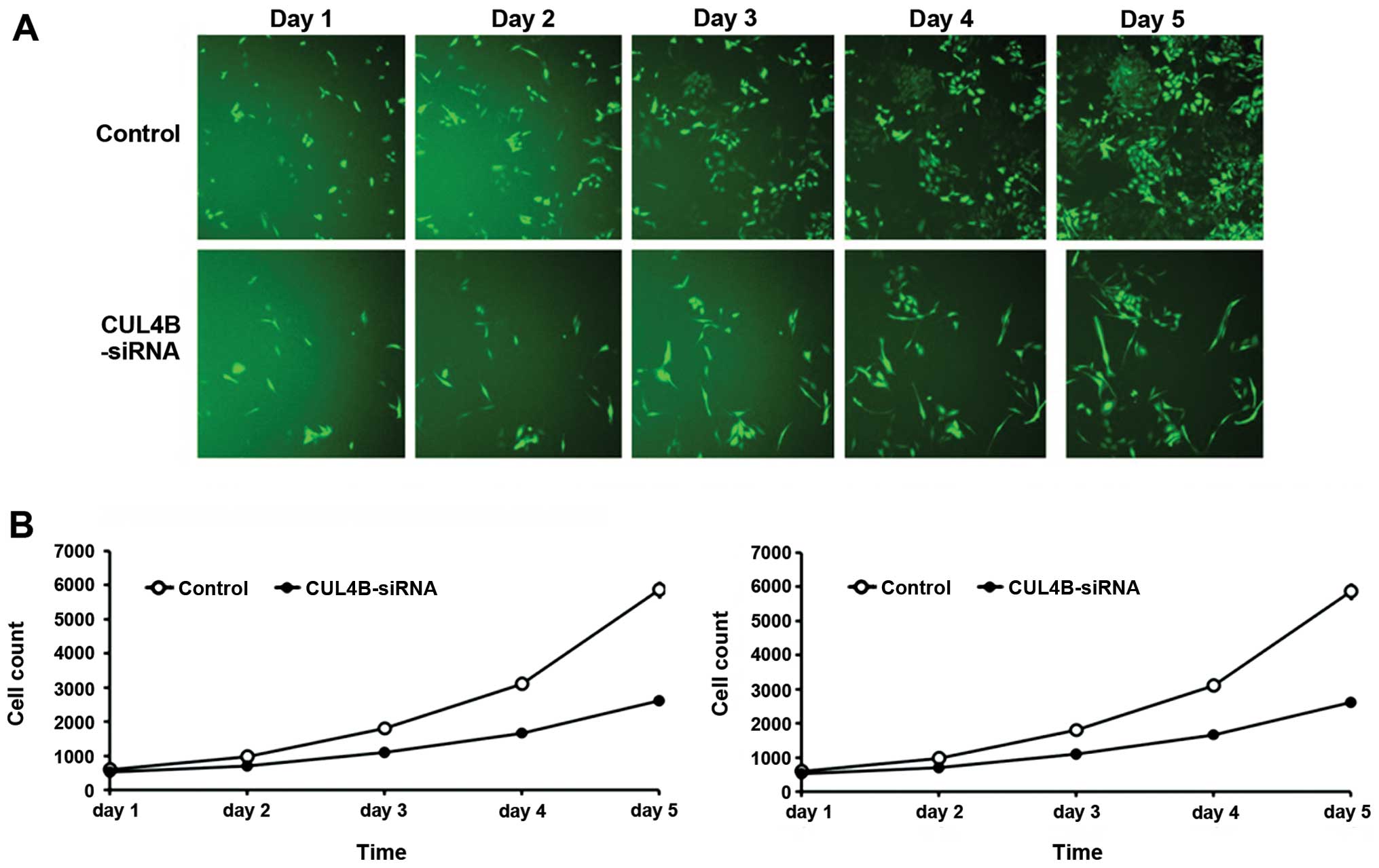
Cellomic analysis of inhibition of the
proliferation of osteosarcoma cells following downregulation of the
CUL4B gene
After transfection with the siRNA lentivirus, the
proliferation rate of SAOS-2 cells in the knockdown group was
inhibited together with the negative control from the third day
(P<0.01). The results suggested that downregulation of the CUL4B
gene inhibited the proliferation of SAOS-2 (Fig. 4).
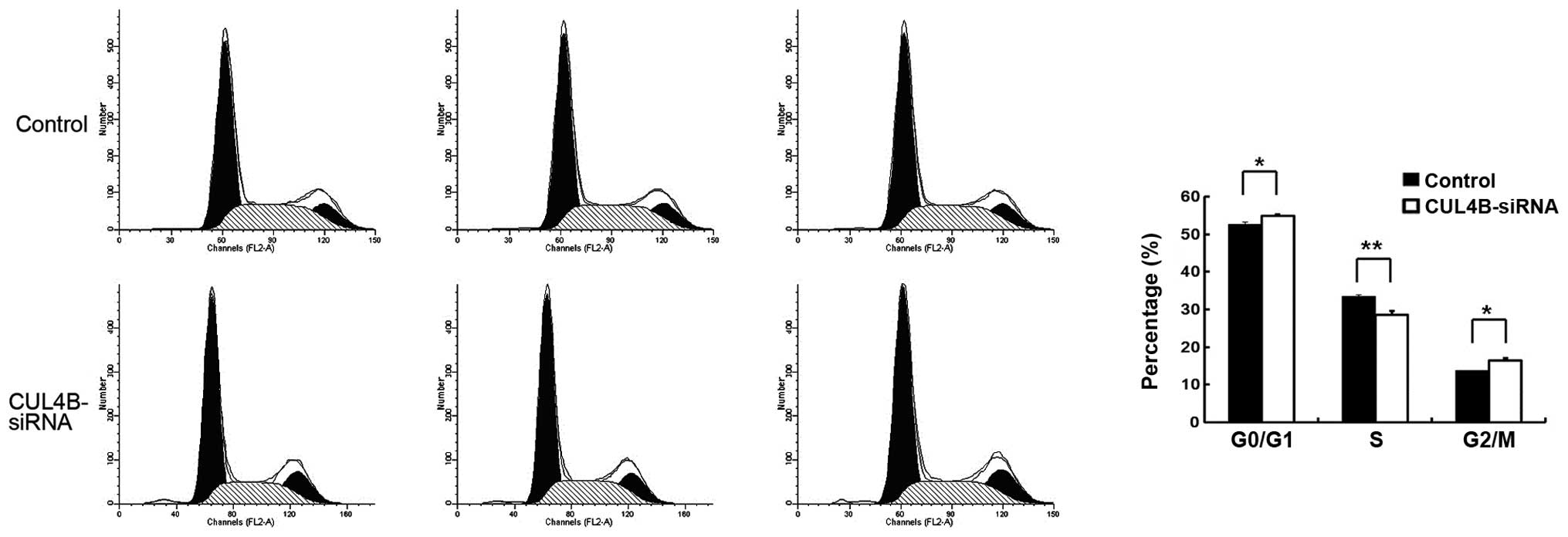
FACS analysis of cell cycle distribution
following CUL4B gene silencing
Following transfection with the siRNA lentivirus,
the percentage of SW1116 cells in the G1 phase was significantly
decreased (P<0.01), while cells in the S (P<0.01) and G2
phases increased significantly (P<0.05) in the knockdown group
compared with the negative control group. This indicates that
downregulation of the CUL4B gene was apparently related to regular
distribution of the SW1116 cells (Fig.
5).
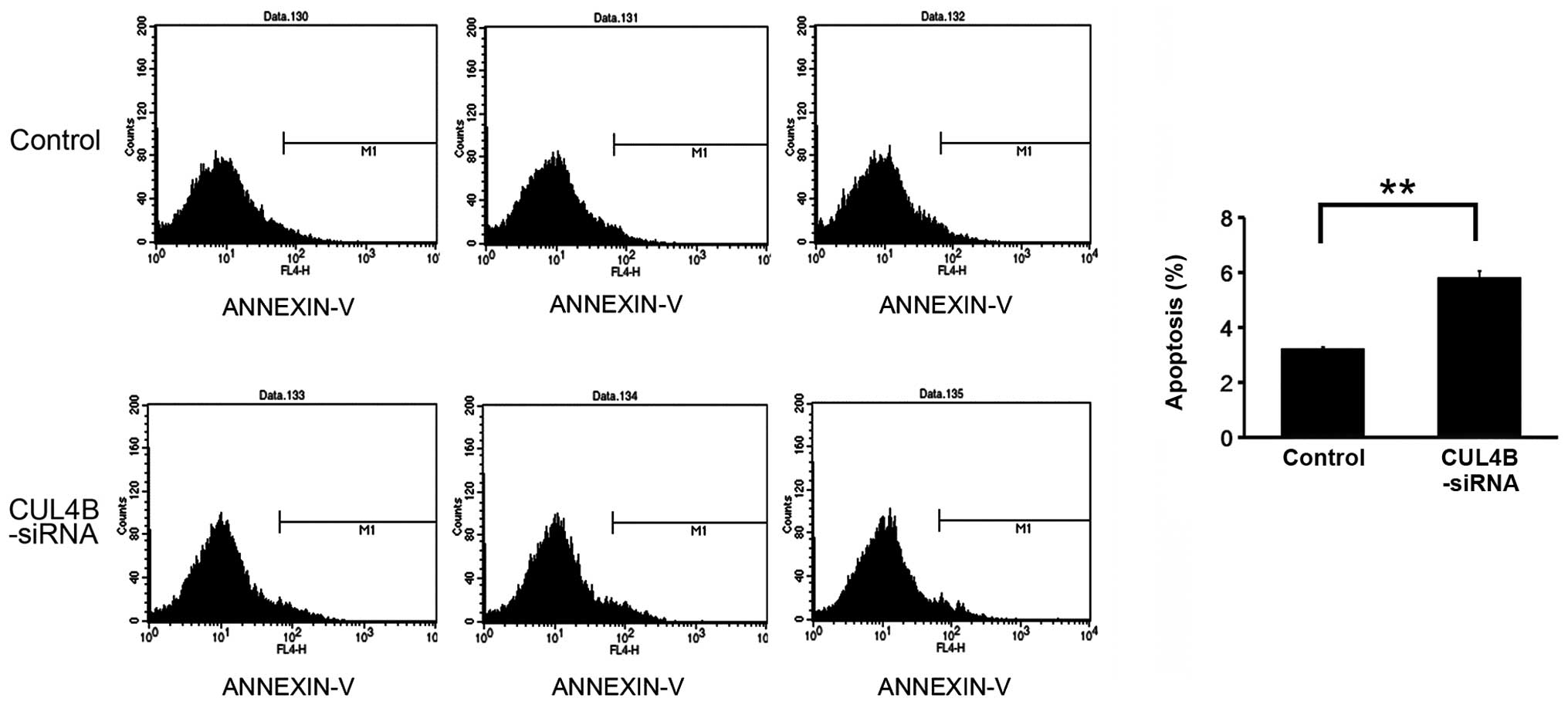
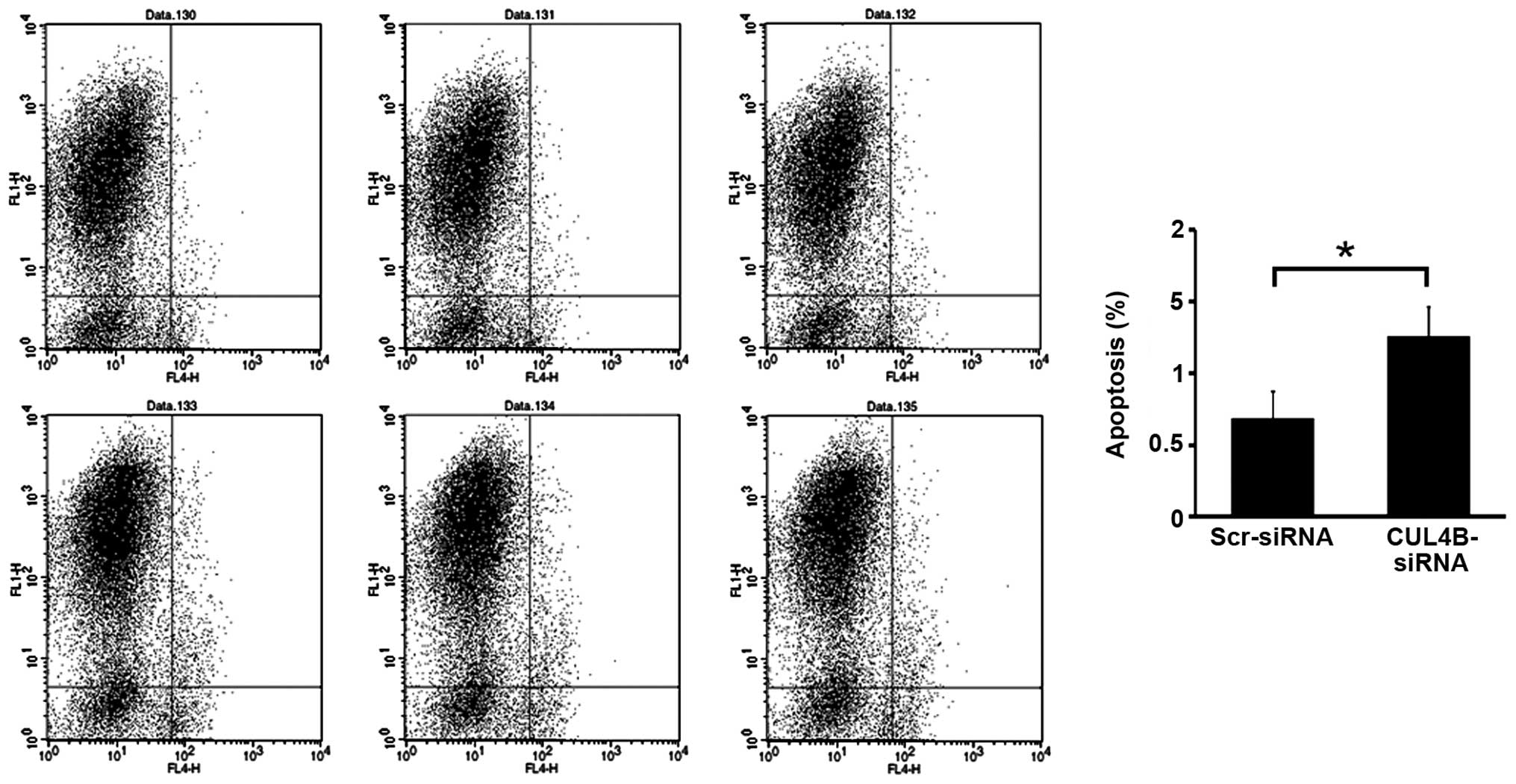
FACS analysis of apoptosis following
CUL4B gene silencing
Transfection by siRNA lentivirus increased the rate
of apoptosis significantly (P<0.05) in the knockdown group
compared with the negative control group, suggesting that the CUL4B
gene silencing stimulated the apoptosis of SAOS-2 cells (Figs. 6 and 7).
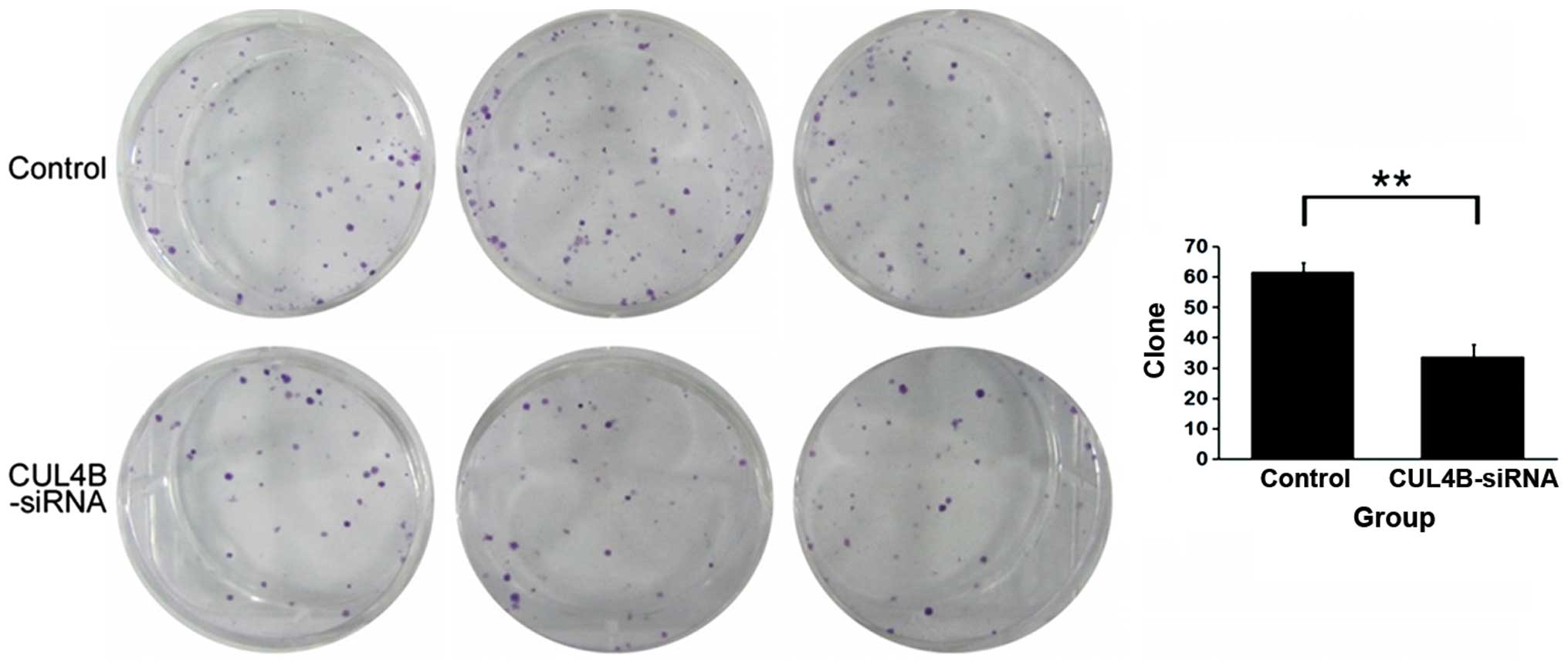
Analysis of inhibition of clonability
following CUL4B gene silencing
We noticed a decrease in the number of SW1116 cell
colonies in the knockdown group following interference by siRNA
lentivirus. Furthermore, the number of cells in the colonies that
had already formed decreased, which was significantly different
from the negative control group (P<0.01). The results suggest
that downregulation of the CUL4B gene largely inhibited the
clonability of the SAOS-2 cells (Fig.
8).
Discussion
In the present study, using RNA interference with a
lentiviral vector containing the CUL4B gene, we knocked down
the expression of the CUL4B gene in osteosarcoma SAOS-2
cells. Our results demonstrated that the knockdown of CUL4B
inhibited osteosarcoma SAOS-2 cell proliferation and clonability
and significantly induced apoptosis. Our findings suggest that
silencing of CUL4B gene may be used in the treatment of human
osteosarcoma.
Cancer cells commonly develop mechanisms by which
they resist cell death either through disruption of apoptotic
processes or activation of survival signals. Within a growing tumor
mass, the sequential acquisition of a number of genetic and
epigenetic alterations during tumor progression also enable cancer
cells to gain the ability to escape apoptosis, induce angiogenesis
and metastasize to distant organs. Notably, CUL4B carries a nuclear
localization signal in its N terminus and is also localized in the
nucleus (15,16), suggesting that CUL4B might be
involved in nuclear-based functions. Recently, one study indicated
that CUL4B is a transcriptional corepressor that regulates
transcription by recruiting PRC2. It was demonstrated that CUL4B
functions as a transcription corepressor and a potential oncogene,
supporting the use of CUL4B as a target for cancer therapy
(17).
Although the mechanism of CUL4B in cancer
development and progression remains unclear, studies have shown
that CUL4B may function together with other proteins, such as
cyclin D1 and p53 (7,18). The cyclin D1 level was low in
quiescent cells, and it increased as cells progressed into the G1
phase and served as a control protein (19). P53 also known as cellular tumor
antigen p53 or tumor-suppressor p53 is a protein that in humans is
encoded by the TP53 gene. The p53 protein is crucial in
multicellular organisms, where it regulates the cell cycle and,
thus, functions as a tumor suppressor, preventing cancer (20).
CUL4-mediated ubiquitination, which is involved in
other types of histone modifications and epigenetic mechanisms,
such as heterochromatin formation, parental imprinting or
X-chromosome inactivation (21–23),
may play an important role in cancer development. CUL4B expression
was found to be significantly higher in tumor samples compared to
adjacent normal tissue, and the level of CUL4B expression was
negatively correlated with the level of IGFBP3 expression.
Moreover, IGFBP3 is induced by wild-type p53 (24) and enhances the p53-dependent
apoptotic response of tumor cells to DNA damage (25). Aberrant promoter methylation of
IGFBP3 at the p53 regulatory element causes gene silencing
resistant to p53 (26).
Interestingly, it has been reported that CRL4A can degrade p53 to
promote cell cycle progression and immortalization (7,27).
Thus, it is reasonable to speculate that CRL4 negatively controls
the p53-IGFBP3 axis. Collectively, in addition to the hypothesis
that CRL4B promotes tumorigenesis through degradation of several
cyclin-dependent kinase inhibitors (8,9) and
coordinates with PRC2 in H3K27me3-mediated transcriptional
silencing (17), recent research
has also shown that CUL4B controls DNA methylation-based
transcriptional repression adding a new element to the
understanding of the oncogenic potential of CRL4B.
Progression of human cancers is known to involve
multiple genetic changes that lead to upregulation or
downregulation of expression of critical genes (28). Gene amplification is a common
mechanism whereby tumor cells increase expression of genes that are
critical for malignancy. However, not all genes present in multiple
copies are necessarily relevant to malignancy. Amplicons may
contain irrelevant genes that are present by virtue of their
physical proximity to target genes. In this study, we investigated
the biological function of the CUL4B gene in tumorigenesis
and proposed the possible mechanism based on previous research. We
provide a new prospective for the role of CUL4B in tumorigenesis
which warrants further research.
In summary, in our study, using RNA interference
with a lentiviral vector containing the CUL4B gene, we
knocked down the expression of the CUL4B gene in
osteosarcoma SAOS-2 cells. Our results demonstrated that the
knockdown of CUL4B can inhibit osteosarcoma SAOS-2 cell
proliferation and clonability and significantly induce apoptosis.
Our findings suggest that silencing of the CUL4B gene may be
valuable for the treatment of human osteosarcoma.
Acknowledgements
This study was supported by grants from The National
Natural Science Foundation of China General Program (no. 120038),
National Key Clinical Specialty Construction Project of China,
Shanghai Municipality Key Clinical Specialty (ZK2012A28) and
Sailing East Development Program of Shanghai East Hospital.
References
|
1
|
Ferrari S and Palmerini E: Adjuvant and
neoadjuvant combination chemotherapy for osteogenic sarcoma. Curr
Opin Oncol. 19:341–346. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sasaki N, Toda T, Kaneko T, Baba N and
Matsuo M: Protective effects of flavonoids on the cytotoxicity of
linoleic acid hydroperoxide toward rat pheochromocytoma PC12 cells.
Chem Biol Interact. 145:101–116. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jackson S and Xiong Y: CRL4s: the
CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 34:562–570.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kopanja D, Roy N, Stoyanova T, Hess RA,
Bagchi S and Raychaudhuri P: Cul4A is essential for spermatogenesis
and male fertility. Dev Biol. 352:278–287. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu L, Lee S, Zhang J, et al: CUL4A
abrogation augments DNA damage response and protection against skin
carcinogenesis. Mol Cell. 34:451–460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yin Y, Lin C, Kim ST, et al: The E3
ubiquitin ligase Cullin 4A regulates meiotic progression in mouse
spermatogenesis. Dev Biol. 356:51–62. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Banks D, Wu M, Higa LA, et al: L2DTL/CDT2
and PCNA interact with p53 and regulate p53 polyubiquitination and
protein stability through MDM2 and CUL4A/DDB1 complexes. Cell
Cycle. 5:1719–1729. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Higa LA, Yang X, Zheng J, et al:
Involvement of CUL4 ubiquitin E3 ligases in regulating CDK
inhibitors Dacapo/p27Kip1 and cyclin E degradation. Cell Cycle.
5:71–77. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nishitani H, Shiomi Y, Iida H, Michishita
M, Takami T and Tsurimoto T: CDK inhibitor p21 is degraded by a
proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway
during S phase and after UV irradiation. J Biol Chem.
283:29045–29052. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schindl M, Gnant M, Schoppmann SF, Horvat
R and Birner P: Overexpression of the human homologue for
Caenorhabditis elegans cul-4 gene is associated with poor
outcome in node-negative breast cancer. Anticancer Res. 27:949–952.
2007.
|
|
11
|
Singhal S, Amin K, Kruklitis R, et al:
Alterations in cell cycle genes in early stage lung adenocarcinoma
identified by expression profiling. Cancer Biol Ther. 2:291–298.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tarpey PS, Raymond FL, O’Meara S, et al:
Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit,
cause an X-linked mental retardation syndrome associated with
aggressive outbursts, seizures, relative macrocephaly, central
obesity, hypogonadism, pes cavus, and tremor. Am J Hum Genet.
80:345–352. 2007. View
Article : Google Scholar
|
|
13
|
Zou Y, Liu Q, Chen B, et al: Mutation in
CUL4B, which encodes a member of cullin-RING ubiquitin ligase
complex, causes X-linked mental retardation. Am J Hum Genet.
80:561–566. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang Y, Liu R, Qiu R, et al: CRL4B
promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in
DNA methylation-based epigenetic silencing. Oncogene. Dec
2–2013.(Epub ahead of print). doi: 1038/onc.2013.522.
|
|
15
|
Nakagawa T and Xiong Y: X-linked mental
retardation gene CUL4B targets ubiquitylation of H3K4
methyltransferase component WDR5 and regulates neuronal gene
expression. Mol Cell. 43:381–391. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zou Y, Mi J, Cui J, et al:
Characterization of nuclear localization signal in the N terminus
of CUL4B and its essential role in cyclin E degradation and cell
cycle progression. J Biol Chem. 284:33320–33332. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu H, Yang Y, Ji Q, et al: CRL4B catalyzes
H2AK119 monoubiquitination and coordinates with PRC2 to promote
tumorigenesis. Cancer Cell. 22:781–795. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aggarwal P, Lessie MD, Lin DI, et al:
Nuclear accumulation of cyclin D1 during S phase inhibits
Cul4-dependent Cdt1 proteolysis and triggers p53-dependent DNA
rereplication. Genes Dev. 21:2908–2922. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Koepp DM, Harper JW and Elledge SJ: How
the cyclin became a cyclin: regulated proteolysis in the cell
cycle. Cell. 97:431–434. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Levine AJ, Momand J and Finlay CA: The p53
tumour suppressor gene. Nature. 351:453–456. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jia S, Kobayashi R and Grewal SI:
Ubiquitin ligase component Cul4 associates with Clr4 histone
methyltransferase to assemble heterochromatin. Nat Cell Biol.
7:1007–1013. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Horn PJ, Bastie JN and Peterson CL: A
Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential
for heterochromatin formation. Genes Dev. 19:1705–1714. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dumbliauskas E, Lechner E, Jaciubek M, et
al: The Arabidopsis CUL4-DDB1 complex interacts with MSI1 and is
required to maintain MEDEA parental imprinting. EMBO J. 30:731–743.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Buckbinder L, Talbott R, Velasco-Miguel S,
et al: Induction of the growth inhibitor IGF-binding protein 3 by
p53. Nature. 377:646–649. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Williams AC, Collard TJ, Perks CM, et al:
Increased p53-dependent apoptosis by the insulin-like growth factor
binding protein IGFBP-3 in human colonic adenoma-derived cells.
Cancer Res. 60:22–27. 2000.
|
|
26
|
Hanafusa T, Shinji T, Shiraha H, et al:
Functional promoter upstream p53 regulatory sequence of IGFBP3 that
is silenced by tumor specific methylation. BMC Cancer. 5:92005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nag A, Bagchi S and Raychaudhuri P: Cul4A
physically associates with MDM2 and participates in the proteolysis
of p53. Cancer Res. 64:8152–8155. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dairkee SH and Smith HS: Genetic analysis
of breast cancer progression. J Mammary Gland Biol Neoplasia.
1:139–151. 1996. View Article : Google Scholar : PubMed/NCBI
|