Introduction
Colorectal cancer (CRC) is one of the leading causes
of cancer-related mortality worldwide. In spite of screening
examination, CRC incidence and the associated mortality have
increase rapidly in the past several decades (1,2). The
prognosis of CRC is associated with stage, and CRC can be
completely cured if detected early; the 5-year survival rate is
93.2% for stage I, and 8.1% for stage IV (3). Thus, early detection of CRC is crucial
to reduce mortality, and CRC-related deaths can be prevented
through early detection and early treatment. Several CRC screening
tests including fecal occult blood testing (FOBT) and colonoscopy
have been available for years (4).
However, these methods are associated with issues such as low
adherence rates, high cost or low sensitivity. An ideal screening
method should have a high sensitivity and specificity for early
stage CRC; it should also be safe and accepted by patients
(5–7).
MicroRNAs (miRNAs) are 22-nucleotide non-coding RNA
molecules that regulate a variety of cellular processes including
cell differentiation, cell cycle progression and apoptosis. miRNAs
have been demonstrated to play an important role in the multistep
processes of carcinogenesis either by oncogenic or
tumor-suppressive function. The study of miRNAs has been extended
to many types of tumors, including CRC (8–10).
These studies have revealed that miRNAs may be potential diagnostic
or prognostic tools for human cancers.
Tumor-associated RNAs have been described in the
serum/plasma of cancer patients for more than a decade. More
recently, several studies have also demonstrated that circulating
miRNAs exist in serum/plasma (11,12).
Accordingly, several subsequent studies have proven that miRNAs can
serve as potential biomarkers for various diseases including
cancer. It has been revealed that miR-92 and miR-21 are
significantly elevated in the plasma of CRC patients and can be
potential non-invasive molecular markers for CRC detection
(13–16). In the present study, we aimed to
identify a novel circulating miRNA in CRC patients and to evaluate
its feasibility as a noninvasive diagnostic test for efficient
detection of CRC.
Materials and methods
Patients and samples
Informed consent was obtained from CRC patients and
non-cancer patients for the use of their blood samples. From April
2011 to June 2013, venous blood samples were collected from the CRC
patients (n=114) and non-cancer patients who suffered inguinal
hernia or gall bladder stone (n=32). In 30 of the CRC patients,
blood samples were obtained before and on the 7th day after
surgery. No cancer patients received chemotherapy or radiotherapy
before blood sampling. The blood samples were obtained from Osaka
University, Osaka Rosai Hospital, Suita Municipal Hospital and
Saiseikai Senri Hospital. Whole blood was collected, centrifuged at
f1,000 rpm and 4°C for 15 min. The supernatant fluids were
centrifuged at 15,000 rpm and 4°C for 10 min. The supernatant
fluids were stored at −80°C until RNA extraction. This study was
conducted under the supervision of the Ethics Board of Osaka
University Hospital.
RNA extraction
Small RNA was enriched from all serum samples using
the mirVana Paris RNA isolation kit (Ambion, Austin, TX, USA),
following the manufacturer’s instructions. Briefly, 400 μl of serum
was thawed on ice and centrifuged at 15,000 rpm for 15 min to
remove cell debris. Next, 300 μl of the supernatant was lysed with
an equal volume of 2X denaturing solution. For normalization of
sample-to-sample variation during the RNA isolation procedures, 20
fmol of synthetic C. elegans miRNA cel-miR-39 was added to
each denatured sample. Small RNAs were then enriched and purified
following the manufacturer’s protocol. The concentration of all RNA
samples were quantified by NanoDrop ND-1000 (Nanodrop, Wilmington,
DE, USA).
miRNA microarray analysis
miRNA microarray experiments were carried out using
Agilent human miRNA microarray catalogued in the Sanger database
ver. 12.0 (design ID 021827). Approximately 10 ng of aliquots of
total RNA with cel-miR-39 was used for making the miRNA probes
according to the Agilent protocol (ver. 2.3). Microarrays were
performed for paired pre-operative and post-operative serum from 10
CRC patients. Briefly, total RNA was dephosphorylated with calf
intestine alkaline phosphatase, denatured with dimethyl sulfoxide,
and labeled with pCp-Cy3 using T4 RNA ligase using an miRNA
labeling reagent and hybridization kit. Probes were hybridized at
55°C for 20 h with rotation. Then the slides were washed by Gene
Expression Wash Buffer 1 at room temperature for 5 min and by Gene
Expression Wash Buffer 2 at 37°C for 5 min. After hybridization and
washing, the slides were scanned using an Agilent scanner (G2505C).
Images were extracted using Agilent Feature Extraction software
(ver. 10.7.3.1) and Agilent GeneSpring GX software (ver. 10.0.2).
Differences in miRNA expressions between the 10 pairs was
determined when the fold-change of cel-miR-39 normalized expression
values was >2.0 and the P-value was <0.05 using paired t-test
for further analysis. The microarray raw data are available in Gene
Expression Omnibus (GEO;http://www.ncbi.nlm.nih.gov/geo) under accession no.
GSE*****.
qRT-PCR
For the microRNA-based RT-PCR assays, 2.5 μl of
enriched small RNAs from serum samples was reverse transcribed
using the TaqMan MicroRNA Reverse Transciption kit (Applied
Biosystems, San Diego, CA, USA) according to the manufacturer’s
instructions in a total reaction volume of 7.5 μl. A 1:20 dilution
of RT products was used as template for the PCR stage. PCR reaction
was performed in triplicates using TaqMan 2X Universal PCR Master
Mix according to the manufacturer’s instructions. Each reaction was
performed in a final volume of 20 μl containing 1.33 μl of the cDNA
and 1 μl of TaqMan miRNA Assay Mix. The amplification profile
consisted of denaturation at 95°C for 10 min, followed by 40 cycles
of 95°C for 15 sec and 60°C for 60 sec. Each sample was run in
triplicates for analysis. The cycle threshold (Ct) is defined as
the number of cycles required for the fluorescent signal to cross
the threshold in qPCR. The 7900 Sequence Detection System 2.3
(Applied Biosystems) software was used to compute the relative
change in RNA expression by the 2−ΔΔCt method with 95%
confidence intervals.
Primers
The miRNA-specific primer sequences, including
miRNA-199a-3p, let-7a, cel-miR-39 and RNU6B, were designed based on
the miRNA sequences obtained from the miRBase. The primer sequences
were: hsa-miR-199a-3p, 5′-ACAGUAGUCUGCACAUUGGUUA-3′; hsa-let-7a,
5′-UGAGGUAGUAGGUUGUAUAGUU-3′; hsa-miR-21,
5′-UAGCUUAUCAGACUGAUGUUGA-3′; cel-miR-39,
5′-UCACCGGGUGUAAAUCAGCUUG-3′ and RNU6B,
5′-CGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTTT-3′.
Statistical analysis
The significance of the serum miRNA level was
determined by Mann-Whitney, Wilcoxon and χ2 tests where
appropriate using the GraphPad Prism 6 (San Diego, CA, USA). The
sensitivity, specificity and accuracy were calculated according to
standard formulas. Receiver operating characteristic (ROC) curve
and area under the ROC curve (AUC) were established for
discriminating patients with CRC. P-values of <0.05 were
considered to indicate a statistically significant result.
Results
Results of the miRNA microarray
analysis
Following comparison between the pre-operative and
post-operative CRC patient serum (n=10; 4 stage II CRCs and 6 stage
III CRCs) by the miRNA array, we identified miRNAs, the majority of
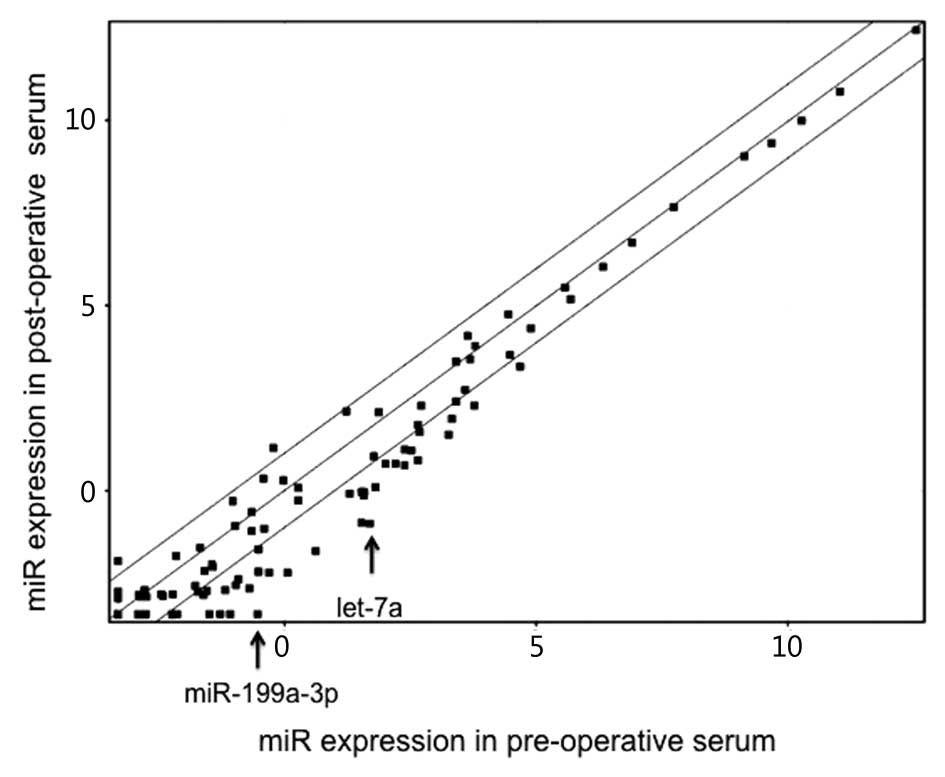
which showed a decrease in the post-operative serum (Fig. 1). Among them, we focused on two
miRNAs, miR-199a-3p and let-7a, whose expression showed the largest
decrease after surgery with significant P-values (6.82- and
5.92-fold decrease; P=0.015 and 0.029, respectively, Fig. 1).
Confirmation of the results obtained from
the miRNA array by qRT-PCR
We then attempted to confirm the results obtained
from the miRNA array by qRT-PCR in extended samples of CRC patients
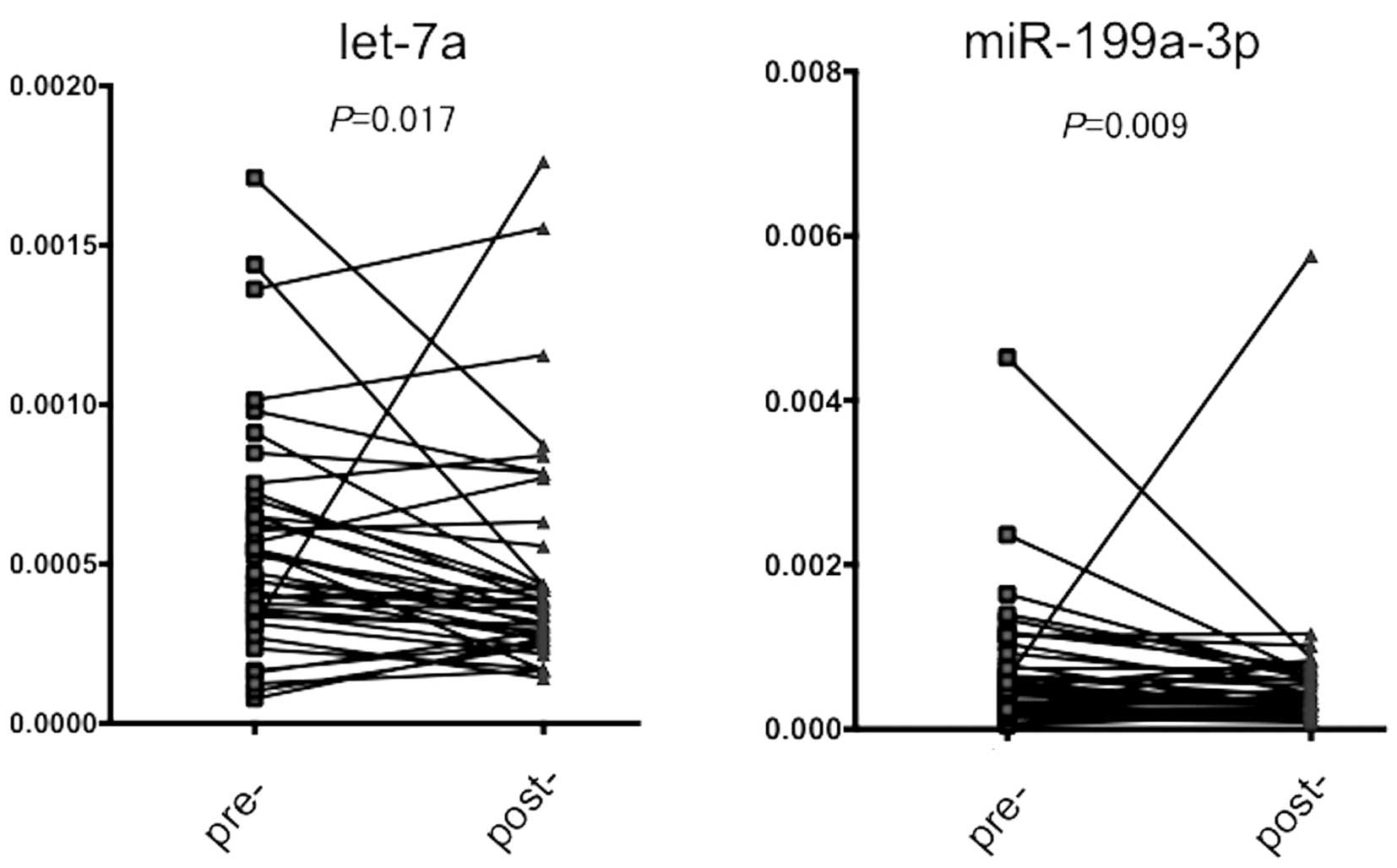
(n=30). As shown in Fig. 2, a
significant decrease in miRNA levels was noted in the
post-operative serum for both miRNAs (P=0.017 for let-7a and 0.009
for miR-199a-3p, respectively), suggesting that the expression
levels of these miRNAs were reduced after removal of the main tumor
by surgery.
Expression of miRNAs in the serum of
normal and CRC patients
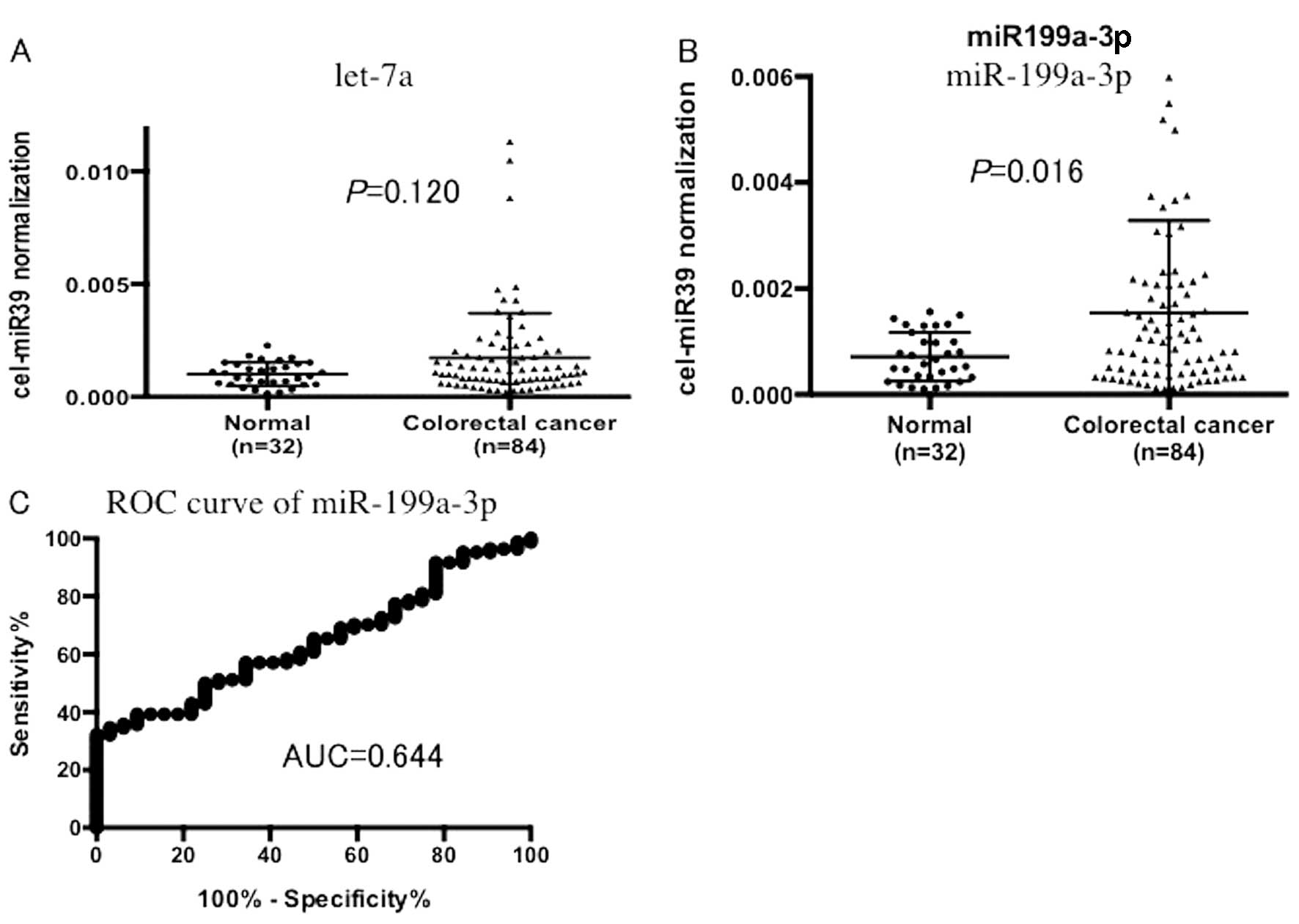
We examined the serum let-7a level in 32 non-cancer
patients and 84 CRC patients and no significant difference was
noted (P=0.120, Fig. 3A). In
contrast, miR199a-3p expression was significantly higher in the CRC
patients than that in the non-cancer patients (P=0.016, Fig. 3B). An ROC curve was drawn for serum
miR-199a-3p, which yielded 0.644 as a value of AUC. When a cut-off
point was set at 0.0010, the sensitivity was 47.6% and the
specificity was 75.0% in discriminating CRC from the non-tumor
control subjects.
Serum miR-21 expression in non-cancer and
CRC patients
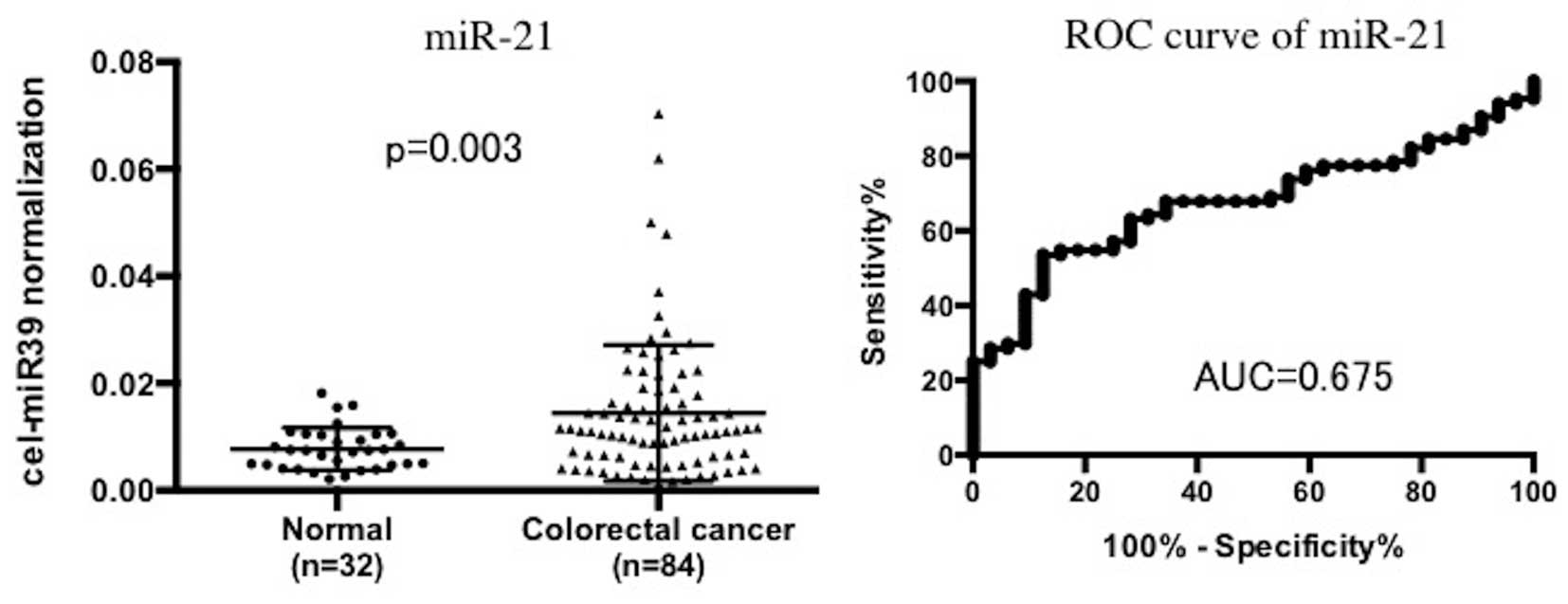
As a reference, we examined miR-21 as a putative
circulating miRNA. Using the same serum sets of normal and CRC
patients, we found that the serum miR-21 levels were significantly
increased in the CRC patients when compared to levels in the
non-tumor control patients (P=0.003). AUC of the ROC curves was
0.675. When a cut-off point was set at 0.0107, the sensitivity was
54.7% and the specificity was 84.4% in discriminating CRC from the
non-tumor control subjects (Fig.
4).
miR-199a-3p expression in patient serum
and clinicopathological characteristics
Clinical and pathological survey indicated that
miR-199-3p was significantly associated with deep wall invasion in
the high miR-199a-3p patient group when compared to the low
miR-199a-3p expression group (P=0.022), when CRC patients were
divided into two groups by median expression (Table I).
 | Table IRelationship between miR-199a-3p and
clinicopathological features. |
Table I
Relationship between miR-199a-3p and
clinicopathological features.
| miR199 |
|---|
|
|
|---|
| High | Low | P-value |
|---|
| Gender | | | 0.354 |
| Male | 30 | 26 | |
| Female | 12 | 16 | |
| Differentiation | | | 0.502 |
| Well | 15 | 18 | |
| Mod, por, muc | 27 | 24 | |
| Tumor size (mm) | | | 0.189 |
| ≥35 | 25 | 19 | |
| <35 | 17 | 23 | |
| Serosal invasion | | | 0.022a |
| T1, T2 | 10 | 20 | |
| T3, T4 | 32 | 22 | |
| Lymph node
metastatis | | | 0.826 |
| Positive | 19 | 18 | |
| Negative | 23 | 24 | |
| Lymphatic
invasion | | | 0.275 |
| Positive | 24 | 19 | |
| Negative | 18 | 23 | |
| Venous invasion | | | 0.232 |
| Positive | 10 | 15 | |
| Negative | 32 | 27 | |
Expression of miR-199a-3p in normal
mucosa and CRC tissue samples
We additionally examined miR-199a-3p expression in
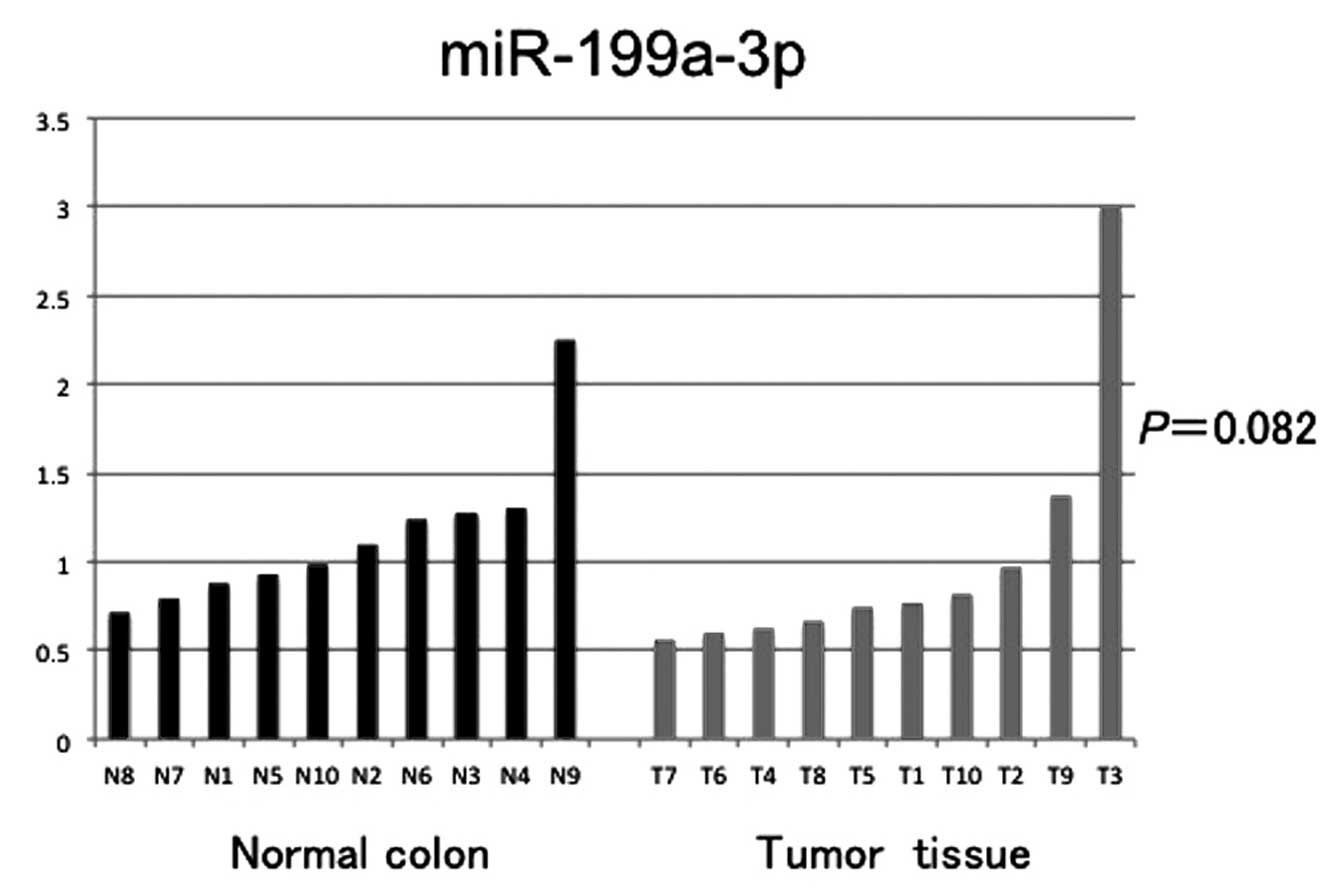
normal colonic mucosa (n=10) and CRC tissue samples (n=10). No
significant difference in miR-199a-3p expression was noted
(P=0.082, Fig. 5).
Discussion
In the present study we aimed to identify a novel
serum marker for colorectal cancer (CRC) by comparison of the
patient serum before and after surgery, using the miRNA array. The
miRNAs were mostly reduced after surgery to various extents. Among
them, we focused on miR-199a-3p and let-7a since the reduction rate
of the miRNAs was relatively large. qRT-PCR verified the results
obtained by the miRNA array in an extended number of CRC patients.
These findings suggest that the two miRNAs may be derived from the
main CRC tumors, either directly or indirectly and that they may be
useful to monitor disease progression.
To investigate whether the two miRNAs may be
informative as serum tumor markers when compared to non-tumor
patients, we compared the serum miRNA expression between non-cancer
and CRC patients. We found that expression of miR-199a-3p, but not
let-7a, was increased in the CRC patients when compared with the
non-tumor patients. Based on these findings, we suggest that
miR-199a-3p could be a superior marker for detection of CRC by
blood tests. Indeed, miR-199a-3p levels increased going from
non-cancer patients to CRC patients and were markedly decreased
after tumor resection.
Clinicopathological survey showed that the serum
miR-199a-3p level was associated with serosal invasion of the
primary CRC tumor, suggesting that miR-199a-3p may be associated
with tumor invasion. In support of this hypothesis, Wan et
al provided evidence that high miR-199a-3p expression in CRC
tissue samples contributes to lymph node and liver metastases and
advanced TNM stage (18).
In analysis of the tissue-derived miR-199a-3p, we
found no significant difference between normal colonic mucosa and
CRC tissue samples. These findings suggest that tumor cells may
secrete miR-199a-3p to a greater extent than normal epithelial
cells, or certain host cells may also contribute to secretion of
miR-199a-3p in the tumor-bearing patients. We postulate that the
former is more probable since other investigators have reported
miR-199a-3p expression in the stool and tumor tissues of CRC
patients (18,19). Moreover, it has been shown that
anti-miRNA against miR-199a-3p suppressed the growth of colon
cancer cells, suggesting that miR-199a-3p is an oncogenic miRNA
(18). In spite of these earlier
discoveries, we are not aware of any studies reporting that serum
miR-199a-3p is a useful biomarker for CRC. Considering the
convenience of blood samples that allow monitoring at numerous time
points by the relatively easy detection system, our finding of
circulating miR-199a-3p in CRC is of clinical importance.
The ROC curve of miR-199a-3p to distinguish cancer
patients from non-cancer patients appeared to have limitations in
sensitivity and specificity. To estimate its value, we examined
serum miR-21 expression in the same series of non-tumor and CRC
patients since miR-21 is known as a putative serum biomarker for
CRC (17). As a result, we found
that the ROC curve of miR-21 was not markedly different of that of
miR-199a-3p in our patient series.
In conclusion, we identified miR-199a-3p from the
differential expression profile between pre-operative and
post-operative serum as a novel serum biomarker for CRC. Further
investigation of serum miR-199a-3p in regards to patient prognosis
and further monitoring of the therapeutic efficacy of chemotherapy
are essential.
Acknowledgements
This study was supported by a Grant-in-Aid for
Scientific Research (B) (24390315 to H.Y.)
Abbreviations:
|
CRC
|
colorectal cancer
|
|
miR/miRNA
|
microRNA
|
|
qRT-PCR
|
quantitative reverse
transcription-polymerase chain reaction
|
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar
|
|
2
|
Jemel A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
3
|
O’Connell JB, Maggard MA and Ko CY: Colon
cancer survival rates with the new American Joint Committee on
Cancer sixth edition staging. J Natl Cancer Inst. 6:1420–1425.
2004.
|
|
4
|
Hewitson P, Glasziou P, Watson E, Towler B
and Irwig L: Cochrane systematic review of colorectal cancer
screening using the fecal occult blood test (hemoccult): an update.
Am J Gastroenterol. 103:1541–1549. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Walsh JM and Terdiman JP: Colorectal
cancer screening: scientific review. JAMA. 289:1288–1296. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baxter NN, Warren JL, Barrett MJ, Stukel
TA and Doria-Rose VP: Association between colonoscopy and
colorectal cancer mortality in a US cohort according to site of
cancer and colonoscopist specialty. J Clin Oncol. 30:2664–2669.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Winawer S, Fletcher R, Rex D, et al:
Colorectal cancer screening and surveillance: clinical guidelines
and rationale - Update based on new evidence. Gastroenterology.
124:544–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He L, Thomson JM, Hemann MT, et al: A
microRNA polycistron as a potential human oncogene. Nature.
435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schetter AJ, Leung SY, Sohn JJ, et al:
MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mitchell PS, Parkin RK, Kroh EM, et al:
Circulating microRNAs as stable blood-based markers for cancer
detection. Proc Nat Acad Sci USA. 105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schwarzenbach H, Hoon DS and Pantel K:
Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev
Cancer. 11:426–437. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pu XX, Huang GL, Guo HQ, et al:
Circulating miR-221 directly amplified from plasma is a potential
diagnostic and prognostic marker of colorectal cancer and is
correlated with p53 expression. J Gastroenterol Hepatol.
25:1674–1680. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ng EK, Chong WW, Jin H, et al:
Differential expression of microRNAs in plasma of patients with
colorectal cancer: a potential marker for colorectal cancer
screening. Gut. 58:1375–1381. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang Z, Huang D, Ni S, Peng Z, Sheng W
and Du X: Plasma microRNAs are promising novel biomarkers for early
detection of colorectal cancer. Int J Cancer. 127:118–126. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng H, Zhang L, Cogdell DE, et al:
Circulating plasma miR-141 is a novel biomarker for metastatic
colon cancer and predicts poor prognosis. PLoS One. 6:e177452010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Toiyama Y, Takahashi M, Hur K, et al:
Serum miR-21 as a diagnostic and prognostic biomarker in colorectal
cancer. J Natl Cancer Inst. 105:849–859. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wan D, He S, Xie B, et al: Aberrant
expression of miR-199a-3p and its clinical significance in
colorectal cancers. Med Oncol. 30:3782013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ahmed FE, Ahmed NC, Vos PW, et al:
Diagnostic microRNA markers to screen for sporadic human colon
cancer in stool: I. Proof of principle. Cancer Genomics Proteomics.
10:93–113. 2013.PubMed/NCBI
|