Introduction
The Wilms’ tumor 1 (WT1) gene is one of the
important regulators of cellular growth and development. It was
originally identified as a gene that is deleted or rearranged in
many cases of hereditary Wilms’ tumor, a childhood kidney neoplasm.
Located at 11p13, the wt1 gene encodes a zinc finger-containing
nuclear protein with DNA- and RNA-binding activities (1,2). Its
target genes are involved in cell growth, metabolism,
differentiation and death, including CDKN1A, a negative regulating
gene in cell cycle progression; MAPK signaling genes; and Wnt
pathway genes (3–7). In many hematological malignancies,
particularly in most acute leukemias, WT1 mRNA expression is high
and is associated with poor prognosis (8–10).
In the present study, WT1 mRNA and protein
expression levels were detected in several leukemic cell lines,
particularly in K562 cells, a blast phase chronic myeloid leukemia
(CML) cell line, where a higher expression was observed. Thus, we
chose K562 cells as the main subject for further study. Curcumin is
one of the WT1 protein inhibitors and was used to treat K562 cells
(2). Correlation between expression
of the WT1 gene and cell biological characteristics was analyzed.
As curcumin is not a specific inhihitor of WT1 protein, we designed
WT1-specific short hairpin RNA (shRNA) nucleotides. The WT1 gene in
K562 cells was downregulated by WT1-specific shRNA nucleotides, and
cell growth was inhibited, the numbers of colonies were obviously
decreased and the cell cycle was arrested at the
G0/G1 phase. These results indicate that the
WT1 gene may play an important role in leukemia cell proliferation
and cell cycle regulation. Yet, the mechanisms involved in the
effects of WT1 on K562 cells remain unknown. Thus, CHIP-DSL was
performed. Classes of the WT1 targets identified consisted of
several vital signaling pathways, such as the Wnt pathway, the MAPK
pathway, cell cycle, apoptosis, differentiation, cell adhesion and
cell migration. The Wnt/β-catenin pathway plays an important role
in tumorigenesis and progression of leukemia. Therefore, we
detected the levels of major genes in the traditional canonical and
non-canonical pathways. We observed that when WT1 was
downregulated, expression of several important genes such as
β-catenin, SUZ12 and Wnt11 was decreased at the mRNA and protein
levels, while expression of Wnt3a and Wnt5a was slightly increased.
Meanwhile, the subcellular location of β-catenin was altered, and
expression levels of the targets of β-catenin, CCND1 and MYC genes,
were obviously decreased. Collectively, these data suggest that WT1
functions as an oncogene in leukemia cells, and one important
mechanism is regulation of the Wnt/β-catenin pathway.
Materials and methods
Cell culture
Human leukemia cell lines K562, HL60, NB4, U937 and
Kasumi-1 were cultured in RPMI-1640 medium supplemented with 10%
fetal calf serum at 37°C in a humidified environment of 5%
CO2. Proper cell densities were planted to ensure
logarithmic growth. To assess the effect of colony formation under
different conditions, aliquots containing 1,000 cells were plated
into 48-well culture plates. At different time points, colonies
containing approximately 50 cells were counted using an inverted
phase contrast microscope.
Western blot assays
Cell pellets were lysed in lysis buffer [50 mM KCl,
2 mM MgCl2, 1 mM EDTA, 5 mM DTT, 25 mM Tris (pH 7.5), 1
μg/ml leupeptin, 10 μg/ml laprotinin, and 1 mM phenylmethylsulfonyl
fluoride] on ice for 30 min and centrifuged for 5 min at 20,000 × g
at 4°C. The protein concentration of the supernatant was determined
using BCA protein assay reagents (Pierce, USA). Equal amounts of
protein (100 μg) were separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE),
electroblotted on nitrocellulose membranes, and immunostained with
the primary antibodies followed by horseradish
peroxidase-conjugated anti-rabbit or anti-mouse secondary antibody.
Finally, protein densities were analyzed using the AlphaEaseFC
software according to the manufacturer’s instructions. Antibodies
included rabbit polyclonal WT1 antibody, β-catenin monoclonal
antibody, wnt3a, wnt5a, wnt11 (Abcam Bio Company) and SUZ12
(CST).
Lentivirus production and cell
transduction
The WT1 shRNA sequence (obtained by the TRC
consortium) was sense, 5′-CCGGGCAGCTAACAATGTCTGGTTACTCGAGTAAC
CAGACATTGTTAGCTGCTTTTTG-3′ and antisense,
5′-AATTCAAAAAGCAGCTAACAATGTCTGGTTACTCG AGTAACCAGACATTGTTAGCTGC-3′;
targeting sequence, GCAGCTAACAATGTCTGGTTA, and the control-shRNA
sense, 5′-CCGGCCTAAGGTTAAGTCGCCCTCGCTCG
AGCGAGGGCGACTTAACCTTAGGTTTTT-3′ and anti-sense,
5′-AATTAAAAACCTAAGGTTAAGTCGCCCTCG CTCGAGCGAGGGCGACTTAACCTTAGGGG-3′.
By ligation, transformation, plasmid extraction and sequencing,
plKO.1 vectors were obtained with the puromycin-selection marker.
Then lentiviral vector production was performed by transient
transfection of 293T cells using calcium phosphate. The viral
supernatant was concentrated 200-fold by ultracentrifugation. K562
cells were plated at a density of 106/ml in serum-free
medium, and then transduction was carried out for 12 h. Puromycin
was then used to select the positive cells (the final concentration
of puromycin was 2–5 μg/ml); using three selecting cycles, the
positive cells were obtained.
MTT assays
The MTT assay was performed as previously described
(11). Briefly, 1×104
cells were plated in each well of 96-well microtiter plates with
100 μl of fresh medium containing curcumin at various
concentrations. After 24, 48 or 72 h of further incubation, 10 μl
methylthiazol tetrazolium (MTT) solution (2.5 mg/ml) was added to
each well, and the cells were incubated for another 4 h. The medium
was then removed, and the formed formazan crystals were dissolved
in 150 μl of dimethyl sulfoxide (Sigma, USA). The plates were
placed on a plate shaker for 10 min, and the absorbance of the
resulting solution was immediately measured at 546 nm using a
microplate reader (SLT-Lab, Salzburg, Austria). The growth
inhibition ratio was calculated using the formula: Growth
inhibitory rate = (1 − T/C) × 100%, where T is the absorbance rate
of the treatment group with curcumin and C is the absorbance rate
of the control group. the IC50 value of curcumin was
calculated using SPSS software (11). These experiments were repeated three
times.
Cell cycle analysis
K562 cells treated with curcumin at different time
points or cells which were silenced for WT1 were washed twice with
cold PBS and then resuspended with ice-cold 70% ethanol overnight.
After washing with cold PBS, the cells were treated with 10 mg/ml
of RNase at 37°C for 30 min and then stained using 40 mg/ml of
propidium iodide (PI) for 30 min. DNA content was measured by flow
cytometry (Beckman Coulter).
Real-time quantitative RT-PCR
Total RNA was extracted from 1×106 cells
using RNAsio (Takara, Japan) according to the protocol supplied
with the reagent. Isolated RNA was resolved in 0.1% diethyl
pyrocarbonate (DEPC)-treated water. Complementary DNA (cDNA) was
synthesized from 2 μg RNA with N6 primers using murine
myeloleukemia virus reverse transcriptase (M-MLV, Promega, USA),
following the procedure provided by the manufacturer.
Real-time quantitative RT-PCR (RQ-PCR) was performed
using SYBR-Premix Ex Taq (Takara, Japan) on a 7500 Thermo Cycler
(Applied Biosystems, USA). Each sample was run in triplicate.
Relative quantification of WT1, BCR/ABL, wnt3a, wnt5a, wnt11,
SUZ12, β-catenin, CCND1 and MYC mRNA expression was conducted by
the comparative Ct method established by Livak and Schmittgen
(12). SDS 2.1 software (Applied
Biosystems, USA) was used. The output relative expression value was
normalized to the endogenous reference GAPDH. The primer sequences
used are as follows: WT1 forward, 5′-CACGAGGAGCAGTGCCTGAG-3′ and
reverse, 5′-AACCCTGATTGCGAATAGCG-3′; GAPDH forward,
5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAA GATGGTGATGGGATTTC-3′;
BCR/ABL forward, 5′-gggctc tatgggtttctgaatg-3′ and reverse,
5′-cgctgaagggcttttgaact-3′; wnt3a forward,
5′-TGGTGTCTCGGGAGTTCGC-3′ and reverse, 5′-CCGTGGCACTTGCACTTGA-3′;
wnt5a forward, 5′-CATACCTTGAGCACGAC-3′ and reverse, 5′-TCTTAA
CGTCCATGTCTAT-3′; wnt11 forward, 5′-GACCTCAAG ACCCGATACCT-3′ and
reverse, 5′-GGAGCCCACCTT CTCATTC-3′; SUZ12 forward,
5′-CCTATTGCCAAAC CTC-3′ and reverse, 5′-TTGCCTTGTATTGTTGT-3′;
β-catenin forward, 5′-GGCAGCAACAGTCTTA-3′ and reverse,
5′-GTCTCAGGGAACATAGC-3′.
Immunofluorescence assay
Cells were washed 3 times with PBS buffer and then
fixed with 4% paraformaldehyde solution at RT for 10 min. The cells
were washed 3 times with PBS buffer, and permeabilized for 10 min
at RT with 0.1% Triton and 0.2% BSA-PBS. Cell were then washed 3
times with PBS buffer, and non-specific protein binding was blocked
with 10% goat serum-PBS or with 2% BSA-PBS for 30 min. Goat serum
or BSA was removed and without washing a coverslip was added. The
primary antibody was diluted in 2% goat serum-PBS or 1% BSA-PBS for
45 min at RT. Washing was carried out 5 times with PBS, added to
coverlip. The secondary antibody was diluted in 2% goat serum-PBS
or 1% BSA-PBS for 30 min at RT. Washing was carried out 5 times
with PBS. The coverslip was placed on the slide very carefully and
observed under a confocal laser scanning scope immediately.
Statistical analysis
Three repetitions were performed for each
experiment. The significance of the differences in the relative
mRNA expression levels and the percentages of cell growth
inhibition between two groups was determined using the paired
t-test. All analyses were carried out using the SPSS software
package. P<0.05 was deemed to indicate a statistically
significant difference.
Results
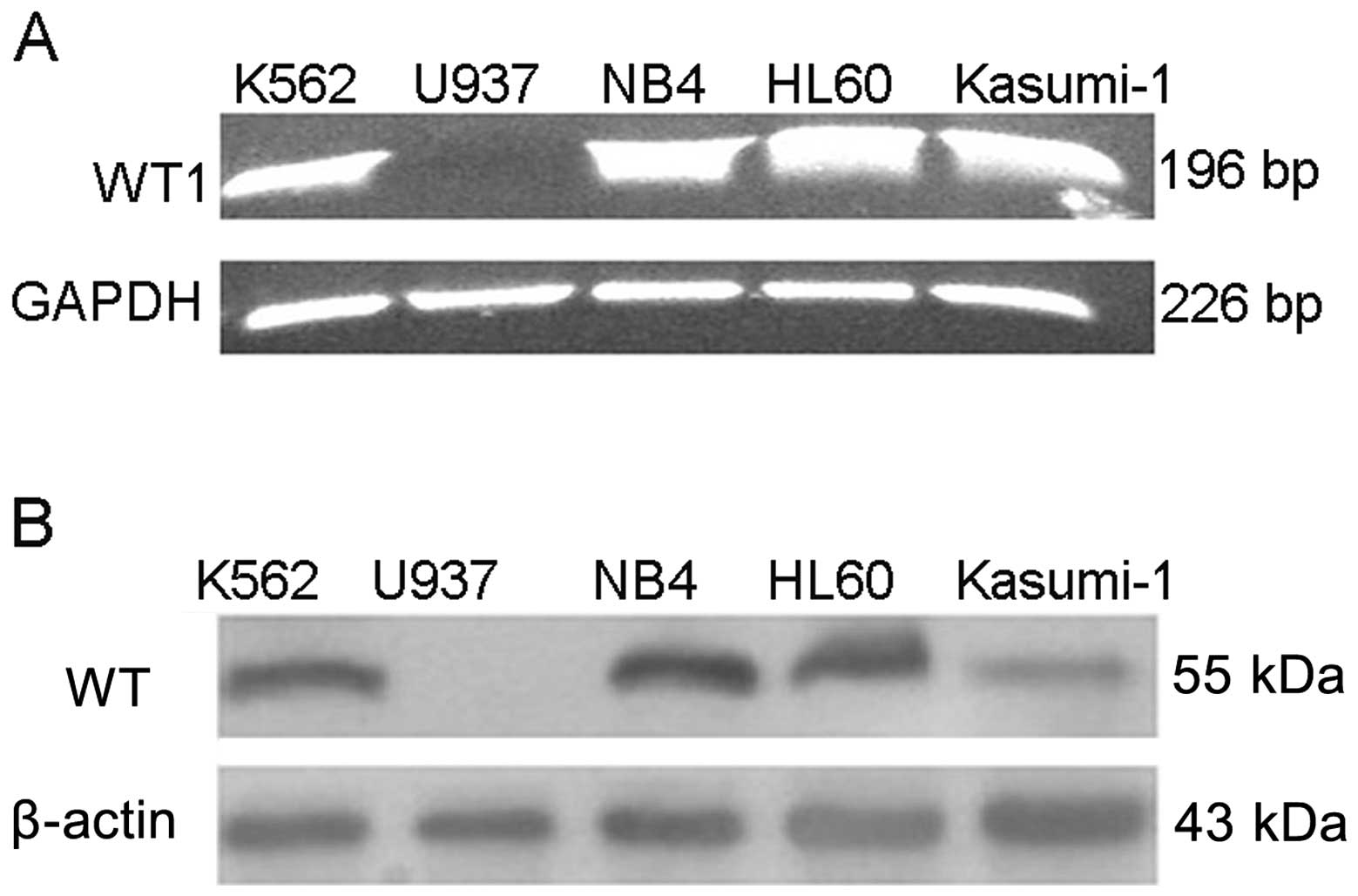
The WT1 gene is highly expressed in most
leukemic cell lines
In order to determine the expression of the WT1 gene
in the AML cell lines, we designed primers of the WT1 gene which
could cover the four main spliceosomes. As shown in Fig. 1, WT1 mRNA was expressed highly in
the K562, HL60, NB4 and Kasumi-1 cells, but was negative in the
U937 cells. Approximately 100 μg of the protein extract was
separated by 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE). As shown in Fig. 1, apart from U937, WT1 was highly
expressed in the other four cell lines.
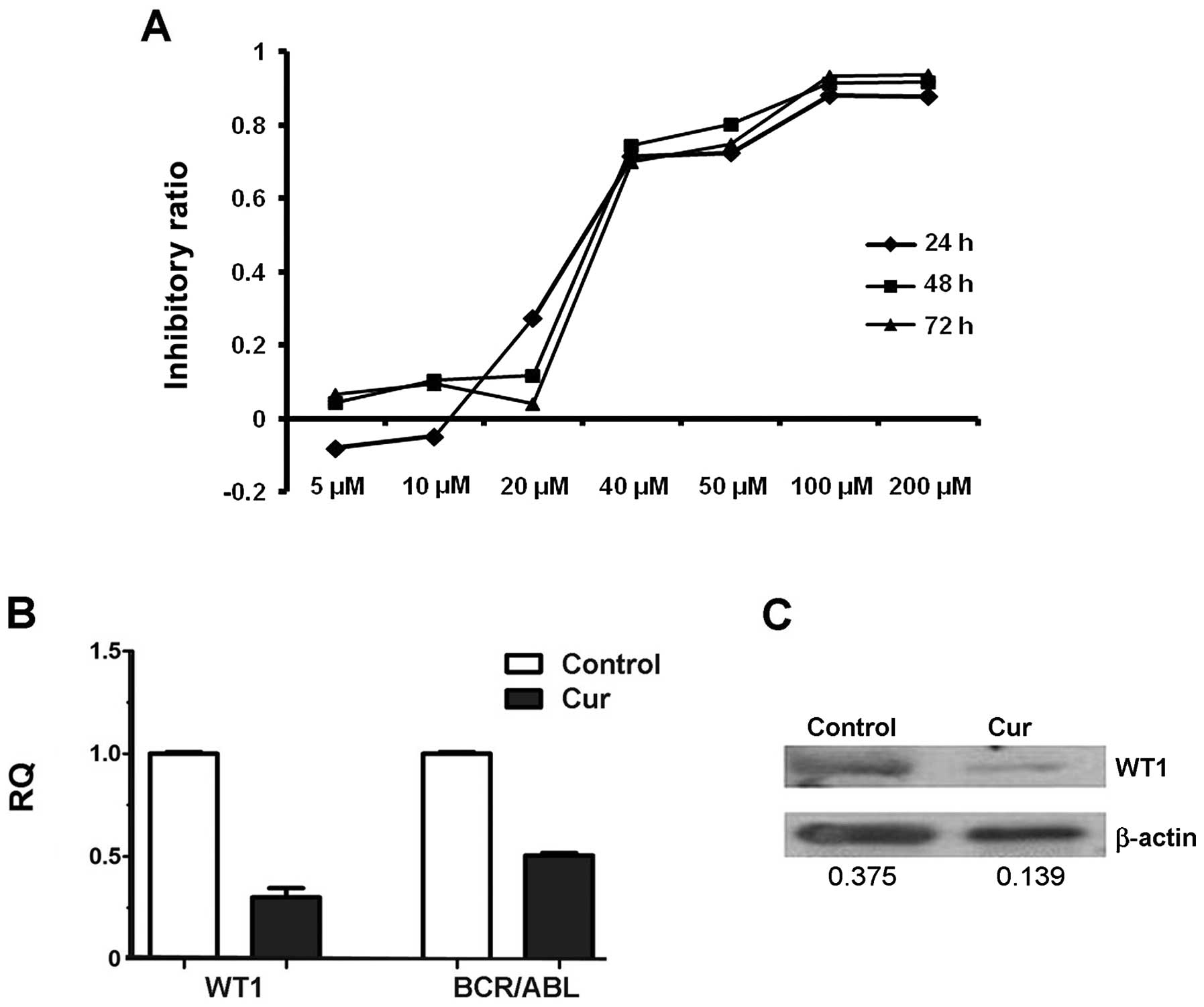
Curcumin, an inhibitor of the WT1 gene,
inhibits cell growth by decreasing WT1 expression
As shown in Fig. 2A,
the cells treated with curcumin showed a dose-dependent decrease in
cell viability. The IC50 values of curcumin in the K562
cells were 28.15, 33.79 and 38.12 μM at time points 24, 48 and 72
h, respectively. For the subsequent studies, we used curcumin at a
concentration of 20 μM.
As shown in Fig. 2B,
WT1 mRNA was decreased by approximately 80% and simultaneously WT1
protein (Fig. 2C) was obviously
reduced after curcumin treatment for 24 h. We also detected the
expression of BCR/ABL and found that BCR/ABL copies were markedly
decreased, which indicated that curcumin also affected the BCR/ABL
pathways in addition to downregulating the WT1 gene.
In addition, the colony formation assays showed a
marked reduction in colony formation (458±50.4 vs. 260±18.7,
P<0.01) 4 days after treatment with 20 μM curcumin, when
compared to the cells treated with DMSO alone. Cell cycle analysis
showed that curcumin also induced cell cycle arrest in the
G2/M phase (data not shown).
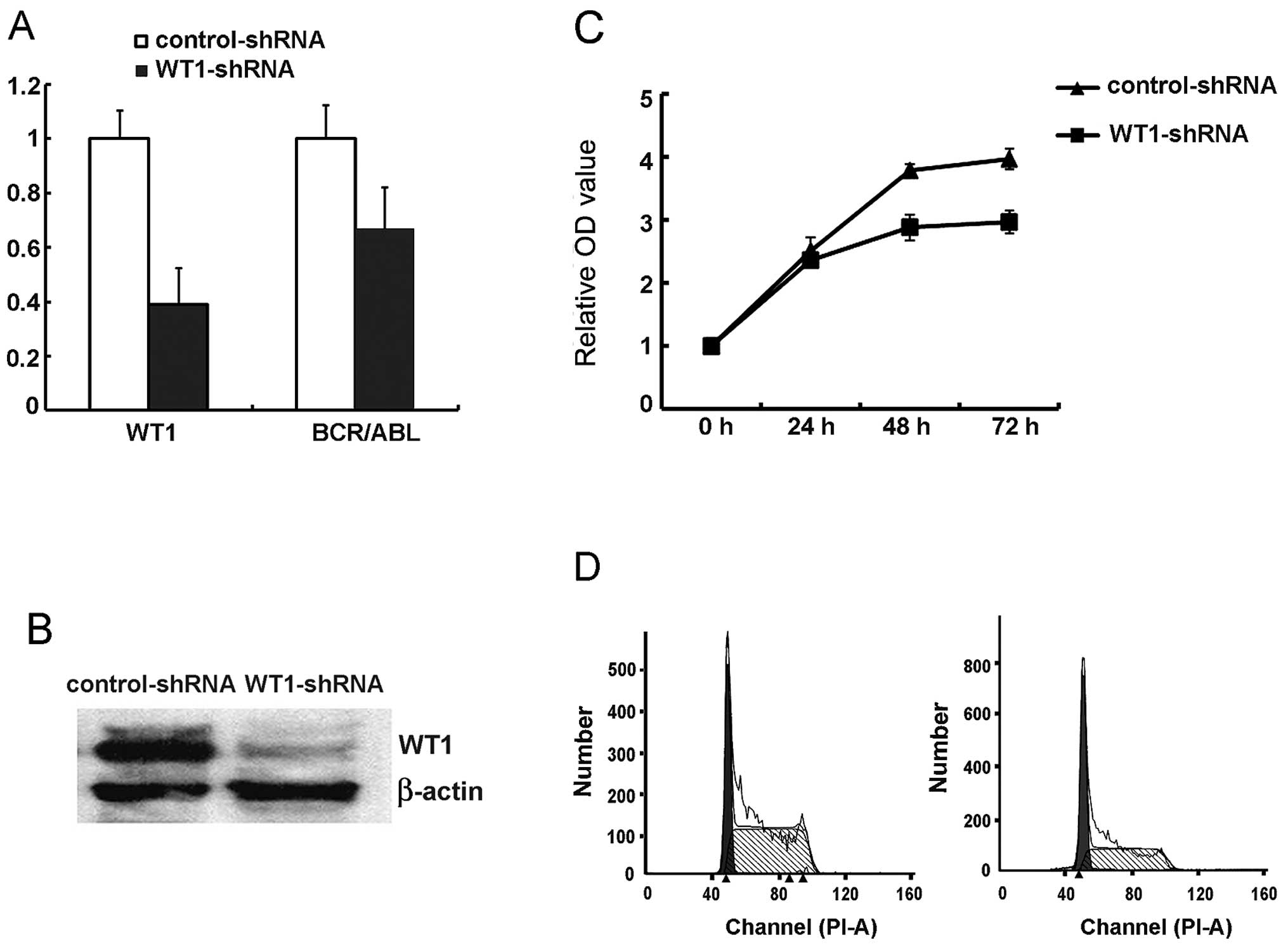
WT1 expression is related to cell
proliferation and cell cycle progression in the AML cell lines
To further analyze the exact role of the WT1 gene in
leukemia cell biology, we downregulated WT1 expression specifically
using RNA interference technique. pLKO.1 lentiviral shRNA
constructs generated by the RNAi consortium with the fragments
expressing puromycin were used to identify and sort the transduced
cells. The efficiency of WT1 downregulation by the different shRNA
vectors was investigated by RNA and protein expression of WT1 in
the leukemic cells. TRC0063 shRNA was confirmed to be the most
efficient oligonucleotide; it downregulated WT1 mRNA and protein by
over 70 and 90% respectively (Fig. 3A
and B) and was selected for further studies (referred to here
as WT1-shRNA).
Cell proliferation, colony formation and cell cycle
following WT1 downregulation were examined. We observed that
WT1-shRNA-transduced K562 cells showed decelerated cell
proliferation in vitro particularly at the 48- and 72-h time
points when compared with cells transduced with the non-specific
sequence (Fig. 3C). To investigate
whether the reduced cell proliferation was due to cell death, the
levels of apoptosis were measured in cells with reduced WT1 levels
at various time points after transduction by flow cytometry. The
percentage of PI−/Annexin V+ cells was not
different when compared to this percentage in the control cells. We
subsequently analyzed the cell cycle distribution of WT1-shRNA- and
control-shRNA-transduced K562 cells and observed an increase in the
proportion of cells in the G0/G1 phase (from
27.7 to 44.7%) while the proportion of S phase cells was decreased
(71.9 vs. 55.3% in control cells; Fig.
3D). These data suggest that WT1 is implicated in the cell
cycle progression, proliferation and self-renewal of AML cells.
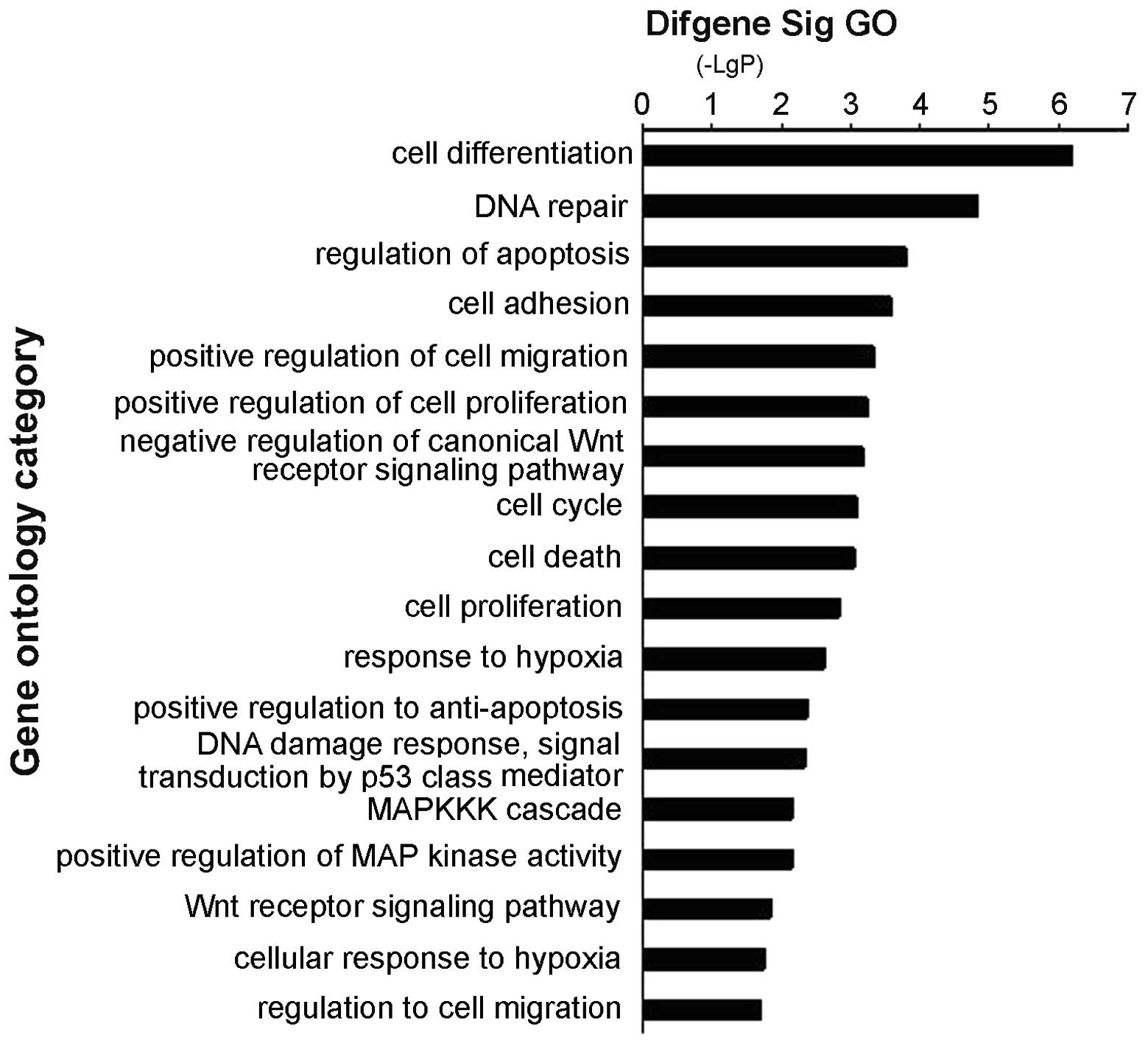
Exploration of the mechanism and function
of WT1 in leukemia
We analyzed the genome-wide transcriptional targets
of the WT1 gene using the chromatin immunoprecipitation-DNA
selection and ligation (ChIP-DSL) approach. DNA from K562 cells was
amplified and marked, and then the data were analyzed using MAS
software (http://bioinfo.capitalbio.com/mas) with a P-value
cutoff of 10−3 (13).
CHIP-CHIP analysis identified 2034 promoters by single array and
207 promoters by 2/2 arrays. As shown in Fig. 4, WT1-bound targets were enriched in
cell apoptosis, Wnt signaling pathway, MAPK signaling, cell
migration, cell proliferation and cell adhesion. The key genes of
the different pathways are shown in Table I.
 | Table IGene ontology of the genes bound by
WT1. |
Table I
Gene ontology of the genes bound by
WT1.
| GO names | Genes |
|---|
| Wnt signaling
pathway | AES, TGFB111, SFRP4,
GSK3A, LRRFIP2, WNT2B, WNT11, GRK5, NKX2-5, CBY1, AXIN1 |
| MAPK signaling
pathway | PDGFB, PDCD10, VEGFA,
EDN1, MAP3K5, SMAD1, CAMKK2, DUSP4, MAPKAPK2, SHC1 |
| Cell migration | CCL26, PIK3R1, VEGFA,
SPAG9, PDPN, WNT11, PLD1, EDN1, FER, FGF1, RRAS2, PDGFB |
As WT1 and the Wnt pathway are important factors in
the progression of CML (14–16),
we speculated that WT1 participates in the pathogenesis of leukemia
mainly by directly regulating genes of the wnt pathway. First, we
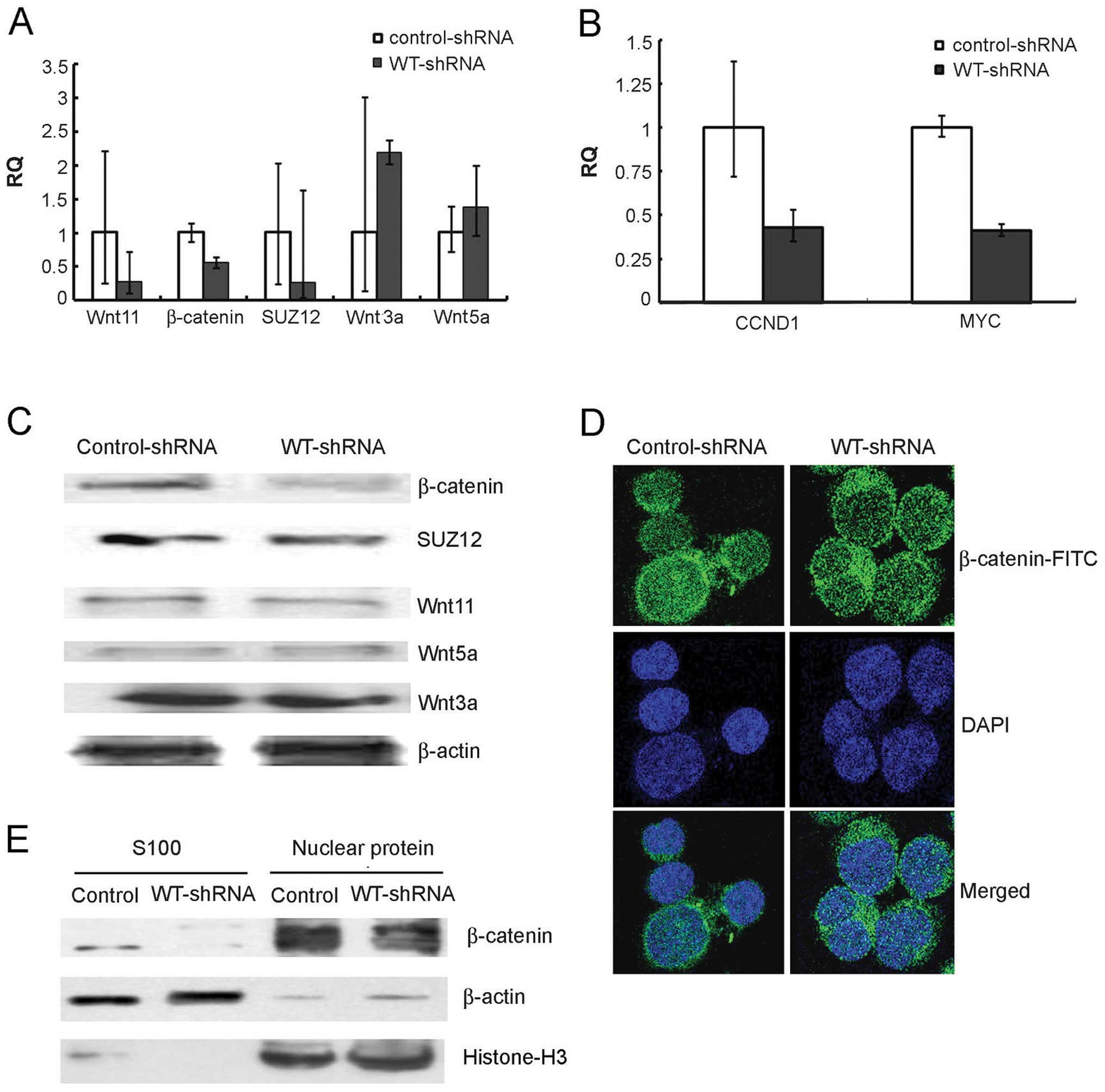
found that β-catenin, an essential gene of the wnt pathway, was
noticeably decreased at the RNA and protein levels in the K562
cells after WT1 was silenced (Fig. 5A
and C). Confocal immunofluorescence and western blot analyses
demonstrated that β-catenin translocated from the nucleus to the
cytoplasm following WT1 downregulation (Fig. 5D and E).
Next, we detected expression of other genes of the
canonical and noncanonical wnt pathway in the WT1-shRNA- and
control-shRNA-transduced K562 cells. As shown in Fig. 5A and B, the typical canonical genes,
such as wnt3a remained unchanged after WT1 interference. However,
expression levels of the noncanonical genes, wnt11 and SUZ12, were
significantly decreased at the RNA and protein levels.
As wnt11 is possibly a transcriptional target of
WT1, we speculated that expression of wnt11 would be directly
affected by the downregulation of WT1, and subsequently the
expression and sublocation of β-catenin changed. In addition, the
expression of β-catenin transcriptional targets, MYC and CCND1, was
also obviously reduced, in the cells transduced with WT1-shRNA
compared to the control cells (Fig.
5B).
Discussion
The WT1 gene is one of the important regulators of
cell growth and development. WT1 mRNA has two important splicing
sites, the fifth exon (which comprises 17 amino acids) and the
ninth exon (which encodes three amino acids, lysine tyrosine and
tyrosine, also called KTS); it can encode four major protein
isomers. The COOH-terminal of WT1 protein contains a Zinc finger
domain and plays an important role in transcription (1–3,5).
The WT1 gene is expressed at a low level in
CD34+ cells of normal bone marrow (9,10). An
animal experiment demonstrated that WT1 was necessary for cell
self-repopulation. Dysregulation of WT1 was found to promote stem
cell proliferation and obstruction of differentration, consequently
increasing the risk of leukemogenesis. As reported, WT1 was highly
expressed in AML patients, and levels of WT1 in chronic myeloid
leukemia patients are related to disease progression (14–17).
In our study, we found that WT1 was highly expressed in different
types of leukemia cell lines. Using the CML blastic cell line K562,
we illustrated that treatment with an inhibitor of WT1, curcumin, a
chemical extracted from plants could decrease the cell growth and
colony formation and arrest the cell cycle at the G2/M
phase. This indicates that curcumin has antitumor activity
partially by inhibiting the WT1 gene and causing cell cycle arrest.
Furthermore, we also noted that curcumin interferes with the
expression of BCR/ABL fusion genes. This phenomenon is extremely
important in the treatment of CML.
To identify the transcriptional targets of the WT1
gene, we performed CHIP-DSL analysis and found that the target
genes included wnt/β-catenin signaling pathway genes, MAPK pathway
genes, cell cycle regulation, cell adhesion and cell apoptosis
genes. These results are consistent with those reported by Kim
et al in Wilms’ tumor cell lines (18). Among these pathways, the wnt pathway
may be a major target of WT1.
shRNAs can specifically and stably downregulate gene
expression. We downregulated the WT1 gene in K562 cells using
lentiviral-mediated expression of shRNAs. Reduction of WT1 levels
in K562 cells decelerated their in vitro proliferative
ability by cell cycle arrest, without affecting cell viability.
Reduction in the levels of WT1 caused an increased proportion of
cells in the G0/G1 phase while a decreased
ratio of S phase cells. To elucidate the mechanisms of WT1
alteration involved in the biology of leukemia cells, we focused on
the wnt/β-catenin pathway. We found that WT1 regulated the wnt
pathway mainly by the wnt11-related non-canonical pathway genes but
not the wnt-related canonical pathway. We demonstrated that
downregulation of WT1 caused wnt11 to decrease at the mRNA and
protein levels, and consequently the changes in wnt11 affected the
expression of protein β-catenin causing it to translocate from the
nucleus to the cytoplasm. Meanwhile, the target genes of β-catenin,
MYC and CCND1, also decreased. As MYC and CCND1 are important genes
in cell growth and cell cycle progession, downregulation of MYC and
CCND1 caused the alteration of the cell cycle and cell growth
(19,20). Finally, we can deduce that by
targeting wnt11, WT1 participates in regulating the wnt/β-catenin
signaling pathway, leading to uncontrolled cell cycle progression,
cell growth and self-repopulation, resulting in leukemogenesis.
Moreover, CHIP-DSL results demonstrated that WT1 may also regulate
cell adhesion and migration, affect the bone marrow
microenvironment and chemosensitivity. All these may contribute to
leukemogenesis.
In conclusion, the WT1 gene is an oncogene and plays
an important role in leukemogenesis. Therapy targeting the WT1 gene
will be a significant strategy for leukemia.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (30870913) and Tianjin Major Science
and Technology Project (12ZCDZSY17500).
References
|
1
|
Yang L, Han Y, Suarez Saiz F and Minden
MD: A tumor suppressor and oncogene: the WT1 story. Leukemia.
21:868–876. 2007.PubMed/NCBI
|
|
2
|
Anuchapreeda S, Thanarattanakorn P,
Sittipreechacharn S, Chanarat P and Limtrakul P: Curcumin inhibits
WT1 gene expression in human leukemic K562 cells. Acta Pharmacol
Sin. 27:360–366. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Drummond IA, Madden SL, Rohwer-Nutter P,
Bell GI, Sukhatme VP and Rauscher FJ III: Repression of the
insulin-like growth factor II gene by the Wilms tumor suppressor
WT1. Science. 257:674–678. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang ZY, Qiu QQ, Huang J, Gurrieri M and
Deuel TF: Products of alternatively spliced transcripts of the
Wilms’ tumor suppressor gene, wt1, have altered DNA binding
specificity and regulate transcription in different ways. Oncogene.
10:415–422. 1995.
|
|
5
|
Laity JH, Chung J, Dyson HJ and Wright PE:
Alternative splicing of Wilms’ tumor suppressor protein modulates
DNA binding activity through isoform-specific DNA-induced
conformational changes. Biochemistry. 39:5341–5348. 2000.
|
|
6
|
Han Y, San-Marina S, Liu J and Minden MD:
Transcriptional activation of c-myc proto-oncogene by WT1 protein.
Oncogene. 23:6933–6941. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Smith SI, Down M, Boyd AW and Li CL:
Expression of the Wilms’ tumor suppressor gene, WT1, reduces the
tumorigenicity of the leukemic cell line M1 in C.B-17 scid/scid
mice. Cancer Res. 60:808–814. 2000.
|
|
8
|
Haber DA, Park S, Maheswaran S, et al:
WT1-mediated growth suppression of Wilms tumor cells expressing a
WT1 splicing variant. Science. 262:2057–2059. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hosen N, Sonoda Y, Oji Y, et al: Very low
frequencies of human normal CD34+ haematopoietic
progenitor cells express the Wilms’ tumour gene WT1 at levels
similar to those in leukaemia cells. Br J Haematol. 116:409–420.
2002.PubMed/NCBI
|
|
10
|
Baird PN and Simmons PJ: Expression of the
Wilms’ tumor gene (WT1) in normal hemopoiesis. Exp Hematol.
25:312–320. 1997.
|
|
11
|
Zhi L, Zhang J, Jia Y, et al: Effect of
G-rich oligonucleotides on the proliferation of leukemia cells and
its relationship with p53 expression. Oligonucleotides. 21:21–27.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
|
|
13
|
Wang Y, Zhang H, Chen Y, et al: LSD1 is a
subunit of the NuRD complex and targets the metastasis programs in
breast cancer. Cell. 138:660–672. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Svensson E, Vidovic K, Lassen C, et al:
Deregulation of the Wilms’ tumour gene 1 protein (WT1) by BCR/ABL1
mediates resistance to imatinib in human leukaemia cells. Leukemia.
21:2485–2494. 2007.
|
|
15
|
Radich JP, Dai H, Mao M, et al: Gene
expression changes associated with progression and response in
chronic myeloid leukemia. Proc Natl Acad Sci USA. 103:2794–2799.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Varma N, Anand MS, Varma S and Juneja SS:
Role of hTERT and WT1 gene expression in disease progression and
imatinib responsiveness of patients with BCR-ABL positive chronic
myeloid leukemia. Leuk Lymphoma. 52:687–693. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Østergaard M, Nyvold CG, Jovanovic JV, et
al: Development of standardized approaches to reporting of minimal
residual disease data using a reporting software package designed
within the European LeukemiaNet. Leukemia. 25:1168–1173.
2011.PubMed/NCBI
|
|
18
|
Kim MK, McGarry TJ, O Broin P, Flatow JM,
Golden AA and Licht JD: An integrated genome screen identifies the
Wnt signaling pathway as a major target of WT1. Proc Natl Acad Sci
USA. 106:11154–11159. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Siapati EK, Papadaki M, Kozaou Z, et al:
Proliferation and bone marrow engraftment of AML blasts is
dependent on β-catenin signalling. Br J Haematol. 152:164–174.
2011.PubMed/NCBI
|
|
20
|
Nakagawa M, Tsuzuki S, Honma K, Taguchi O
and Seto M: Synergistic effect of Bcl2, Myc and Ccnd1 transforms
mouse primary B cells into malignant cells. Haematologica.
96:1318–1326. 2011. View Article : Google Scholar : PubMed/NCBI
|