Introduction
Cervical cancer is the second most common cancer and
the fifth most deadly malignancy in females worldwide, affecting
500,000 individuals each year. It is the leading cause of cancer
mortality among women in developing countries (1). Invasion and metastasis are the major
causes of cancer-related death. Persistent infection with high-risk
types of human papillomavirus (HPV) is known to cause cervical
cancer (2). However, additional
genetic and epigenetic alterations are required for progression
from precancerous disease to invasive cancer. Genomic imbalances
can contribute to the deregulated expression of oncogenes and
tumor-suppressor genes in cancer cells, and the accumulation of
such altered genes has been correlated with tumor progression
(3–5). DNA methylation is an early and
frequent molecular alteration in cervical carcinogenesis. The
dysregulated activation of genes, such as CD44 and
SOX9 plays a role in cervical cancer (6–8). Genes
amplified and suggested to be involved in cervical cancer, such as
SKP2, TERT, TRIO, RNASEN and
PRKAA1, are overexpressed in tumor samples (9) and cell lines (10,11).
The inactivation of tumor-suppressor genes and the activation of
oncogenes play a significant role in carcinogenesis. However, the
etiology of cervical carcinoma remains poorly understood.
CD38 was originally defined as a T-cell
activation/proliferation molecule (12). However, CD38 is currently defined as
an ectoenzyme and a receptor (13,14),
while the expression of CD38 is not dependent on cell lineage or
activation (15). Human CD38 is a
multifunctional protein that triggers proliferation and
differentiation. The enzyme shares extensive sequence similarity
with Aplysia californica ADP-ribosyl cyclase (ADPRC). CD38
is known to be involved in activities typical of cell surface
receptors, such as signaling for activation and proliferation
events and heterotypic cell adhesion (16–18).
CD38 contribution to disease progression and relapse in acute
myeloid and chronic lymphocytic leukemia is well established and
the expression of the enzyme is considered an important prognostic
marker in leukemia (19–22).
The phosphatidylinositol 3-kinase (PI3K)/Akt pathway
is known to play key roles in cell proliferation, apoptosis, cell
survival in various cell types (23), and PIP3 is a lipid-signaling second
messenger that further activates its downstream effectors such as
Akt, inducing a conformational change in Akt that exposes the
critical Thr308/309 residue to phosphorylation, and subsequently
phosphorylated at Ser473/474 for full-length activation (24,25).
It has been previously demonstrated that PI3K/Akt signaling
pathways regulate metastasis in a variety of cancer cells (26,27).
In this study, we examined the expression levels of
CD38 and the key molecular of PI3K/Akt signaling pathway in
cervical cancer tissues. At the same time, the effects of CD38 were
studied in vivo. Our results showed that CD38 was highly
expressed in cervical carcinoma tissues and plays an important role
in dysregulation of the PI3K/Akt signaling pathway.
Materials and methods
Cells culture
The Caski human cervical cancer cell line was
cultured in RPMI-1640 supplemented with 10% fetal bovine serum
(FBS) (both from Gibco by Life Technologies, Grand Island, NY,
USA), 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in the
presence of 5% CO2.
Patient samples
Ten participants were recruited at the Third Xiangya
Hospital, Central South University (Changsha, Hunan, China).
Consent forms were obtained from individual patients, and
experimental protocols were approved by the Institutional Review
Board of the Third Xiangya Hospital. All 10 participants were
female with histologically-confirmed cervical cancer (Table I). All the subjects enrolled in the
study were Chinese. Cervical cancer and corresponding non-tumor
normal tissues were collected. Each biopsy sample was submitted to
routine histological diagnosis, or quantitative polymerase chain
reaction (qPCR), western blot analysis and immunohistochemistry
(IHC).
 | Table ICharacteristics of female cervical
cancer patients diagnosed with squamous cell cancer. |
Table I
Characteristics of female cervical
cancer patients diagnosed with squamous cell cancer.
| Samples | Age (years) | HPV type | Laborersa |
|---|
| A | 57 | 16,53,58 | Yes |
| B | 49 | 16,58 | No |
| C | 53 | 18,35 | No |
| D | 51 | 16 | No |
| E | 45 | 33,58 | No |
| F | 41 | 16 | Yes |
| G | 50 | 16,58 | Yes |
| H | 43 | 16 | No |
| I | 48 | 16 | No |
| J | 62 | 52 | Yes |
Total RNA extraction and quantitative
real-time polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from the cervical cancer and
corresponding non-tumor normal tissues with TRIzol reagent (Qiagen,
Carlsbad, CA, USA). cDNA synthesis was carried out using the
RevertAid First Strand cDNA Synthesis kit (CWBio, Beijing, China)
according to the manufacturer’s instructions. RT-qPCR was performed
with GoTaq qPCR Master Mix (CWBio). The primers used for the
RT-qPCR are shown in Table II.
RT-qPCR was carried out with the Bio-Rad CFK96TM Real-Time system
(Bio-Rad, Hercules, CA, USA). The data were analyzed by Bio-Rad CFK
manager 2.0 software. The expression of mRNA was assessed by
evaluating threshold cycle (Ct) values and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as
an internal control GAPDH was calculated using the
2−ΔΔCt equation previously adopted by Livak et al
(29): ΔΔCt = (CtTarget
− CtGAPDH) cervical cancer − (CtTarget −
CtGAPDH) control.
 | Table IIList of human-specific primer
sequences used in the study. |
Table II
List of human-specific primer
sequences used in the study.
| Target | Primer forward | Primer reverse |
|---|
| CD20 |
gggatctatgcacccatctg |
tggagtttttctccgttgct |
| CD21 |
caaggcacaattccttggtt |
tctaaggaactcccggtgtg |
| CD26 |
caaattgaagcagccagaca |
cacacttgaacacgccactt |
| CD34 |
caagccaccagagctattcc |
tccaccgttttccgtgtaat |
| CD38 |
tgctgatgacctcacatggt |
ccattgagcatcacatggac |
| CD73 |
gccgctttagagaatgcaac |
caggttttcgggaaagatca |
| CD90 |
cccagtgaagatgcaggttt |
gacagcctgagagggtcttg |
| CD123 |
ggacgtccagtacgacctgt |
actttgagaaccgctggaga |
| CD133 |
ttgtggcaaatcaccaggta |
tcagatctgtgaacgccttg |
IHC and evaluation of staining
IHC was performed using the peroxidase
anti-peroxidase technique following a microwave antigen retrieval
procedure. Antibody for CD38 was purchased from Proteintech
Biotechnology (Chicago, IL, USA). Antibody against CD38
(1:100) was overlaid on cervical cancer and corresponding non-tumor
normal tissue sections and incubated overnight at 4°C. Secondary
antibody incubation (Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA) was performed at room temperature for 30 min.
The sections evaluated by two investigators in a
blind manner in an effort to provide a consensus on staining
patterns by light microscopy (Olympus, Tokyo, Japan). CD38 staining
was assessed according to the methods described by Hara and Okayasu
(28) with minor modifications.
Each case was rated according to a score that added a scale of
intensity of staining to the area of staining. At least 10
high-power fields were chosen randomly, and >1,000 cells were
counted for each section. The intensity of staining was graded on
the following scale: 0, no staining; 1+, mild staining; 2+,
moderate staining; 3+, intense staining. The area of staining was
evaluated as follows: 0, no staining of cells in any microscopic
fields; 1+, <30% of tissue stained positive; 2+, between 30 and
60% stained positive; 3+, >60% stained positive. The minimum
score when summed (extension + intensity) was, therefore, 0, and
the maximum, 6. A combined staining score (extension + intensity)
of ≤2 was considered to be a negative staining (low staining),
while a score between 3 and 4 was considered to be a moderate
staining; whereas a score between 5 and 6 was considered to be a
strong staining.
Expression analysis of miR-634, miR-664
and miR-140-5p in cervical cancer
Total RNA was extracted from the cervical cancer and
corresponding non-tumor normal tissues with TRIzol reagent (Qiagen)
according to the manufacturer’s instructions. cDNA was synthesized
from 2 mg of total RNA with M-MLV Reverse Transcriptase (Promega,
Fitchburg, WI, USA) in a 25 ml volume {2 mg total RNA, 400 mM
reverse transcription primer [oligo(dT)18 for random primers for U6
rRNA and miR-634-, miR-664- and miR-140-5p-specific primers
(Bulge-Loop™ miRNA qPCR primers from RiboBio, China) for miRNA], 4
U/ml M-MLV, 1 U/ml inhibitor, 0.4 mM dNTP mix}. qPCR was carried
out with the reagents of a SYBR-Green I mix (Takara, Dalian, China)
in a 20 ml reaction volume (10 ml SYBR-Green I mix, 200 mM forward
and reverse primer, 2 ml cDNA template) on an MJ Opticon Monitor
Chromo4 instrument (Bio-Rad) using the following protocol: 95°C for
20 sec; 40 cycles of 95°C for 10 sec, 60°C for 20 sec and 70°C for
1 sec. Data analysis was performed using the 2−ΔΔCt
method (6,29).
Construction of pEGFP-N1-CD38 vector
The coding region of CD38 gene was generated by PCR
with the primer pair 5′-ATACTCGAGATGGCCAACTGCGAGTTCAG-3′ and
5′-GCGAAGCTTTCAGATCTCAGATGTGCAAG-3′. The PCR was performed under
the following conditions: one cycle for 5 min at 94°C; 30 cycles
for 45 sec at 94°C, 45 sec at 55°C, and 90 sec at 72°C, and ended
with 10 min at 72°C. The fragments were cloned into the TA vector
(Promega) and used to transform E. coli JM109 (Takara).
Following selection and propagation, the pure plasmid DNA was
prepared by standard methods. The DNA fragments were removed from
the TA vector by restriction enzyme digestion with XhoI and
HindIII (Promega) to subclone into the pEGFP-N1 vector. The
fusion sequences were verified by DNA sequencing using ABI
3730.
Cell transfection
Cell transfection was achieved by using
Lipofectamine, according to the manufacturer’s instructions (Life
Technologies, Grand Island, NY, USA). Cells (2×105) were
placed in each well of a 6-well plate 24 h prior to the
transfection. For each transfection, 2 μg of pEGFP-N1-CD38 plasmid
and pEGFP-N1 vector plasmid was transfected into Caski cells,
respectively. The plasmids were diluted with 100 μl of serum-free
media and 4 μl Lipofectamine was added into 100 μl serum-free
media. The two solutions were combined, mixed gently and incubated
at room temperature for 30 min. The 200 μl mixture and 200 μl of
serum-free media were added into each well. The cells were then
incubated at 37°C for 24 h, followed by replacing the transfection
media with fresh complete culture media. After an additional 48 h
culture, the cells were harvested for the western blot
analysis.
Western blot analysis
The cervical cancer and corresponding non-tumor
normal tissues, and Caski cells were lysed in RIPA buffer (CWBio)
and total protein concentration was determined using
Pierce® BCA Protein assay kit (Thermo Scientific, Inc.,
Rockford, IL, USA). Extracts containing 50 μg of proteins were
separated in 10% SDS-PAGE gels and electroblotted onto
nitrocellulose membranes (HyClone Laboratories, Logan, UT, USA).
The membranes were inhibited using Tris-buffered saline/Tween-20
(25 mM Tris-HCl, 150 mM NaCl, pH 7.5 and 0.05% Tween-20) containing
5% non-fat milk followed by overnight incubation at 4°C with
primary antibodies [rabbit anti-PI3K antibody, 1:500; and rabbit
anti-Akt antibody, 1:300 (Cell Signalling Technology, USA); rabbit
anti-MDM2 antibody, 1:200 and rabbit anti-p53 antibody, 1:200
(Wuhan Boster, Wuhan, China)]. Following three washes, the
membranes were incubated with horse-radish peroxidase-conjugated
secondary antibodies (Santa Cruz Biotechnology, Inc.) and the
specific signals were visualized using an ECL detection system.
Anti-GAPDH antibody (1:3,000; Santa Cruz Biotechnology, Inc.) was
used as a loading control.
Statistical analysis
Differences of non-parametric variables were
analyzed by the Mann-Whitney U test. Differences of the
quantitative variables between groups were analyzed by the
Student’s t-test using SPSS 11.0 program (SPSS, Chicago, IL, USA).
P<0.05 was considered statistically significant.
Results
Detection of mRNA expression levels of
the CD molecules in cervical cancer
To detect the mRNA expression levels of the CD
molecules in cervical cancer and the adjacent non-cancerous
tissues, 10 samples of each were selected to perform qPCR of the
CD20, CD21, CD26, CD34, CD38,
CD73, CD90, CD123 and CD133 genes. Data
were analyzed using the 2−ΔΔCt method. The fold-change
in the expression of these genes relative to the internal control
gene, GAPDH, was also analyzed. The expression of the
CD38, CD34 and CD90 genes was higher in the
cervical cancer samples compared with the adjacent non-cancerous
tissues and the normalized CD38, CD34 and CD90
gene expression in the cervical cancer samples was upregulated by
4.40-, 2.71- and 2.64-fold, respectively (Table III). The expression levels were
not significantly different for CD20, CD26, CD73 and CD133 between
the cervical cancer and adjacent non-cancerous tissues (Table III). Thus, we selected to
determine the funciton of CD38 and its underlying mechanism in
cervical cancer.
 | Table IIIIdentification of the mRNA expression
level of the CD molecules in cervical cancer and adjacent
non-cancerous tissues by qPCR. |
Table III
Identification of the mRNA expression
level of the CD molecules in cervical cancer and adjacent
non-cancerous tissues by qPCR.
| Gene | Sample | n | Gene Ct (mean ±
SD) | GAPDH Ct (mean ±
SD) | ΔCt (mean ±
SD) | ΔΔCt (mean ±
SD) | Folda |
|---|
| CD20 | Cervical
cancer | 10 | 31.56±1.46 | 19.43±0.76 | 12.13±1.17 | 0.07±0.04 | 0.95
(0.93–0.98) |
| Non-cancerous
tissues | 10 | 33.97±0.71 | 21.91±0.94 | 12.06±0.85 | |
| CD21 | Cervical
cancer | 10 | 29.07±1.69 | 19.43±0.76 | 9.64±1.12 | −0.31±0.13 | 1.23
(1.13–1.35) |
| Non-cancerous
tissues | 10 | 31.86±0.61 | 21.91±0.94 | 9.95±0.74 | |
| CD26 | Cervical
cancer | 10 | 31.69±1.73 | 19.43±0.76 | 12.26±1.04 | −0.10±0.06 | 1.07
(1.02–1.12) |
| Non-cancerous
tissues | 10 | 34.27±0.78 | 21.91±0.94 | 12.36±0.81 | |
| CD34 | Cervical
cancer | 10 | 33.22±1.92 | 19.43±0.76 | 13.79±1.28 | −1.44±0.47 | 2.71
(1.96–3.76) |
| Non-cancerous
tissues | 10 | 37.14±1.12 | 21.91±0.94 | 15.23±1.04 | |
| CD38 | Cervical
cancer | 10 | 30.42±1.77 | 19.43±0.76 | 10.99±1.37 | −2.14±0.85 | 4.40
(2.45–7.94) |
| Non-cancerous
tissues | 10 | 35.04±1.42 | 21.91±0.94 | 13.13±1.22 | |
| CD73 | Cervical
cancer | 10 | 31.69±1.73 | 19.43±0.76 | 12.26±1.39 | −0.45±0.19 | 1.37
(1.20–1.57) |
| Non-cancerous
tissues | 10 | 34.62±0.78 | 21.91±0.94 | 12.71±0.84 | |
| CD90 | Cervical
cancer | 10 | 30.93±0.98 | 19.43±0.76 | 11.50±0.86 | −1.40±0.42 | 2.64
(1.97–3.53) |
| Non-cancerous
tissues | 10 | 34.81±1.05 | 21.91±0.94 | 12.90±0.97 | |
| CD123 | Cervical
cancer | 10 | 31.70±1.79 | 19.43±0.76 | 12.27±1.34 | −0.84±0.38 | 1.79
(1.37–2.31) |
| Non-cancerous
tissues | 10 | 35.02±0.42 | 21.91±0.94 | 13.11±0.59 | |
| CD133 | Cervical
cancer | 10 | 32.61±2.91 | 19.43±0.76 | 13.18±1.90 | −0.04±0.02 | 1.02
(1.01–1.04) |
| Non-cancerous
tissues | 10 | 35.13±1.85 | 21.91±0.94 | 13.22±1.45 | |
Western blot analysis of protein
expression levels of CD38 in cervical cancer
To determine whether the CD38 gene was
expressed at a higher level in the cervical cancer compared with
the adjacent non-cancerous tissues, the protein expression levels
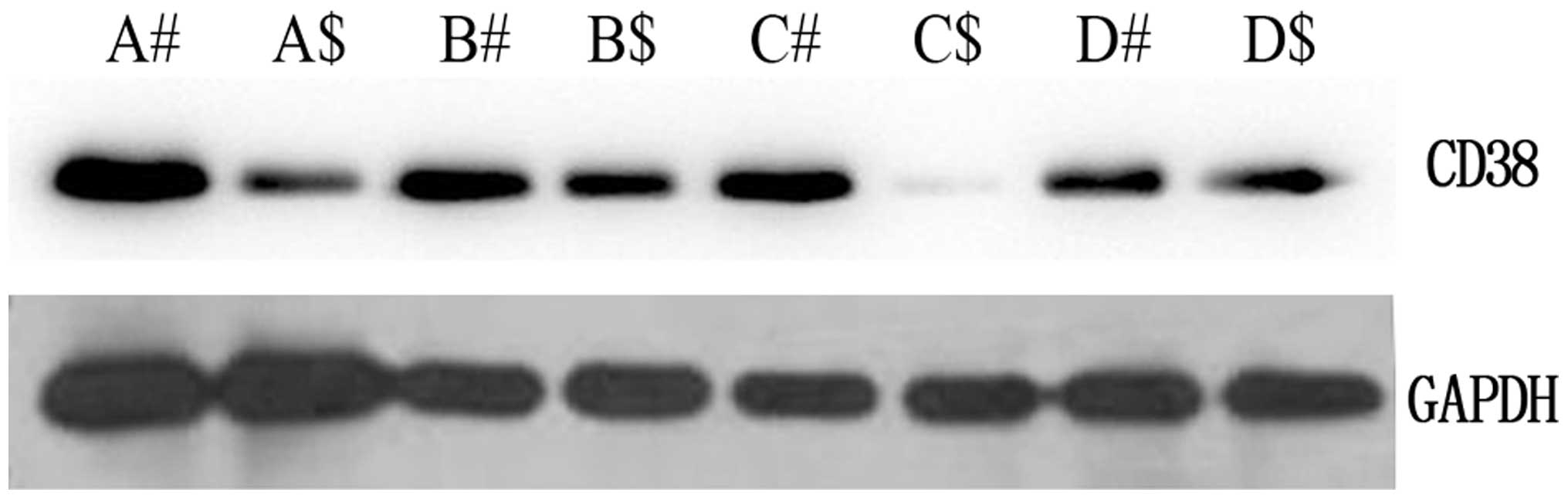
of CD38 were further examined by western blot analysis (Fig. 1). In comparison with the adjacent
non-cancerous tissues, the expression level was identified to be
greater in cervical cancer tissues, which corresponded with the
qPCR results. These results suggested that CD38 is highly expressed
in cervical cancer.
IHC analysis of protein expression levels
of CD38 in cervical cancer
IHC was carried out with antibodies against CD38
protein in the cervical cancer and adjacent non-cancerous tissues.
CD38 was identified as differentially expressed between the
cervical cancer and adjacent non-cancerous tissues. IHC showed a
similar pattern in protein expression, which was similar to that of
the western blot results. There was a 40% (12/30) high score of
CD38 in the cervical cancer and 16.6% (5/30) in the adjacent
non-cancerous tissues. The distribution of the low score was 23.3%
(7/30) and 56.7% (17/30) in the cervical cancer and adjacent
non-cancerous tissues, respectively (P=0.02<0.05) (Fig. 1) (Table
IV). The IHC results corresponded with those of the qPCR
results.
 | Table IVThe difference of CD38 expression
between cervical cancer and the adjacent non-cancerous tissues. |
Table IV
The difference of CD38 expression
between cervical cancer and the adjacent non-cancerous tissues.
| | Score |
|---|
| |
|
|---|
| Tissues | n | Low (0–2)
n (%) | Moderate
(3–4)
n (%) | High
(5–6)
n (%) | P-value |
|---|
| Cervical
cancer | 30 | 7 (23.3) | 11 (36.7) | 12 (40.0) | =0.02<0.05 |
| Non-cancerous | 30 | 17 (56.7) | 8 (26.7) | 5 (16.6) | |
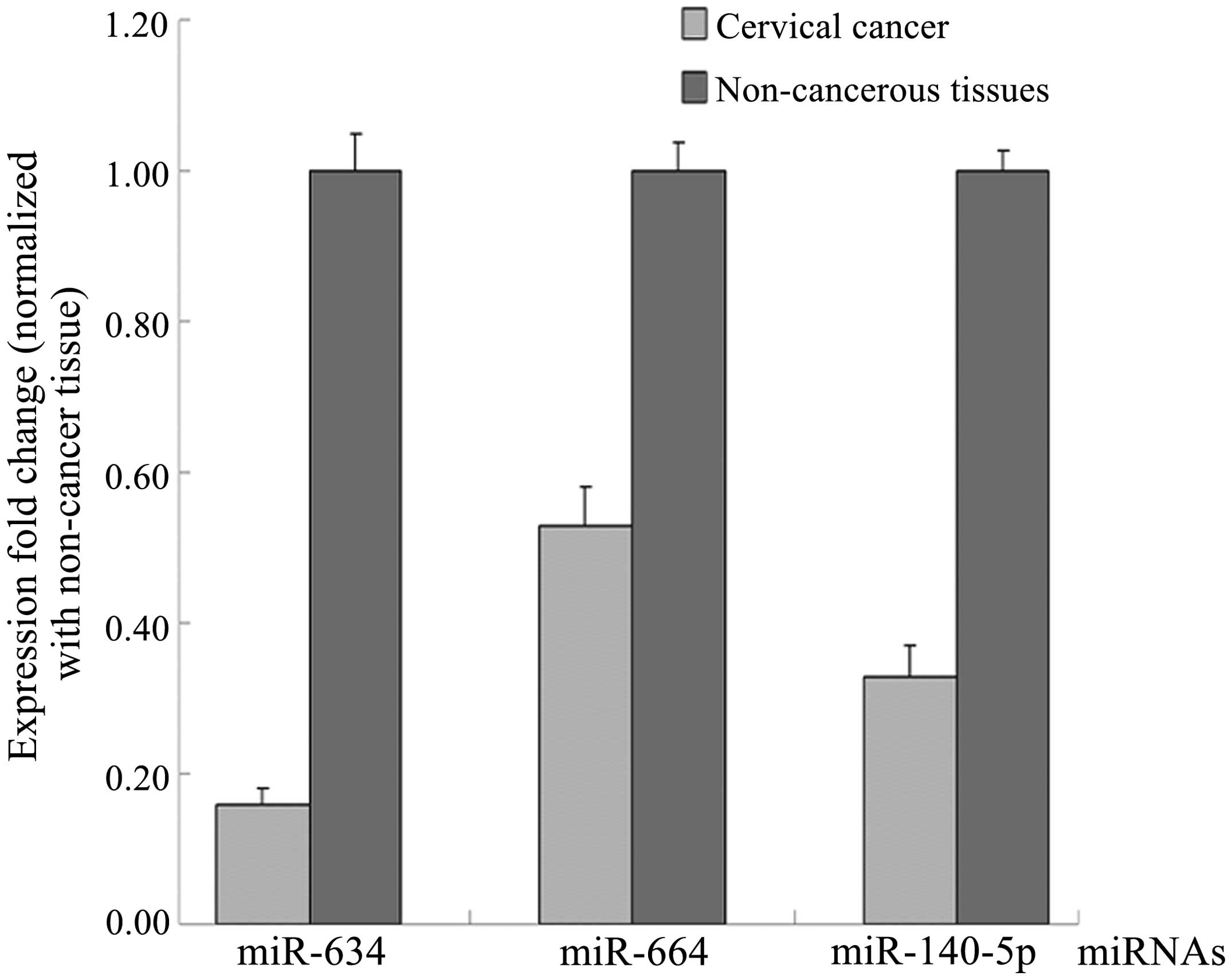
miR-634, miR-664 and miR-140-5p which
regulate CD38 exhibit a low expression in cervical cancer
tissues
As CD38 is a potential miR target, the open access
programs, TargetScan (http://www.targetscan.org/), PicTar (http://pictar.mdc-berlin.de/) and miRBase (http://mirbase.org/index.shtml), were used to
predict the targets of miR-634, miR-664 and miR-140-5p. The
endogenous expression of miR-634, miR-664 and miR-140-5p was
compared between the cervical cancer and adjacent non-cancerous
tissues by qPCR. The expression of miR-634, miR-664 and miR-140-5p
was downregulated in the cervical cancer tissues (Fig. 3). These results suggested that CD38
was upregulated in the cervical cancer as compared with the
non-cancerous tissues.
CD38 is correlated with dysregulation of
the PI3K/Akt signaling pathway in cervical cancer tissues in
vitro
To determine the possible mechanism of CD38 in
cervical cancer, we examined the expression levels of key molecules
in the PI3K/Akt signaling pathway by western blot analysis. PI3K
and Akt were upregulated in cervical cancer compared with the
adjacent non-cancerous tissues. MDM2 had the same tendency as PI3K
and Akt. However, p53 was downregulated in cervical cancer
(Fig. 4). Combined with the above
result showing CD38 was highly expressed in cervical cancer, the
results suggested that CD38 is correlated with dysregulation of the
PI3K/Akt signaling pathway in cervical cancer tissues in
vitro.
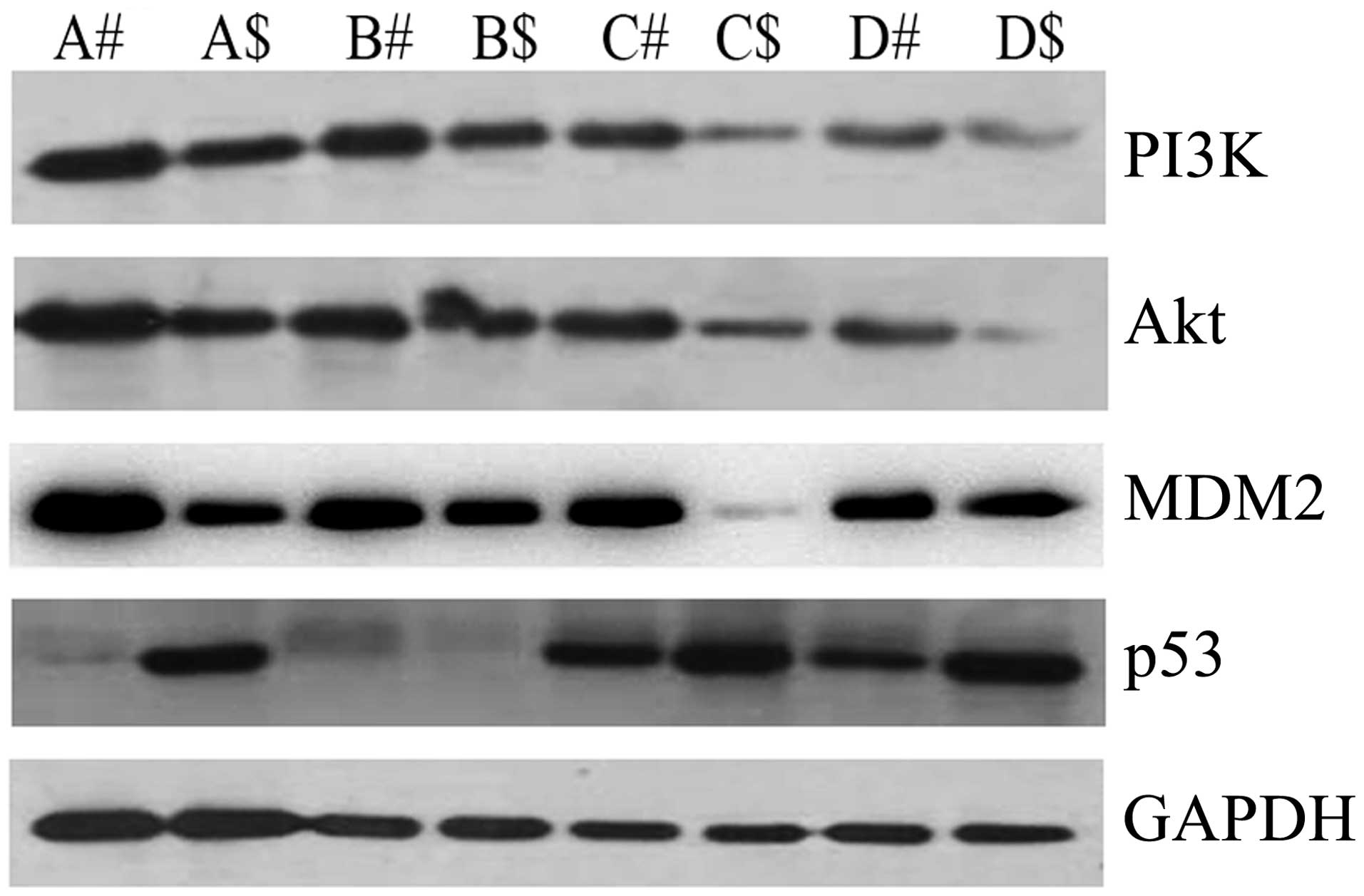 | Figure 4Expression levels of
phosphatidylinositol 3-kinase (PI3K), Akt, MDM2, and p53 protein in
cervical cancer and the adjacent non-cancerous tissues. In total,
A, B, C and D tissues which were used in the detection of mRNA
expression levels by qPCR were selected to detect the expression
levels of PI3K, Akt, MDM2 and p53 protein by western blot analysis.
#, denotes cervical cancer and $, denotes the adjacent
non-cancerous tissues. Data are representative of three independent
experiments. |
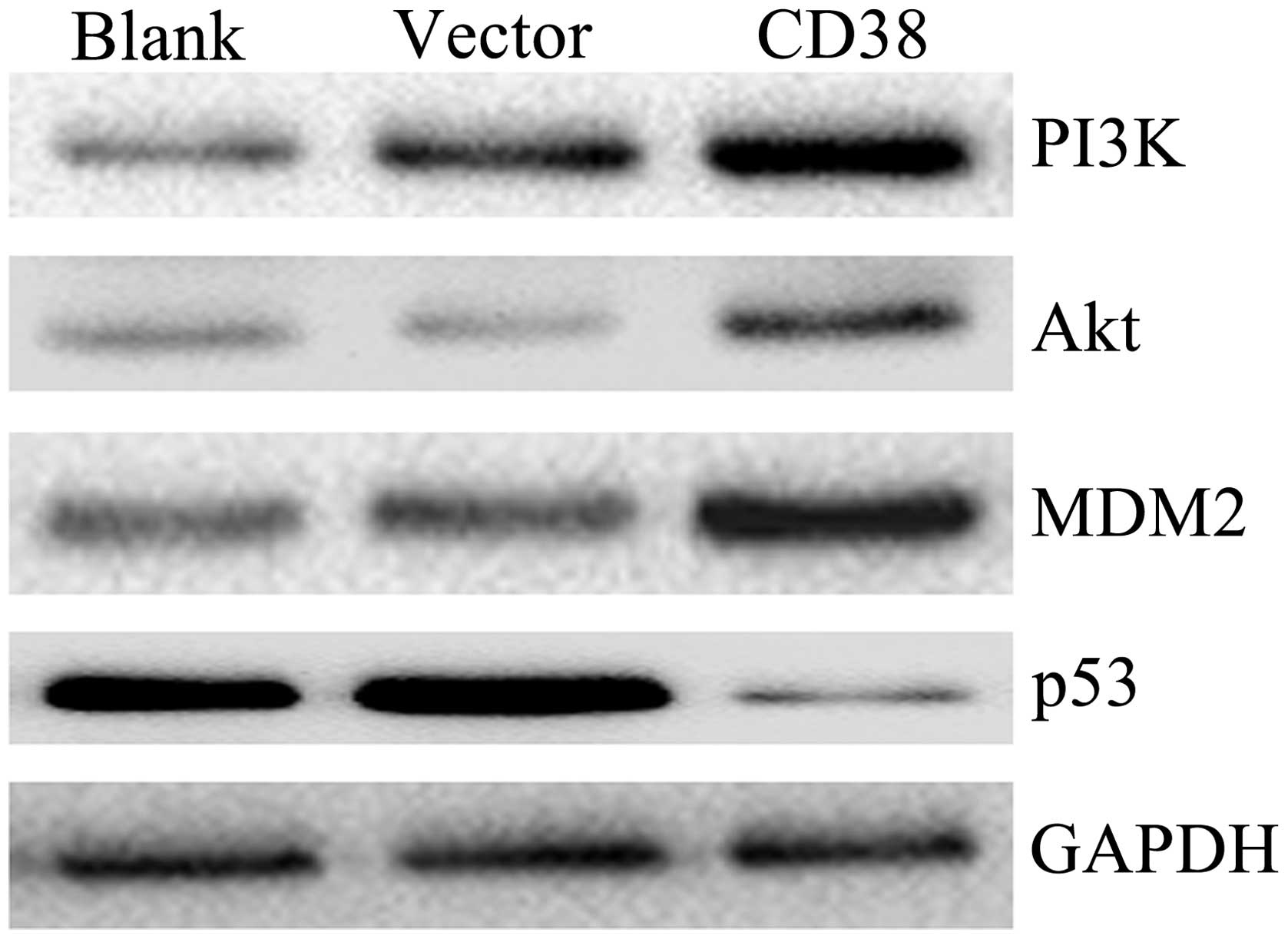
CD38 overexpression affects the
expression of PI3K, Akt, MDM2 and p53 in vivo
To confirm whether CD38 affects the expression of
PI3K, Akt, MDM2 and p53 in vivo, we constructed the plasmid
of pEGFP-N1-CD38. The plasmid of pEGFP-N1-CD38 and pEGFP-N1 was
transfected into the Caski cervical cancer cell line. The cells
were collected after 48 h with transfection and the expression
levels of PI3K, Akt, MDM2 and p53 proteins were examined in
vivo. The PI3K, Akt and MDM2 were upregulated in Caski cells in
which CD38 was overexpressed. At the same time, p53 was
downregulated in Caski cells in which CD38 was overexpressed
(Fig. 5). The results suggested
that CD38 overexpression affected the expression of PI3K, Akt, MDM2
and p53 in vivo.
Discussion
Approximately 530,000 new cervical cancer cases are
diagnosed annually worldwide, with 275,000 individuals succumbing
from the disease (30). The disease
is largely preventable. Persistent infection with high-risk types
of HPV is known to cause cervical cancer. However, the inactivation
of tumor-suppressor genes and activation of oncogenes play a
significant role in the progression from precancerous disease to
invasive cancer, caused by the genetic and epigenetic alterations
(31–34). The etiology of cervical carcinoma
remains poorly understood.
Human CD38 is a multifunctional protein that
triggers proliferation and differentiation, contributing to disease
progression and relapse in certain types of cancer (20–22).
To the best of our knowledge, no studies have been conducted on the
relationship between CD38 and cervical cancer. In this study, we
detected the mRNA expression levels of CD38 in cervical cancer and
found that CD38 was upregulated by 4.40-fold in cervical cancer
compared with the adjacent non-cancerous tissues. The protein
expression level of CD38 exhibited the same tendency as the mRNA
level in cervical cancer as indicated by the western blot analysis
results. At the same time, the results of IHC showed that there was
a 40% (12/30) high score of CD38 in cervical cancer tissues and
16.6% (5/30) in the adjacent non-cancerous tissues
(p=0.02<0.05). Perenkov et al (35) found that the expression of the
CD38 gene is heterogeneous in the tumor cells of patients
with colorectal cancer. CD38 expression is triggered at least in
part by a certain cytokine(s) secreted by cancer cells (36). Our data are consistent with those
observations and suggest that CD38 maybe play an important role in
cervical cancer.
To verify CD38 is highly expressed in cervical
cancer, we detected the expression of miR-634, miR-664 and
miR-140-5p, which were the predicted target genes of CD38. Our data
show that the expression of miR-634, miR-664 and miR-140-5p were
downregulated in the cervical cancer tissues. Yang et al
(37) found that the upregulation
of miR-664, miR-485-3p and miR-495 contributed to a lower MAT1A
expression in hepatocellular carcinoma (HCC), and enhanced
tumorigenesis may provide potential targets for HCC therapy.
MicroRNA-140-5p was significantly decreased in HCC tissues and its
expression levels were correlated with multiple nodules, vein
invasion, capsular formation, and differentiation of HCC (38). Our results demonstrate that the
expression level of CD38 was high in cervical cancer as compared
with that of the miRNA level.
To explore the possible mechanism of CD38 in
cervical cancer, we detected the expression levels of key molecules
in the PI3K/Akt signaling pathway by western blot analysis in
vitro and found that PI3K, Akt and MDM2 were upregulated in
cervical cancer, but p53 was downregulated in cervical cancer.
Furthermore, we studied whether CD38 affected the expression of
PI3K, Akt, MDM2 and p53 in vivo. CD38 overexpression is able
to increase the expression of PI3K, Akt and MDM2, and suppress the
expression of p53 in vivo. It has been demonstrated that
PI3K/Akt signaling pathways regulate metastasis in a variety of
cancer cells (23,26,27).
Through the regulation of the expression of downstream target
genes, p53 regulates cell cycle arrest, apoptosis, senescence,
cellular energy metabolism and anti-oxidant defense.
In conclusion, our results have shown that CD38 was
highly expressed in cervical carcinoma tissues and play an
important role in the dysregulation of the PI3K/Akt signaling
pathway.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (81272975, 81402270), the Key Project
of Hunan Provincial Natural Science Foundation (12JJ2044), the
Project of Hunan Provincial Natural Science Foundation (12JJ3121),
the Project of Hunan Provincial Development and Reform Commission,
the Planned Science and Technology Project of Hunan Province
(2010FJ3088 and 2012FJ2014), and the Key Project of Hunan
Provincial Natural Science Foundation (12JJ2044).
Abbreviations:
|
CD38
|
CD38 molecule
|
|
Akt
|
v-akt murine thymoma viral oncogene
homolog 1
|
|
PI3K
|
phosphatidylinositol 3-kinase
|
|
MDM2
|
MDM2 oncogene, E3 ubiquitin protein
ligase
|
|
TP53
|
tumor protein p53
|
|
GADPH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
|
IHC
|
immunohistochemistry
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Yugawa T and Kiyono T: Molecular
mechanisms of cervical carcinogenesis by high-risk human
papillomaviruses: novel functions of E6 and E7 oncoproteins. Rev
Med Virol. 19:97–113. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bozic I, Antal T, Ohtsuki H, Carter H, Kim
D, Chen S, Karchin R, Kinzler KW, Vogelstein B and Nowak MA:
Accumulation of driver and passenger mutations during tumor
progression. Proc Natl Acad Sci USA. 107:18545–18550. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kirchhoff M, Rose H, Petersen BL, Maahr J,
Gerdes T, Lundsteen C, Bryndorf T, Kryger-Baggesen N, Christensen
L, Engelholm SA and Philip J: Comparative genomic hybridization
reveals a recurrent pattern of chromosomal aberrations in severe
dysplasia/carcinoma in situ of the cervix and in advanced-stage
cervical carcinoma. Genes Chromosomes Cancer. 24:144–150. 1999.
View Article : Google Scholar
|
|
5
|
Heselmeyer K, Macville M, Schröck E,
Blegen H, Hellström AC, Shah K, Auer G and Ried T: Advanced-stage
cervical carcinomas are defined by a recurrent pattern of
chromosomal aberrations revealing high genetic instability and a
consistent gain of chromosome arm 3q. Genes Chromosomes Cancer.
19:233–240. 1997. View Article : Google Scholar
|
|
6
|
Xiao S, Zhou Y, Jiang J, Yuan L and Xue M:
CD44 affects the expression level of FOS-like antigen 1 in cervical
cancer tissues. Mol Med Rep. 9:1667–1674. 2014.
|
|
7
|
Ibrahim EM, Stewart RL, Corke K, Blackett
AD, Tidy JA and Wells M: Upregulation of CD44 expression by
interleukins 1, 4, and 13, transforming growth factor-beta1,
estrogen, and progestogen in human cervical adenocarcinoma cell
lines. Int J Gynecol Cancer. 16:1631–1642. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wobus M, Kuns R, Wolf C, Horn LC, Köhler
U, Sheyn I, Werness BA and Sherman LS: CD44 mediates constitutive
type I receptor signaling in cervical carcinoma cells. Gynecol
Oncol. 83:227–234. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Scotto L, Narayan G, Nandula SV,
Subramaniyam S, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M,
Schneider A, Arias-Pulido H and Murty VV: Integrative genomics
analysis of chromosome 5p gain in cervical cancer reveals target
over-expressed genes, including Drosha. Mol Cancer.
7:582008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dowen SE, Neutze DM, Pett MR, Cottage A,
Stern P, Coleman N and Stanley MA: Amplification of chromosome 5p
correlates with increased expression of Skp2 in HPV-immortalized
keratinocytes. Oncogene. 22:2531–2540. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kloth JN, Oosting J, van Wezel T, Szuhai
K, Knijnenburg J, Gorter A, Kenter GG, Fleuren GJ and Jordanova ES:
Combined array-comparative genomic hybridization and
single-nucleotide polymorphism-loss of heterozygosity analysis
reveals complex genetic alterations in cervical cancer. BMC
Genomics. 8:532007. View Article : Google Scholar
|
|
12
|
Funaro A, Spagnoli GC, Ausiello CM,
Alessio M, Roggero S, Delia D, Zaccolo M and Malavasi F:
Involvement of the multi-lineage CD38 molecule in a unique pathway
of cell activation and proliferation. J Immunol. 145:2390–2396.
1990.PubMed/NCBI
|
|
13
|
Howard M, Grimaldi JC, Bazan JF, Lund FE,
Santos-Argumedo L, Parkhouse RM, Walseth TF and Lee HC: Formation
and hydrolysis of cyclic ADP-ribose catalyzed by lymphocyte antigen
CD38. Science. 262:1056–1059. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Malavasi F, Funaro A, Roggero S,
Horenstein A, Calosso L and Mehta K: Human CD38: a glycoprotein in
search of a function. Immunol Today. 15:95–97. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Malavasi F, Deaglio S, Funaro A, Ferrero
E, Horenstein AL, Ortolan E, Vaisitti T and Aydin S: Evolution and
function of the ADP ribosyl cyclase/CD38 gene family in physiology
and pathology. Physiol Rev. 88:841–886. 2008. View Article : Google Scholar
|
|
16
|
Karimi-Busheri F, Zadorozhny V, Shawler DL
and Fakhrai H: The stability of breast cancer progenitor cells
during cryo-preservation: maintenance of proliferation,
self-renewal, and senescence characteristics. Cryobiology.
60:308–314. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karimi-Busheri F, Zadorozhny V, Li T, Lin
H, Shawler DL and Fakhrai H: Pivotal role of CD38 biomarker in
combination with CD24, EpCAM, and ALDH for identification of H460
derived lung cancer stem cells. J Stem Cells. 6:9–20.
2011.PubMed/NCBI
|
|
18
|
Karimi-Busheri F, Zadorozhny V, Carrier E
and Fakhrai H: Molecular integrity and global gene expression of
breast and lung cancer stem cells under long-term storage and
recovery. Cell Tissue Bank. 14:175–186. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Malavasi F, Deaglio S, Damle R, Cutrona G,
Ferrarini M and Chiorazzi N: CD38 and chronic lymphocytic leukemia:
a decade later. Blood. 118:3470–3478. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hamblin TJ: CD38: what is it there for?
Blood. 102:1939–1940. 2003. View Article : Google Scholar
|
|
21
|
Jawad M, Yu N, Seedhouse CH, Tandon K,
Russell NH and Pallis M: Targeting of
CD34+CD38− cells using Gemtuzumab ozogamicin
(Mylotarg) in combination with tipifarnib (Zarnestra) in acute
myeloid leukaemia. BMC Cancer. 12:4312012.
|
|
22
|
Dürig J, Naschar M, Schmücker U,
Renzing-Köhler K, Hölter T, Hüttmann A and Dührsen U: CD38
expression is an important prognostic marker in chronic lymphocytic
leukaemia. Leukemia. 16:30–35. 2002.
|
|
23
|
Qiao M, Sheng S and Pardee AB: Metastasis
and Akt activation. Cell Cycle. 7:2991–2996. 2008. View Article : Google Scholar
|
|
24
|
Kreisberg JI, Malik SN, Prihoda TJ,
Bedolla RG, Troyer DA, Kreisberg S and Ghosh PM: Phosphorylation of
Akt (Ser473) is an excellent predictor of poor clinical outcome in
prostate cancer. Cancer Res. 64:5232–5236. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Malik SN, Brattain M, Ghosh PM, Troyer DA,
Prihoda T, Bedolla R and Kreisberg JI: Immunohistochemical
demonstration of phospho-Akt in high Gleason grade prostate cancer.
Clin Cancer Res. 8:1168–1171. 2002.PubMed/NCBI
|
|
26
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-kinase Akt pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wagner EF and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hara A and Okayasu I: Cyclooxygenase-2 and
inducible nitric oxide synthase expression in human astrocytic
gliomas: correlation with angiogenesis and prognostic significance.
Acta Neuropathol. 108:43–48. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
|
|
30
|
IARC Globocan. Cervical Cancer Incidence
and Mortality Worldwide in 2008. http://globocan.iarc.fr/factsheets/cancers/cervix.asp.
|
|
31
|
Alameda F, Espinet B, Corzo C, Muñoz R,
Bellosillo B, Lloveras B, Pijuan L, Gimeno J, Salido M, Solé F,
Carreras R and Serrano S: 3q26 (hTERC) gain studied by fluorescence
in situ hybridization as a persistence-progression indicator in
low-grade squamous intraepithelial lesion cases. Hum Pathol.
40:1474–1488. 2009. View Article : Google Scholar
|
|
32
|
Ma YY, Wei SJ, Lin YC, Lung JC, Chang TC,
Whang-Peng J, Liu JM, Yang DM, Yang WK and Shen CY: PIK3CA as an
oncogene in cervical cancer. Oncogene. 19:2739–2744. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vazquez-Mena O, Medina-Martinez I,
Juárez-Torres E, Barrón V, Espinosa A, Villegas-Sepulveda N,
Gómez-Laguna L, Nieto-Martínez K, Orozco L, Roman-Basaure E, Muñoz
Cortez S, Borges Ibañez M, Venegas-Vega C, Guardado-Estrada M,
Rangel-López A, Kofman S and Berumen J: Amplified genes may be
overexpressed, unchanged, or downregulated in cervical cancer cell
lines. PLoS One. 7:e326672012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Andersson S, Wallin KL, Hellström AC,
Morrison LE, Hjerpe A, Auer G, Ried T, Larsson C and
Heselmeyer-Haddad K: Frequent gain of the human telomerase gene
TERC at 3q26 in cervical adenocarcinomas. Br J Cancer. 95:331–338.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Perenkov AD, Novikov DV, Sakharnov NA,
Aliasova AV, Utkin OV, Baryshnikov AIu and Novikov VV:
Heterogeneous expression of CD38 gene in tumor tissue in patients
with colorectal cancer. Mol Biol (Mosk). 46:786–791. 2012.(In
Russian).
|
|
36
|
Albeniz I, Demir-Coşkun O, Türker-Şener L,
Baş A, Asoğlu O and Nurten R: CD38 expression as response of
hematopoietic system to cancer. Oncol Lett. 2:659–664.
2011.PubMed/NCBI
|
|
37
|
Yang H, Cho ME, Li TW, Peng H, Ko KS, Mato
JM and Lu SC: MicroRNAs regulate methionine adenosyltransferase 1A
expression in hepatocellular carcinoma. J Clin Invest. 123:285–298.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang H, Fang F, Chang R and Yang L:
MicroRNA-140-5p suppresses tumor growth and metastasis by targeting
transforming growth factor β receptor 1 and fibroblast growth
factor 9 in hepatocellular carcinoma. Hepatology. 58:205–217.
2013.PubMed/NCBI
|