Introduction
Gastric carcinoma remains the fourth most common
tumor and the second most common cause of cancer death world-wide
(1). Despite gradual use of new
therapeutic approaches, the mortality rate caused by gastric
carcinoma remains high. Surgery is a mainstay for gastric cancer
treatment, however, it presents a poor outcome since a majority of
patients with gastric cancer are diagnosed at an advanced stage.
Therefore, treatment depends mainly on chemotherapy. However, there
are no normative chemotherapeutic schemes for gastric cancer. The
5-year survival rate of patients diagnosed at an advanced stage is
<30% and 70% for those diagnosed at an early stage (2). Therefore, it is important to identify
the molecular mechanism underlying the genesis and development of
gastric cancer and to identify reliable biomarkers and therapeutic
targets, consequently extending the lifespan of patients.
Recent studies have revealed that the roles of
microRNAs (miRNAs) in the genesis and development of cancer have
been increasingly understood along with ongoing investigations
(3–5). miRNAs are known to be involved in many
human tumors: in patients with intestinal or pancreatic cancer, for
instance, a high expression of miR-10b correlates significantly
with invasion and poor prognosis (6,7), and a
high miR-21 level is positively correlated with later clinical
stages of rectal carcinoma and poorly differentiated cells
(8). miRNA182 and miRNA205 were
found to be closely associated with the genesis and development of
breast cancer (9,10). Chao et al (11) found that abnormal miR-187 expression
is involved in the genesis of ovarian cancer and its role varies in
different stages. Evidence suggests that the abnormal expression of
miRNAs contributes to resistance of various types of cancer to
chemotherapy: miR-20a can affect the sensibility of SW620 and
SW680, two colorectal adenocarcinoma cell lines, to
chemotherapeutics and the effects have been shown to be associated
with its target gene BNIP2 (12); miR-32 regulates cancer cell
sensibility to chemotherapeutics via its target gene Bim
(13); miR-200c and mir-451 play
roles in drug resistance of MCF-7 breast cancer cells, the former
negatively regulating MDR1, thus increasing chemosensitivity of
MCF-7/ADR to epirubicin (14) and
the latter increasing resistance of tumor cells to
adriamycin-induced apoptosis through the negative regulation of its
target gene P-gp (15).
Findings of previous studies have shown that miRNA21, miR-130b,
miR-650 and miR-150 are involved in the genesis and development of
gastric carcinoma (16–20). Tumor-promoting miR-421 and miR-106a
are potential tumor markers in the diagnosis of gastric cancer
(21,22). Moreover, miRNA-15b, miRNA16 and
miRNA497 may affect the drug resistance of gastric carcinoma cells
by regulating BCL-2 (23,24). Results of the aforementioned studies
suggest that miRNAs are closely associated with the genesis and
development of gastric cancer and play important roles in the
chemoresistance of gastric cancer.
The aim of the present study was to determine
whether miRNA-135a-5p expression was increased in gastric cancer
compared with adjacent non-tumor tissues. The microarray analysis
revealed that miRNA135a-5p is one of the miRNAs that were
significantly differentially expressed in human gastric cancer
samples. We isolated 20 pairs of gastric cancer and para-carcinoma
tissue samples and detected mature miRNA135a-5p levels and its
potential target AP-2α, predicted by bioinformatics analysis. Using
quantitative PCR (qPCR) and western blotting, respectively,
possible correlations were investigated. Furthermore, we verified
the prediction using a luciferase reporter gene system. Chemically
synthetic RNA was used to alter miRNA135a-5p content in BGC-823
cells and its effects on AP-2α and proliferative activity and
resistance to adriamycin-induced apoptosis were investigated. As a
transcription factor, AP-2α plays a role in tumor inhibition
(25–27), although its direct involvement in
chemoresistance remains to be determined. miRNA-135a-5P may
regulate a key gene associated with proliferation or apoptosis via
AP-2α. A possible binding site of AP-2α in BCL-2 promoter through
bioinformatics analysis was identified and verified using a
luciferase reporter gene system. Additionally, we attempted to
elucidate the regulatory mechanism by exploring effects of
miRNA135a-5p on BCL-2, a crucial gene in cell proliferation
and apoptosis.
Materials and methods
Prediction of seed region of miRNA135a-5p
in AP-2α mRNA and the binding site of AP-2α in BCL-2 promoter
TargetScan was used to predict the possible target
(seed region) of miRNA135a-5p in mRNA sequence of AP-2α
(NM_003220.2) and TFSEARCH software was used to identify the
potential binding site of AP-2α in BCL-2 promoter.
Vector construction and RNA
synthesis
AP-2α human 3′-untranslated region (3′-UTR, 314 bp)
was amplified from cDNA obtained through the reverse transcription
of total RNA of 293 cells, using the primers:
5′-GCTCTAGATGTGGAGCCTAAGAGAACAGA-3′ and
5′-GCTCTAGAAATTCGTGTATTTGTGTTC-3′. The amplification parameters
used were: 32 cycles of denaturation at 95°C for 10 sec, annealing
at 58°C for 30 sec and extension at 72°C for 30 sec. The product
was digested with XbaI and inserted into the pGL3-promoter
vector (Promega, Madison, WI, USA). The seed region was mutated
from 5′-AGCCATA-3′ to 5′-ACAGACT-3′ by point mutation. The
resulting vectors were designated as pGL-WT-AP-2α and pGL-MT-AP-2α,
respectively. Sequences of the constructed vector were verified by
sequencing. Human genomic DNA was extracted from 293 cells and the
BCL-2 promoter sequence (694 bp) was amplified using the primers:
5′-GCTCTAGACAGGAGGAGGAGAAAGGGT-3′ and
5′-GCTCTAGAAAACAAATGCATAAGGCAACGATC-3′. The cycling parameters were
32 cycles of denaturation at 95°C for 10 sec, annealing at 60°C for
30 sec and extension at 72°C for 45 sec. The PCR product was
digested and inserted into PGL3-enhancer (Promega), a luciferase
reporter vector. The binding site was mutated from
5′-ACCGGCGGGCC-3′ to 5′-GCAGCGCGCCG-3′. The resulting luciferase
vectors were designated as pGL3-WT-BCL-2 and pGL3-MT-BCL-2,
respectively. Sequences of the constructed vector were verified by
sequencing. Chemically synthesized miRNA135a-5p-mimics, inhibitor
and NC were obtained from Shanghai Sangon (Shanghai, China).
Cell culture and treatment and luciferase
assay
A total of 293 cells at log phase were suspensed and
seeded in 96-well plates. For groups of AP-2α overexpression and
silencing, Lv-AP-2α (Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA) and Lv-shRNA-AP-2α viruses were used for infection at a
MOI of 5. Transfection was carried out 24 h later, using
Lipofectamine 2000 according to the manufacturer’s instructions.
The cells were also transfected with 50 ng pRL-TK (Promega) for
luciferase reference. After 48 h, the cells were collected and the
relative luciferase activities were measured by the dual luciferase
reporter assay system (Promega). BGC-823 cells (Cell bank of
Chinese Academy of China, Shanghai, China) were suspended and
seeded in 6-well plates at 1×105 cells/well and cultured
for 24 h. For AP-2α overexpression and silencing, Lv-AP-2α (Santa
Cruz Biotechnology, Inc.) and Lv-shRNA-AP-2α viruses were used for
infection at a MOI of 10. The infection efficiency was estimated by
fluorescence microscopy. The cells were reseeded and transfected
with miRNA135a-5p-mimics, inhibitor or NC sequence and collected
for BCL mRNA and protein measurement.
Detection of miRNA135a-5p and BCL-2 mRNA
using real-time PCR
Twenty pairs of gastric cancer and adjacent tissue
samples were obtained from the Department of Gastroenterology of
the Changhai Hospital (Shanghai, China). Each sample (~50 mg) was
rinsed with 1 ml cooled DPBS and 1 ml pre-cooled TRIzol was added
before subjecting to RNA extraction. Total RNA (2 μg) was used for
cDNA preparation using the M-MLV reverse transcription kit (Takara
Bio, Dalian, China) and the specific primers used were: U6 snRNA
(NM_001101.3), 5′-TACCTTGCGAAGTGCTTAAAC-3′ and miRNA135a-5p,
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTCACA-3′. RNA
contents were detected using PCR of fluorescent dye (Takara Bio)
according to the manufacturer’s instructions. The primers used for
quantification of human U6 snRNA and miRNA-137 were: U6 snRNA,
5′-GTGCTCGCTTCGGCAGCACAT-3′ and 5′-TACCTTGCGAAGTGCTTAAAC-3′,
producing a segment of 211 bp; and miRNA-137:
5′-GCCGGCGCCCGAGCTCTGGCTC-3′ and 5′-TATGGCTTTTTATTCCTATGTGA-3′,
producing a segment of 214 bp. PCR systems were: Takara SYBR Premix
Ex Tap 10 μl, forward and reverse primers (20 μM) 0.2 μl each and
cDNA 2 μl, followed by the addition of dH2O to 20 μl.
Cycling parameters used were: 40 cycles of denaturation at 95°C for
10 sec, annealing at 60°C for 20 sec and extension at 72°C for 20
sec. U6 snRNA was used as a reference to normalize miRNA-137 levels
using the 2−ΔΔCT method. Each RNA sample was run in
triplicate. Total RNA was isolated from BGC-823 cells infected with
Lv-AP-2α or Lv-shRNA-AP-2α virus and then transfected with
miRNA135a-5p-mimics, inhibitor or NC sequence 48 h after
transfection and reverse transcribed into cDNA. BCL-2 mRNA contents
were subsequently detected using the primers:
5′-TGCACCTGACGCCCTTCACCG-3′ and 5′-TTATCCTGGATCCAGGTGTGC-3′.
Western blotting
The tissue samples mentioned above were also used
for AP-2α protein measurement. Each sample (~100 mg) was rinsed
with 1 ml cooled dPBS and added with 1 ml tissue lysis buffer (50
mM pH 8.0 Tris, 1 mg/ml leupeptin, 150 mM NaCl, 0.5% Nonidet P-40,
5 mM EDTA, 100 mM phenylmethylsulfonyl fluoride, 1 M dithiolthretol
and 1 mg/ml aprotinin) for protein extraction. Protein
concentrations were detected by BCA assay. Protein samples (11 μl)
were separated by SDS-PAGE and transferred to PVDF membranes. Blots
were blocked in TBST containing 5% non-fat milk at room temperature
for 2 h and incubated with the primary antibodies against AP-2α and
β-actin (AP-2α 1:300 and β-actin 1:800; both from Santa Cruz) at
4°C overnight. Bands were detected with ECL chemiluminescence
substrates (Pierce Biotechnology, Inc. Rockford, IL, USA) and the
optical densities were analyzed with the image processing software.
The relative content of AP-2α was calculated as optical density of
AP-2α band/optical density of β-actin band. Western blotting was
also used to detect AP-2α and BCL-2 changes in cells infected with
or without Lv-AP-2α or Lv-shRNA-AP-2α viruses and then transfected
with miRNA135a-5p-mimics, inhibitor or NC sequence. The primary
antibody against BCL-2 was diluted at 1:500.
Cell proliferation and apoptosis
assays
BGC-323 cells transfected with miRNA135a-5p-mimics,
inhibitor or NC sequence were reseeded into 96-well plates at
5×104 cells/well after 24-h transfection. Adriamycin
(Sigma-Aldrich, St. Louis, MO, USA) was then added to a final
concentration of 1, 5 or 25 μg/ml. CCK-8 solution (10 μl; Dojindo,
Osaka, Japan) was added into each well at different time points.
The absorbance at 450 m was determined after an additional 4-h
incubation. The Annexin V-FITC Apoptosis Detection kit II (BD
Biosciences, Pharmingen, CA, USA) was used for apoptosis analysis.
Briefly, the cells were collected, washed with dPBS and suspended
in 500 μl binding buffer and stained with 5 μl Annexin V-FITC in
the dark for 10 min and then stained with 5 μl propidium iodide for
5 min. Flow cytometry (FACSCalibur; BD Biosciences) was conducted
using FL1 channel for Annexin V-FITC and FL2 channel for PI
(FACSCalibur) at an excitation wavelength of 488 nm.
Statistical analysis
SPSS13.0 was used for statistical analysis. Values
are presented as the means ± standard deviation (SD). Factorial
analysis was employed for the inter- and intra-group comparison.
P<0.05 was considered significant.
Results
Analysis on the relative levels of
miRNA135a-5p and AP-2α in gastric cancer and para-carcinoma
tissues
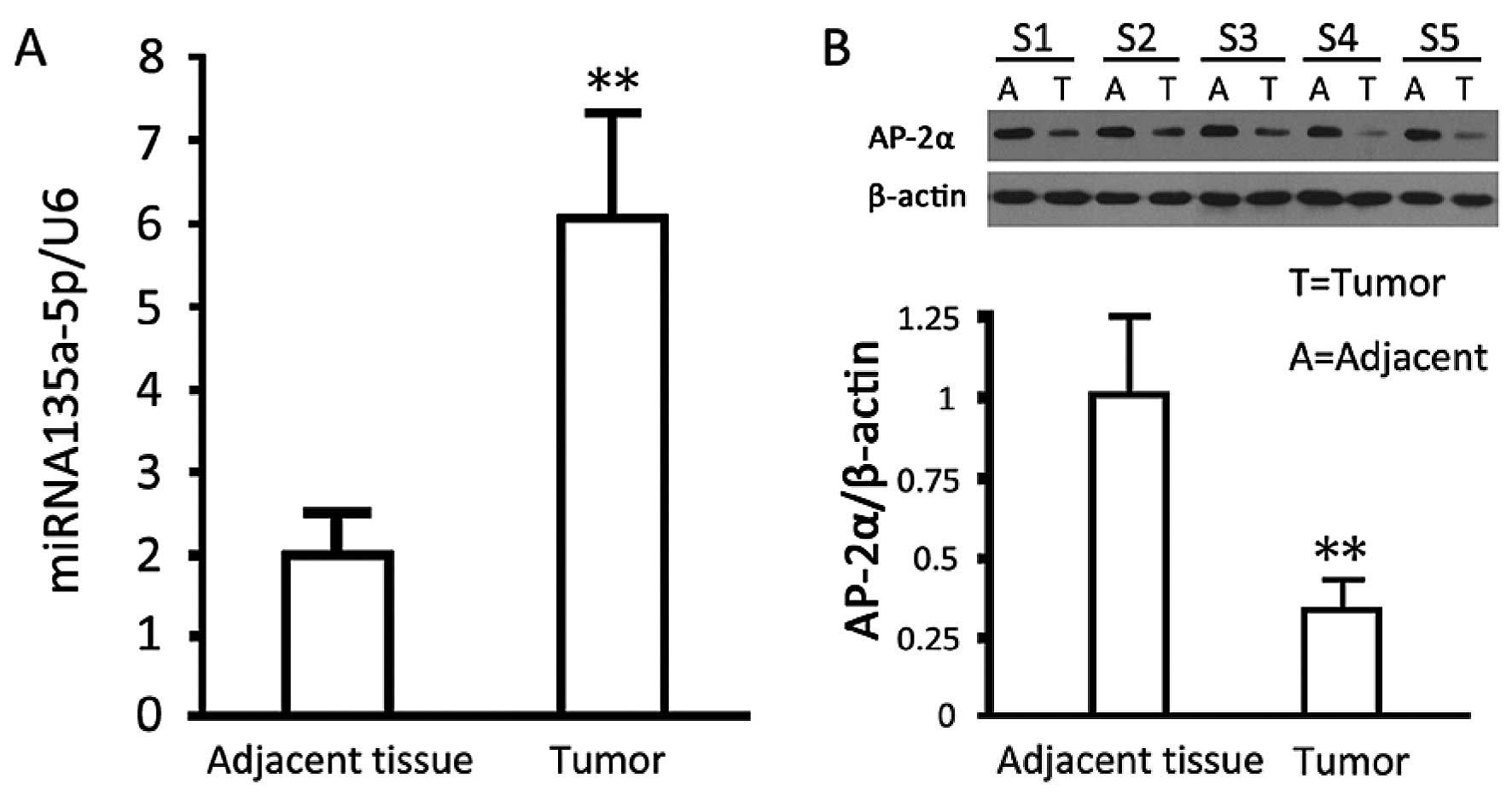
Quantitative results showed that miRNA135a-5p levels
in gastric cancer tissues were significantly higher than those in
para-carcinoma tissues (P<0.05). In comparison with those for
para-carcinoma tissues, western blotting results showed that AP-2α
expressed in cancer tissues was significantly decreased (p<0.05)
(Fig. 1).
Verification of interaction between
miRNA135a-5p and AP-2α
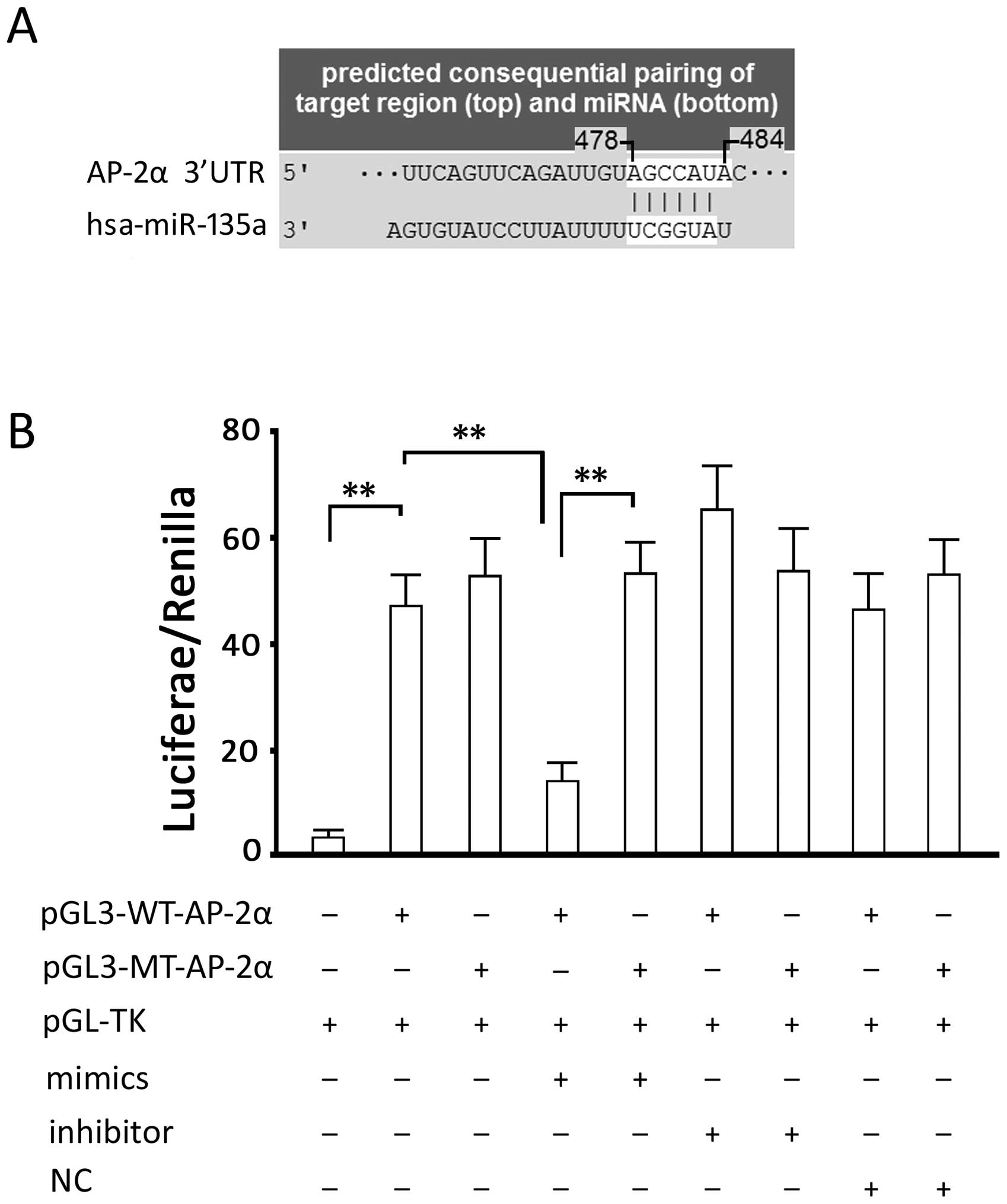
The analysis of TargetScan showed that there is a
possible binding site (seed region), 5′-AGCCAUA-3′, in 3′-UTR of
the AP-2α gene, between bases 478–484 (Fig. 2A). 3′-UTR of AP-2α was cloned into
the pGL-3 luciferase reporter vector for verification. Luciferase
activity detection showed that the miRNA135a-5p-mimic significantly
inhibited intercellular luciferase activity (P<0.05, compared
with the group transfected with the luciferase expression vector
alone) and miRNA135a-5p-inhibitor slightly increased the luciferase
activity without reaching statistical significance. However, the
two vectors did not directly affect the luciferase activity in
cells transfected with the luciferase expression vector carrying a
mutated binding site; in comparison with the group transfected the
luciferase expression vector alone, the cells transfected with
miRNA135a-5p-NC showed a similar luciferase activity, indicating
that RNA transfection had no effect on luciferase activity
(Fig. 2B). These results suggested
that the binding site of has-miRNA135a-5p in AP-2α is in line with
the predicted sequence.
Effects of miRNA135a-5p intervention on
proliferation and drug resistance in BGC-823 cells
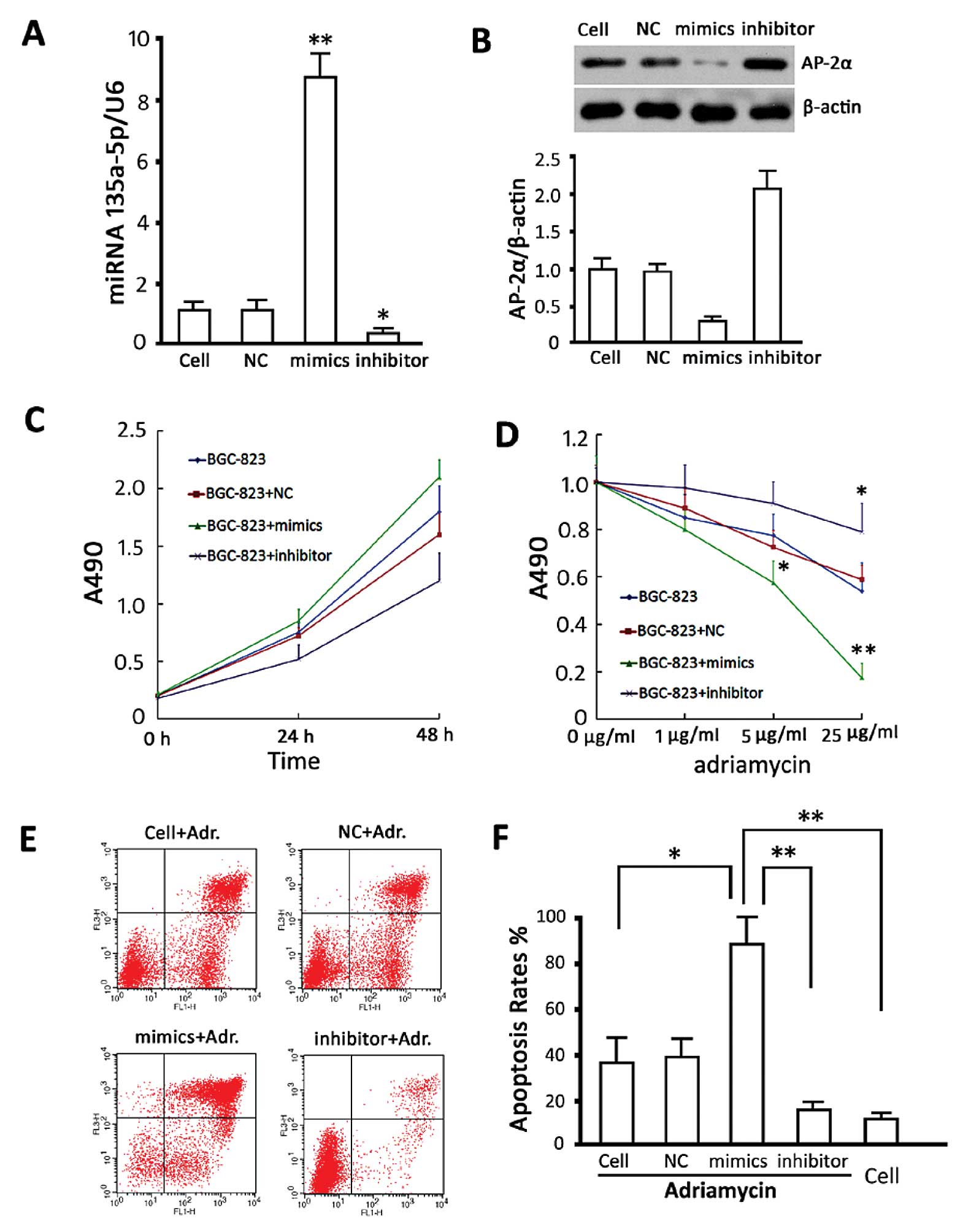
Analysis on relative levels of miRNA135a-5p and
AP-2α in transfected BGC-823 cells was detected. The results of
miRNA135a-5p showed that miRNA135a-5p-mimics significantly
increased miRNA135a-5p content (P<0.05, compared with the
non-transfected group) although the inhibitor significantly
decreased miRNA135a-5p content (P<0.05, compared with the
non-transfected group), and the transfection control group showed
no difference as compared to the non-transfected group (Fig. 3A). Protein content detection showed
that miRNA135a-5p-mimic significantly decreased AP-2α levels
(P<0.05, compared with the untransfected group), whereas the
miRNA135a-5p-inhibitor significantly increased AP-2α expression
(P<0.05, compared with the non-transfected group). No difference
was observed between the transfection control and non-transfected
groups (Fig. 3B).
To investigate the effects of changes in
miRNA135a-5p on the proliferation of BGC-823 cells, CCK-8 was used
to analyze the proliferation of BGC-823 cells transfected with
miRNA135a-5p-mimics, inhibitor and NC 48 h after transfection. The
data suggested that high miRNA135a-5p content promoted cell
proliferation at the logarithmic phase, although no statistically
significant difference was observed (P>0.05, vs. the control
group). The cell viabilities in the group treated with the
transfection agent and the group transfected with NC sequence were
not altered (P>0.05, vs. the control group) (Fig. 3C). The cells were treated with
adriamycin at different concentrations. A cell viability assay was
carried out 24 h later and the results showed that
miRNA135a-5p-mimics increased the sensitivity of BGC-823 cells to
adriamycin, (P<0.05, compared with the non-transfected group)
and miRNA135a-5p-inhibitor decreased this sensitivity (P<0.05,
compared with the non-transfected group). The cell viability of the
transfection control group was not different to the cells treated
with adriamycin alone (Fig.
3D).
To investigate the effects of miRNA135a-5p changes
on adriamycin-induced apoptosis in BGC-823 cells, adriamycin (5
μg/ml) was used to treat BGC-823 cells, 24 h after transfection and
apoptosis was detected. Our results showed that miRNA135a-5p-mimic
transfection increased the sensitivity to adriamycin and the
apoptotic rate induced by adriamycin and miRNA135a-5p-inhibitor
impaired adriamycin-induced apoptosis (P<0.05, vs. the group
without gene intervention) (Fig. 3E and
F).
Verification of binding site of AP-2α in
BCL-2 promoter
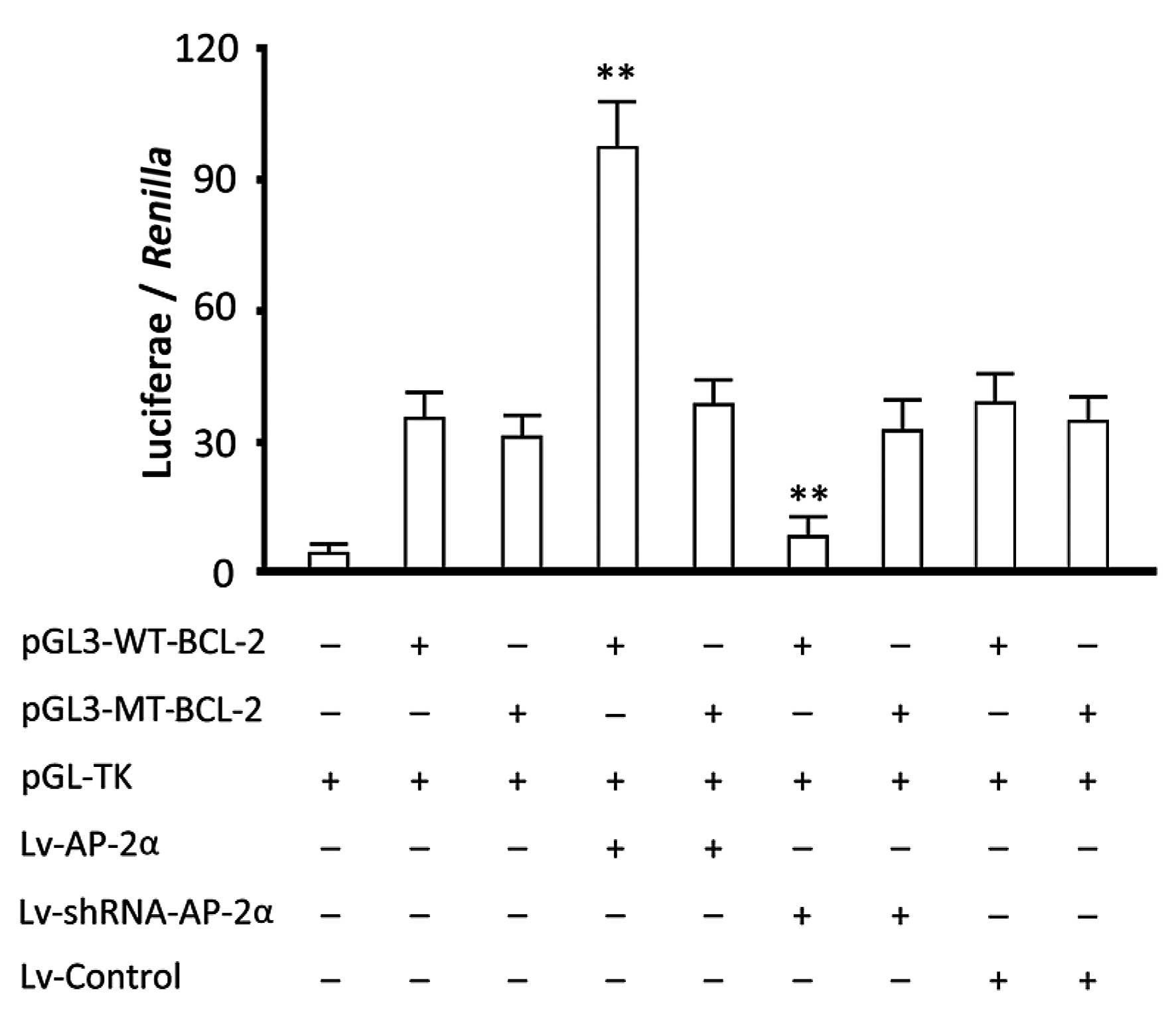
The results of the TFSEARCH analysis revealed a
possible binding site 5′-ACCGGCGGGCC-3′ of AP-2α in the BCL-2
promoter. Luciferase activity analysis showed that the
overexpression of AP-2α increased luciferase activity in the group
transfected with the reporter gene vector carrying a wild-type
BCL-2 promoter (P<0.05, vs. the group transfected with the
report vector alone) and silencing AP-2α decreased the luciferase
activity (P<0.05, vs. the group transfected with report vector
alone). The overexpression or silencing of AP-2α did not have any
obvious effects on luciferase activity in the groups transfected
with the reporter gene vector carrying a mutant type BCL-2 promoter
(Fig. 4). Therefore, a binding site
of AP-2α in BCL-2 promoter exists through which AP-2α may regulate
gene transcription.
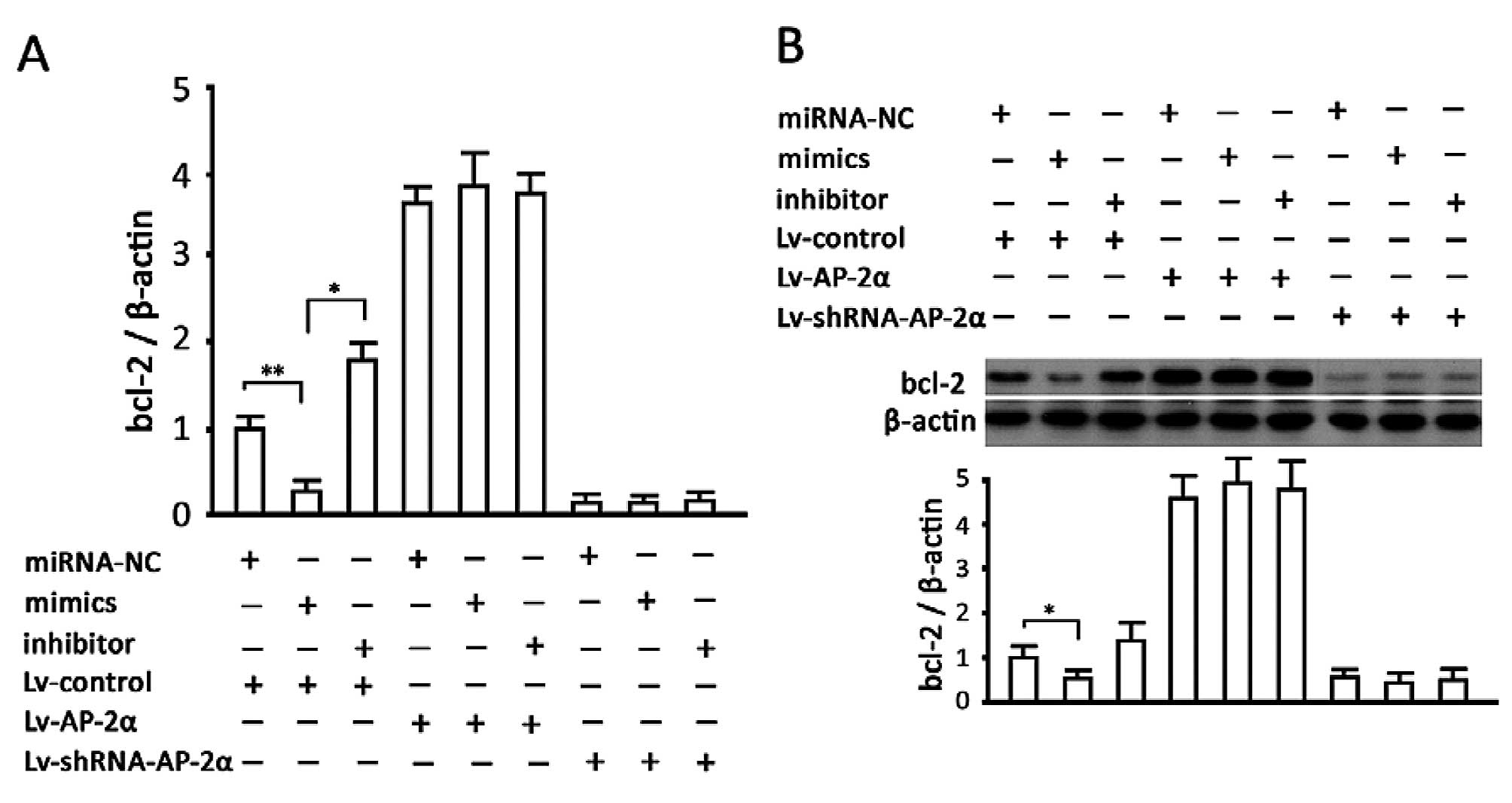
Analysis on the miRNA135a-5p-AP-2α-BCL2
pathway in apoptosis resistance regulation in gastric cells
The results of mRNA content detection showed that
the increase or decrease of miRNA135a-5p inhibited or increased
transcription of the BCL-2 gene (P<0.05, vs. the
non-transfected group), respectively. AP-2α overexpression
increased the BCL-2 mRNA level, while silencing AP-2α decreased
this level directly. In addition, changes in miRNA135a-5p had no
significant effect on BCL-2 mRNA in the cells of overexpressed or
silenced AP-2α. The results of protein measurement were concordant
with teh mRNA results (Fig. 5).
Discussion
miRNAs are a wide range of molecules regulating a
mass of genes, accounting for 2–3% of genes in the human genome. In
terms of structure, miRNAs are a class of short (18–24 nt)
single-stranded non-coding regulatory RNAs that negatively regulate
gene expression through complementary to target sites in the 3′-UTR
of target mRNAs at the post-transcriptional level, resulting in
mRNA degradation or translational repression and consequently
control such biological behaviors as tumor genesis and development.
Recent studies have demonstrated changes of miRNA expression in a
variety of human tumors, which may be a common characteristic of
cancer, indicating that miRNAs play critical roles in
tumorigenesis. For instance, miR-125b-1, located on a fragile site
on chromosome 11q24, has been shown to be associated with
breast, lung, ovarian and uterine cancer and play a tumor
suppressor role (28). The
downregulated expression of miR-15a/miR-16-1 is considered one of
the main causes of leukemia, lymphoma and prostate cancer (29). miR-143/miR-145 expression is
significantly decreased in breast, prostate, and uterine cancer,
and lymphoma (30). Let-7 is
considered to be a tumor-suppressor gene associated with lung
cancer (31). miR-21 is highly
expressed in glioblastoma and breast cancer and gene interference
experiments have shown it has a tumor-promoting function (32). Similarly, miR-155 expression is
increased in Burkitt’s and Hodgkin’s lymphoma, thus it may also
have a cancer-promoting effect (33).
Chemotherapy is the mainstay for the treatment of
malignant tumor, however, it is restricted by primary and acquired
drug resistance. The mutation of key genes in many drug resistance
pathways at the genetic or epigenetic level can lead to the
occurrence of drug resistance in tumor cells. As a possible
regulating way of these key genes, miRNAs are involved in the
regulatory function of tumor sensitivity to chemotherapeutic drugs
(34,35). Abnormal miRNA expression exists in
the majority of cancers (36) and
more than half of miRNAs are located on chromosome regions prone to
change in tumors. Up- or downregulation of miRNA expression
directly results in the abnormal expression of target genes and
alters the drug sensitivity of tumor cells by possible signaling
pathways. It has been reported that miR-21 overexpression exists in
patients with bile duct cancer, thus the patients are not sensitive
to gemcitabine, and reducing miR-21 with antisense
oligodeoxynucleotides enhanced the sensitivity of MCF7 cells to
topotecan and promoted apoptosis, which may be mediated by a low
expression of BCL2 (37,38). Studies have revealed that drug
sensitivity-related miRNAs, also include let-7a, miR-130a, miR-214,
mir-27a, miR-451, miR-221/222, miR-199, miR-15b/16, miR-34 and
miR-328, most of which are associated with apoptosis (39). Let-7a inhibits apoptosis by
downregulating CASP3, an important enzyme in apoptosis and the
overexpression of let-7a in A431 and HepG2 cells increased their
resistance to adriamycin, paclitaxel and interferon-γ, while
inhibiting let-7a increased chemotherapy-induced apoptosis
(40). miR-214 can downregulate
PTEN, leading to excessive activation of Akt pathway and cell
proliferation, thus patients with miR-214 overexpression did not
exhibit sensitivity to cisplatin (41). Low expression of miR-15b and miR-16
in a multidrug resistant gastric cancer cell line, SGC-7901/VCR,
maintains its target gene BCL-2 at a high level,
contributing to the cell resistance to drugs (23). In these studies, BCL-2, a crucial
molecule inhibiting apoptosis, is overexpressed in a variety of
tumors, resulting in multi-drug resistance. Therefore, examining
the relationship between BCL-2 and miRNAs is crucial.
In previous experiments, we screened miRNAs of
differential expression in gastric cancer tissue samples and found
that miRNA135a-5p may be one of those expressing significant
difference (data not shown), so we detected the miR135a-5p
expression by quantitative real-time PCR in gastric cancer tissues
in this study and found, compared to adjacent tissues, its content
in tumor tissues was significantly increased. Subsequently, through
a series of experiments, we demonstrated that AP-2α is one of its
target genes, which plays a role in tumor suppression as reported
in recent studies (42,43). We tried to knock down miRNA135a-5p
to inhibit cell proliferation by increasing AP-2α levels. However,
although AP-2α was increased, proliferation of BGC-823 cells was
not altered substantially. Notably, we treated cells with
adriamycin following genetic intervention and found that changes in
miRNA135a-5p content affected apoptosis in the tumor cells. This
finding suggested that there is a connection between miRNA135a-5p
and tumor drug sensitivity. We excluded the direct link by
bioinformatics analysis and speculated that miRNA135a-5p may affect
crucial drug-sensitive genes through its target gene AP-2α.
Since AP-2α is a transcription factor, we analyzed the genes
regulated by it and found that there is a possible binding site in
the promoter region of BCL-2 gene, which was confirmed by the
luciferase reporter method. To verify the miRNA135a-5p-AP-2α-BCL2
pathway, we altered miRNA135a-5p content in the cells of AP-2α
overexpression and knockdown to observe BCL-2 mRNA and protein
contents. The result also confirmed that miRNA135a-5p affects BCL-2
gene expression mediated by AP-2α.
Acknowledgements
This study was supported by the National Science
Foundation of China (81372293 and 81241088 to Y.K.F., 81273161 to
K.J.F.), New Century Excellent Talents in Heilongjiang Province
University (to Y.K.F.), Post-Doctoral Science Foundation of China
(2012M520762 to Y.K.F.), Department of Education of Heilongjiang
Province of China (12531736 to Y.M.P.), Program for Innovation
Research Team in Science and Technology in Heilongjiang Province
University (to Y.K.F. and Z.S.J.) and Program from China
Scholarship Council (to Y.K.F.)
References
|
1
|
Villanueva MT: Combination therapy: update
on gastric cancer in East Asia. Nat Rev Clin Oncol. 8:6902011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Leung WK, Wu MS, Kakugawa Y, et al:
Screening for gastric cancer in Asia: current evidence and
practice. Lancet Oncol. 9:279–287. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Widschwendter M and Jones PA: DNA
methylation and breast carcinogenesis. Oncogene. 21:5462–5482.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schoof CR, Botelho EL, Izzotti A and dos
Vasques LR: MicroRNAs in cancer treatment and prognosis. Am J
Cancer Res. 2:414–433. 2012.PubMed/NCBI
|
|
5
|
Hu J, Cheng Y, Li Y, et al: microRNA-128
plays a critical role in human non-small cell lung cancer
tumourigenesis, angiogenesis and lymphangiogenesis by directly
targeting vascular endothelial growth factor-C. Eur J Cancer.
50:2336–2350. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nakata K, Ohuchida K, Mizumoto K, et al:
MicroRNA-10b is overexpressed in pancreatic cancer, promotes its
invasiveness, and correlates with a poor prognosis. Surgery.
150:916–922. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nishida N, Yamashita S, Mimori K, et al:
MicroRNA-10b is a prognostic indicator in colorectal cancer and
confers resistance to the chemotherapeutic agent 5-fluorouracil in
colorectal cancer cells. Ann Surg Oncol. 19:3065–3071. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu K, Li G, Fan C, Zhou X, Wu B and Li J:
Increased expression of microRNA-21and its association with
chemotherapeutic response in human colorectal cancer. J Int Med
Res. 39:2288–2295. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang L, Mao P, Song L, et al: miR-182 as
a prognostic marker for glioma progression and patient survival. Am
J Pathol. 177:29–38. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Adachi R, Horiuchi S, Sakurazawa Y,
Hasegawa T, Sato K and Sakamaki T: ErbB2 down-regulates
microRNA-205 in breast cancer. Biochem Biophys Res Commun.
411:804–808. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chao A, Lin CY, Lee YS, et al: Regulation
of ovarian cancer progression by microRNA-187 through targeting
Disabled homolog-2. Oncogene. 31:764–775. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chai H, Liu M, Tian R, Li X and Tang H:
miR-20a targets BNIP2 and contributes chemotherapeutic resistance
in colorectal adenocarcinoma SW480 and SW620 cell lines. Acta
Biochim Biophys Sin (Shanghai). 43:217–225. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gocek E, Wang X, Liu X, Liu CG and
Studzinski GP: MicroRNA-32 upregulation by 1,25-dihydroxyvitamin D3
in human myeloid leukemia cells leads to Bim targeting and
inhibition of AraC-induced apoptosis. Cancer Res. 71:6230–6239.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kovalchuk O, Filkowski J, Meservy J, et
al: Involvement of microRNA-451 in resistance of the MCF-7 breast
cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther.
7:2152–2159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen J, Tian W, Cai H, He H and Deng Y:
Down-regulation of microRNA-200c is associated with drug resistance
in human breast cancer. Med Oncol. 29:2527–2534. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Z, Li Z, Gao C, et al: miR-21 plays
a pivotal role in gastric cancer pathogenesis and progression. Lab
Invest. 88:1358–1366. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chan SH, Wu CW, Li AF, Chi CW and Lin WC:
miR-21 microRNA expression in human gastric carcinomas and its
clinical association. Anticancer Res. 28:907–911. 2008.PubMed/NCBI
|
|
18
|
Lai KW, Koh KX, Loh M, et al:
MicroRNA-130b regulates the tumour suppressor RUNX3 in gastric
cancer. Eur J Cancer. 46:1456–1463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang X, Zhu W, Zhang J, et al:
MicroRNA-650 targets ING4 to promote gastric cancer tumorigenicity.
Biochem Biophys Res Commun. 395:275–280. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu Q, Jin H, Yang Z, et al: MiR-150
promotes gastric cancer proliferation by negatively regulating the
pro-apoptotic gene EGR2. Biochem Biophys Res Commun. 392:340–345.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang Z, Guo J, Xiao B, et al: Increased
expression of miR-421 in human gastric carcinoma and its clinical
association. J Gastroenterol. 45:17–23. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xiao B, Guo J, Miao Y, et al: Detection of
miR-106a in gastric carcinoma and its clinical significance. Clin
Chim Acta. 400:97–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xia L, Zhang D, Du R, et al: miR-15b and
miR-16 modulate multidrug resistance by targeting BCL2 in human
gastric cancer cells. Int J Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu W, Zhu D, Lu S, et al: miR-497
modulates multidrug resistance of human cancer cell lines by
targeting BCL2. Med Oncol. 29:384–391. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Orso F, Penna E, Cimino D, et al:
AP-2alpha and AP-2gamma regulate tumor progression via specific
genetic programs. FASEB J. 22:2702–2714. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jonckheere N, Fauquette V, Stechly L, et
al: Tumour growth and resistance to gemcitabine of pancreatic
cancer cells are decreased by AP-2alpha overexpression. Br J
Cancer. 101:637–644. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Allouche A, Nolens G, Tancredi A, et al:
The combined immunodetection of AP-2alpha and YY1 transcription
factors is associated with ERBB2 gene overexpression in primary
breast tumors. Breast Cancer Res. 10:R92008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thorsen J, Aamot HV, Roberto R, Tjonnfjord
GE, Micci F and Heim S: Myelodysplastic syndrome with a
t(2;11)(p21;q23–24) and translocation breakpoint close to
miR-125b-1. Cancer Genet. 205:528–532. 2012.
|
|
29
|
Liu J, Chen G, Feng L, et al: Loss of p53
and altered miR15-a/16-1→MCL-1 pathway in CLL: insights from
TCL1-Tg:p53(−/−) mouse model and primary human leukemia cells.
Leukemia. 28:118–128. 2014.PubMed/NCBI
|
|
30
|
Kent OA, Chivukula RR, Mullendore M, et
al: Repression of the miR-143/145 cluster by oncogenic Ras
initiates a tumor-promoting feed-forward pathway. Genes Dev.
24:2754–2759. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hertel J, Bartschat S, Wintsche A, Otto C
and Stadler PF: Evolution of the let-7 microRNA family. RNA Biol.
9:231–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wickramasinghe NS, Manavalan TT, Dougherty
SM, Riggs KA, Li Y and Klinge CM: Estradiol downregulates miR-21
expression and increases miR-21 target gene expression in MCF-7
breast cancer cells. Nucleic Acids Res. 37:2584–2595. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jones K, Nourse JP, Keane C, Bhatnagar A
and Gandhi MK: Plasma microRNA are disease response biomarkers in
classical Hodgkin lymphoma. Clin Cancer Res. 20:253–264. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Blower PE, Verducci JS, Lin S, et al:
MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol
Cancer Ther. 6:1483–1491. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Salter KH, Acharya CR, Walters KS, et al:
An integrated approach to the prediction of chemotherapeutic
response in patients with breast cancer. PLoS One. 3:e19082008.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bertino JR, Banerjee D and Mishra PJ:
Pharmacogenomics of microRNA: a miRSNP towards individualized
therapy. Pharmacogenomics. 8:1625–1627. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Meng F, Henson R, Lang M, et al:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
miR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bussing I, Slack FJ and Grosshans H: let-7
microRNAs in development, stem cells and cancer. Trends Mol Med.
14:400–409. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tsang WP and Kwok TT: Let-7a microRNA
suppresses therapeutics-induced cancer cell death by targeting
caspase-3. Apoptosis. 13:1215–1222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang H, Kong W, He L, et al: MicroRNA
expression profiling in human ovarian cancer: miR-214 induces cell
survival and cisplatin resistance by targeting PTEN. Cancer Res.
68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xu M, Chen X, Chen N, et al: Synergistic
silencing by promoter methylation and reduced AP-2α transactivation
of the proapoptotic HRK gene confers apoptosis resistance and
enhanced tumor growth. Am J Pathol. 182:84–95. 2013.PubMed/NCBI
|
|
43
|
Fu L, Chen W, Guo W, et al: Berberine
Targets AP-2/hTERT, NF-κB/COX-2, HIF-1α/VEGF and
Cytochrome-c/Caspase Signaling to Suppress Human Cancer Cell
Growth. PLoS One. 8:e692402013.PubMed/NCBI
|