Introduction
Clinical application of molecular targeted-therapy,
such as epidermal growth factor receptor tyrosine kinase inhibitors
(EGFR-TKIs), has improved the prognosis of lung cancer; the overall
survival of patients with metastatic non-small cell lung cancer
(NSCLC) presenting with EGFR mutations has risen to 27–30.5
months (1–3). Although approximately 70% of lung
cancer patients with EGFR-activating mutations show tumor
response to treatment with EGFR-TKI, they eventually acquire
resistance within 10–12 months (1–3). The
mechanisms of acquired resistance include EGFR secondary
mutation, T790M, MET amplification, small cell
transformation and overexpression of hepatocyte growth factor (HGF)
(4–7). Based on this evidence, irreversible
types of EGFR-TKIs and/or MET inhibitors produce marked tumor
response in vitro and animal experiments (8–10).
However, clinical trials using those agents targeted to patients
who have acquired resistance to EGFR-TKIs have not been
satisfactory (11–13). One of the reasons could be that
biomarkers related to the mechanisms of acquired resistance were
not available for these trials. Since the biological
characteristics of lung cancer could be altered during treatment,
it is necessary to clarify the molecular events in each individual
at the time of acquired resistance to EGFR-TKIs for selection of
the appropriate patient population.
We recently established a novel detection system for
T790M using plasma DNA, named the mutation-biased PCR and quenched
probe (MBP-QP) system, which is a sensitive, fully-automated
system. Using this system, we reported that T790M was detected in
plasma DNA obtained from 53% of lung cancer patients who acquired
resistance to EGFR-TKIs (14). This
system can be repeatedly applied to the same patients because of
the non-invasiveness of collecting plasma DNA. Since most lung
cancer recurrence after treatment with EGFR-TKI occurs as distant
metastases, peripheral blood is appropriate for monitoring
recurrence. Therefore, we chose plasma as the sample for monitoring
molecular events related with acquired resistance. Since T790M and
overexpression of HGF occur in 69–87% of patients who acquire
resistance to EGFR-TKIs (15,16),
we examined HGF and T790M using plasma samples.
HGF, a ligand for receptor tyrosine kinase, MET,
contributes to the promotion of metastasis and angiogenesis
(17–19). HGF is mainly secreted from
fibroblasts and fat-storing cells as an inactive form, named
pro-HGF, which is transformed into the active form by
HGF-converting enzyme, HGF activator (HGFA), injury and
glucocorticoids (18). Cancer cells
such as those from lung and breast cancers have been known to be
major sources of HGF, suggesting that HGF functions in both the
autocrine and paracrine machinery (17–19).
Neutrophils in the local environment of cancer tissue and in
peripheral blood have been reported to produce HGF mRNA and
release mature HGF (20,21). Considering these results, we assume
that HGF levels in peripheral blood should reflect HGF production
both in the localized environment and in the circulation throughout
the whole body.
The present study therefore describes our
investigation of whether quantification of the HGF level in
combination with detection of T790M in peripheral blood is useful
for monitoring mechanisms of acquired resistance to EGFR-TKI.
Materials and methods
Patient selection and blood samples
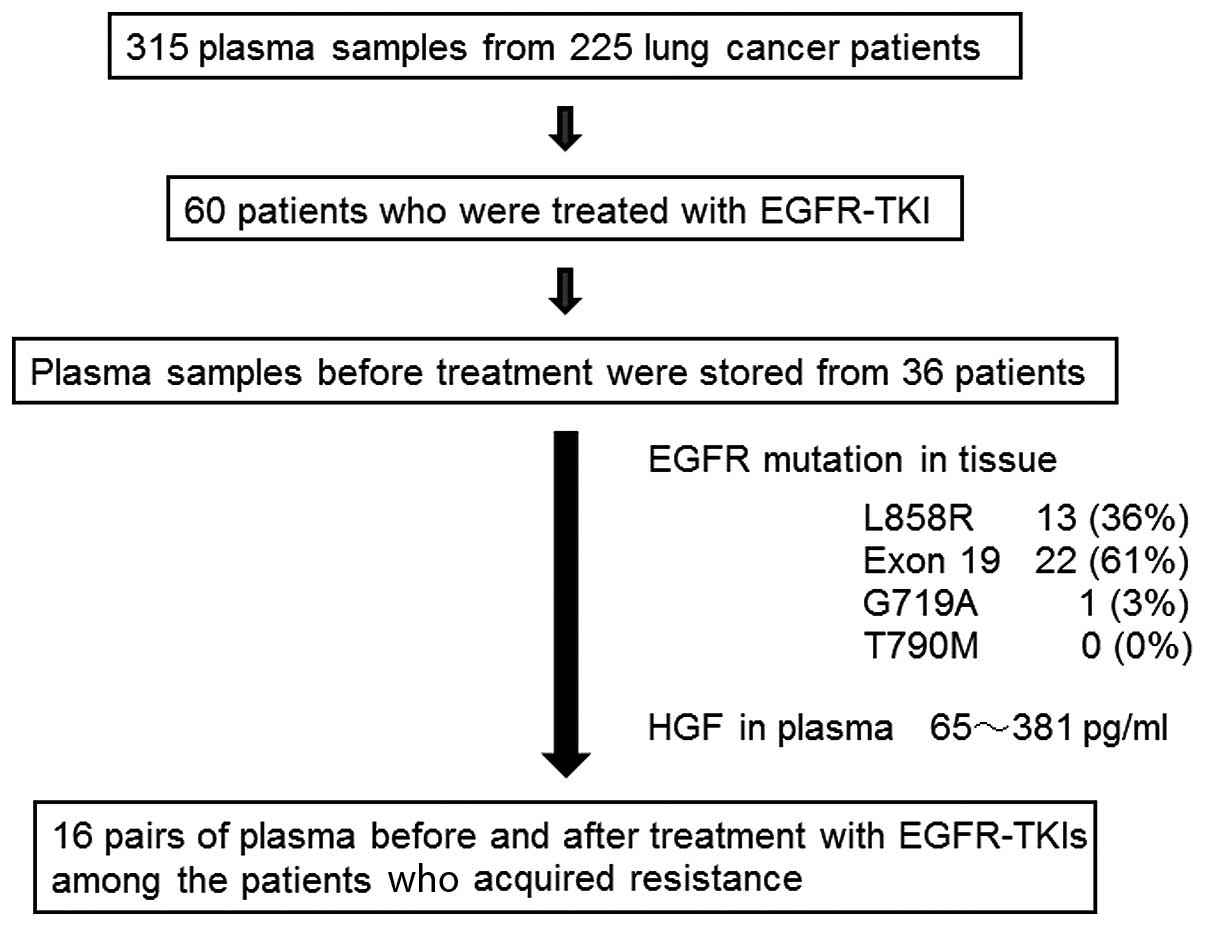
We obtained 315 plasma samples from 225 lung cancer
patients who underwent surgical treatment or chemotherapy at Saga
University Hospital from 2000 to 2013. Plasma samples were
repeatedly collected from 48 patients when lung cancer progressed,
from 2 to 7 times and they were obtained from 177 patients once.
HGF levels in plasma were determined using ELISA as described
below. Among the patients, 60 were treated with EGFR-TKIs, from
whom plasma samples were randomly collected from 36 patients before
treatment (Fig. 1). These samples
were used for quantification of HGF as well as detection of T790M
using plasma DNA. The clinical stage of the cancer was determined
according to criteria in the 7th edition of the
International Union Against Cancer when plasma samples were
obtained (22). The criteria for
acquired resistance were defined according to Jackman et al
as follows: previous treatment with a single EGFR-TKI, a tumor that
harbors an EGFR mutation associated with drug sensitivity or
objective clinical benefit from treatment with an EGFR-TKI, as
systemic progression of the disease while on continuous treatment
with an EGFR-TKI within the last 30 days and no intervening
systemic therapy between cessation of EGFR-TKI and initiation of
new therapy (23). The study
protocol was approved by the Clinical Research Ethics Committee of
Saga University. All patients provided informed consent for
obtaining blood according to the Declaration of Helsinki.
Quantification of the HGF level in
plasma
Peripheral blood samples from lung cancer patients
were collected into tubes containing 3.8% citric acid. Plasma was
immediately separated from blood cells by 3,000 rpm centrifugation
at 4°C for 20 min. Supernatants were collected and stored at −80°C
until assays were performed. The HGF level in plasma was measured
by enzyme-linked immunosorbent assay (Immunis HGF EIA; B-Bridge
International, Mountain View, CA, USA; limit of detection, 100
pg/ml), according to the manufacturer’s recommendations. Fifty
microliters of plasma was applied to the assay system. All samples
were assayed in duplicate. Color intensity was measured at 450 nm
with a spectrophotometric plate reader. HGF concentrations were
determined by comparison with standard curves.
DNA extraction from plasma and detection
of the EGFR T790M mutation
DNA was isolated from 200 μl of patient plasma using
a QIAamp® DNA mini kit (Qiagen, Hilden, Germany)
according to the manufacturer’s instructions. T790M mutation was
detected using the MBP-QP method as described previously (14). Briefly, MBP-QP is a fully automated
system with two steps: mutation-biased PCR (MBP) and quenched probe
(QP) system using i-densy (ARKRAY Inc., Kyoto, Japan). For MBP, the
primers for the wild-type and mutant-type were mixed with genomic
DNA, which leads to high specificity since each primer could be
competitively hybridized to the wild-type and mutant sequences. In
addition, the length of the reverse primer for the mutant was
longer than that for the wild-type and the annealing temperature
was designed to be optimum to the mutant primer, resulting in
higher efficiency of amplification of the mutant sequence. The
presence of T790M in the amplified sequences was determined by
monitoring the fluorescence intensity of a TAMRA-conjugated,
guanine-specific quench fluorophore probe (QProbe; J-Bio21, Tokyo,
Japan), which is complementary to T790M. The dissociation
temperatures were 66°C for the mutant and 59°C for the
wild-type.
Statistical analysis
The association between HGF levels and
clinicopathological characteristics was tested using the
nonparametric Mann-Whitney U test for continuous variables and
Kruskal-Wallis analysis was used for assessing whether the
distribution of HGF differed among the pathological stages.
Survival rate was calculated according to the Kaplan-Meier method
with differences assessed using the log rank test. Cox proportional
hazards regression analysis, with adjustment for potentially
confounding variables (gender, smoking status, histology,
pathological stage and EGR mutations), was used to calculate the
hazard ratio (HR) and 95% confidence internal (CI) of survival
outcome of lung cancer patients. All statistical analyses were
conducted using IBM SPSS Statistics 19 (SPSS Inc., IBM
Company).
Results
Clinicopathological characteristics of
the lung cancer patients with high HGF levels in plasma
The 225 lung cancer patients comprised 91% non-small
cell and 9% small cell lung cancer cases (Table I). Sixty-five percent were
adenocarcinoma cases and 36% expressed EGFR mutations
(L858R, exon 19 deletions, or others). The lower limit of HGF
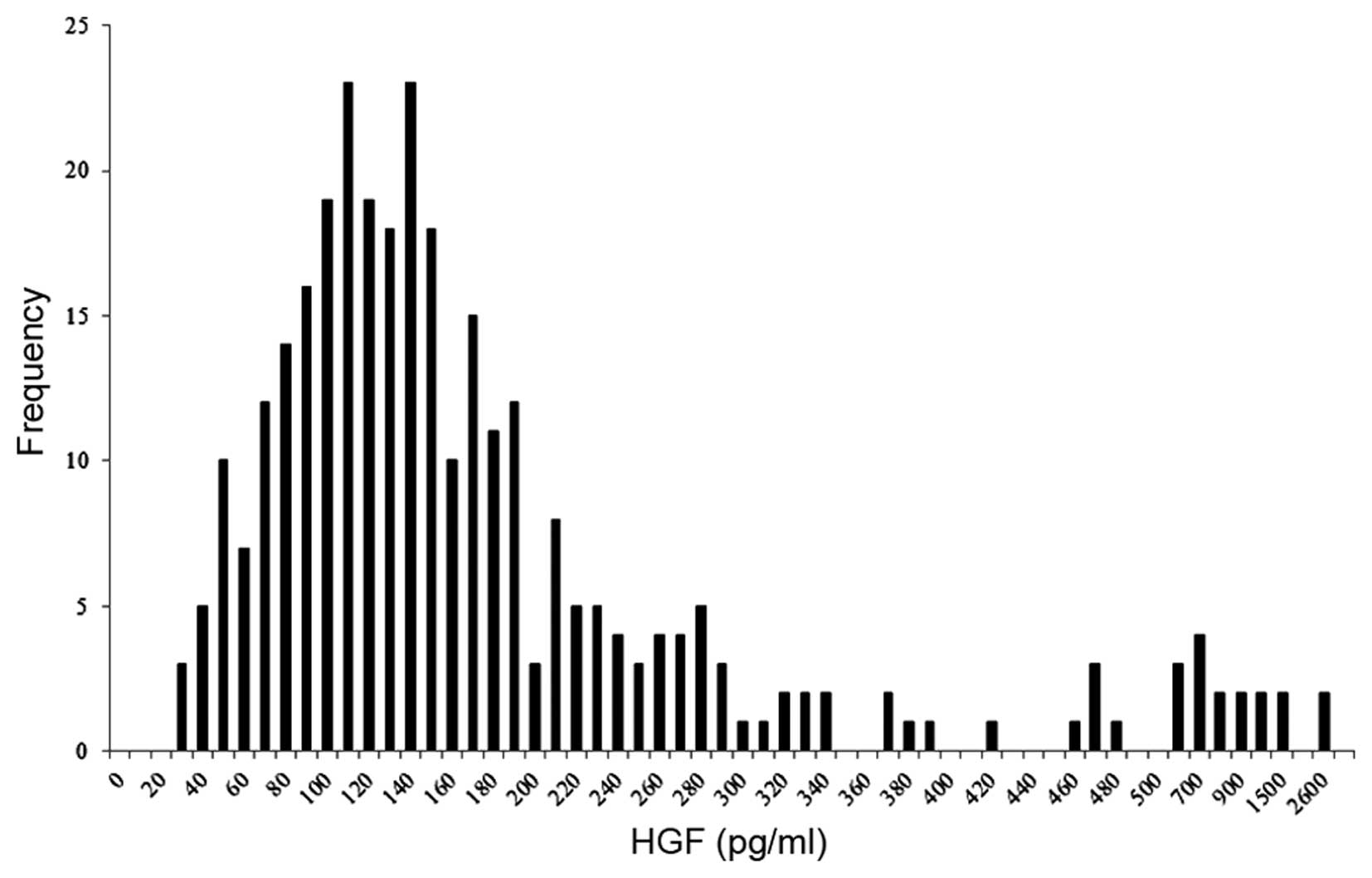
quantification was 100 pg/ml; 31 of the 315 plasma samples had HGF
levels below that limit. According to calculation by a standard
curve, the distribution of HGF levels is shown in Fig. 2. The median HGF level was 140 pg/ml
and the upper end of the range was 2,600 pg/ml. The correlation
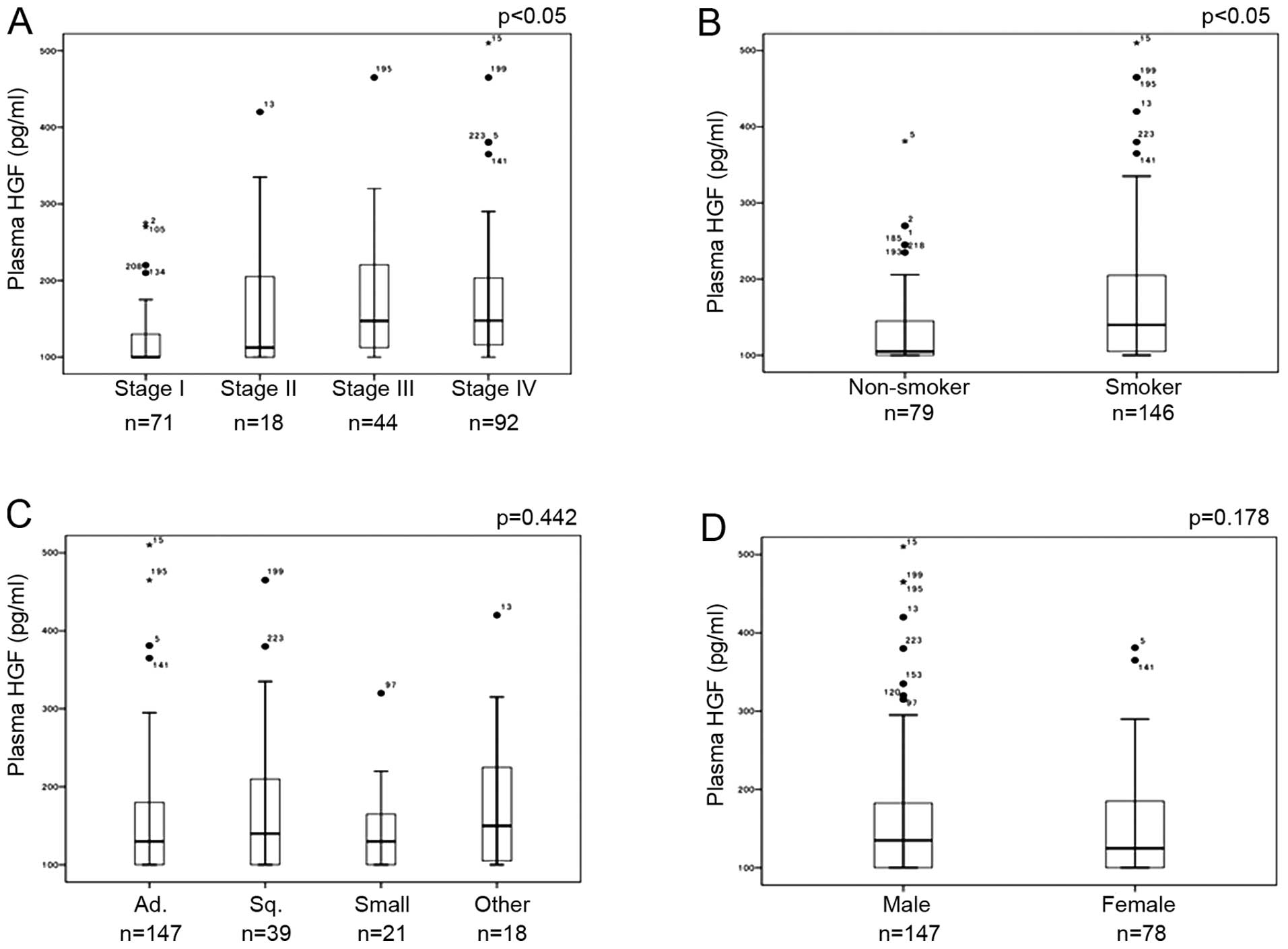
with clinicopathological characteristics showed that the HGF levels
were significantly higher among patients with advanced stage and
among smokers (Fig. 3A and B).
Associations between HGF levels and either histological type
(Fig. 3C) or gender (Fig. 3D) were not observed. Prognosis
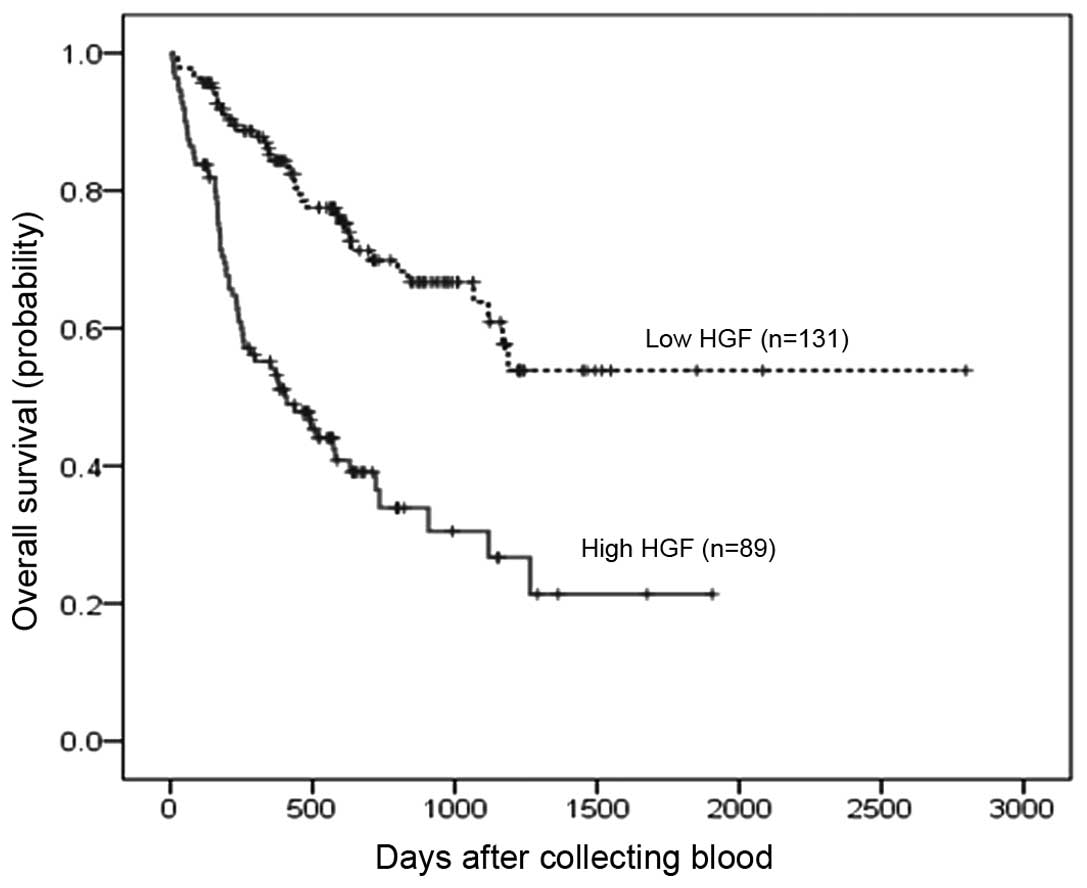
according to HGF level in plasma, comparing HGF greater than the
median and HGF less than or equal to the median, is shown in
Fig. 4. The median survival time
with high HGF was 409 days whereas that with low HGF was not
achieved, so that the high HGF group had significantly shorter
survival (log rank p<0.001). In addition to the HGF level in
plasma, possible prognostic factors, including gender, age,
pathological stage, histology, smoking status and EGFR
mutation, were analyzed. Based on a multivariate Cox proportional
hazards model, Table II shows that
pathological stage, EGFR mutation status and HGF level in
plasma had significant effects on survival even when simultaneously
adjusted. EGFR mutation-positive cases showed favorable
survival, whereas HGF level in plasma, as well as pathological
stage, was a predictor of poor survival. These results suggest that
clinicopathological characteristics of the patients with high HGF
levels in plasma are equivalent to those of patients with high HGF
in lung cancer tissues as previously reported in other laboratories
(24,25).
 | Table ICharacteristics of the study
patients. |
Table I
Characteristics of the study
patients.
| Characteristics | Data, n (%) |
|---|
| Total | 225 |
| Age (years) |
| Median | 68 |
| Range | 41–88 |
| Gender |
| Male | 147 (65) |
| Female | 78 (35) |
| Smoking |
| Smoker | 146 (65) |
| Non-smoker | 79 (35) |
| Histology |
| Squamous cell
carcinoma | 39 (17) |
| Adenocarcinoma | 147 (65) |
| Small cell
carcinoma | 21 (9) |
| Others | 18 (9) |
| Pathological
stage |
| I | 79 (35) |
| II | 20 (9) |
| III | 62 (28) |
| IV | 64 (28) |
| EGFR
mutation |
| L858R | 31 (14) |
| Exon 19
deletion | 44 (19) |
| Others | 7 (3) |
| Negative | 78 (35) |
| Unknown | 65 (29) |
 | Table IISurvival outcome by multivariate Cox
proportional hazards analysis for the lung cancer patients. |
Table II
Survival outcome by multivariate Cox
proportional hazards analysis for the lung cancer patients.
| Factors | HR (95% CI) | P-value |
|---|
| Age | 1.01
(0.99–1.03) | 0.51 |
| Gender (male vs.
female) | 0.89
(0.43–1.82) | 0.74 |
| Smoking status
(non-smoker vs. smoker) | 0.66
(0.31–1.39) | 0.27 |
| Histology (Ad vs.
others) | 1.14
(0.69–1.90) | 0.61 |
| Pathological stage
(IV vs. I, II, III) | 5.70
(3.63–8.95) | <0.001 |
| EGFR
mutation (negative, unknown vs positive) | 2.20
(1.23–3.93) | 0.008 |
| HGF (high vs.
low) | 2.52
(1.67–3.80) | <0.001 |
Elevation of HGF level and/or T790M using
plasma samples were frequently observed among the patients who
acquired resistance to EGFR-TKIs
Among the 225 patients examined in this study, 60
were treated with EGFR-TKIs, and plasma samples were randomly
collected from 36 patients before treatment (Fig. 1). EGFR mutations including
L858R in 13 patients and exon 19 deletions in 22 patients were
observed in lung cancer tissues obtained from 36 patients before
treatment with EGFR-TKIs. T790M was not detected in cancer tissues
in any patients. The range of HGF levels in plasma was from 65 to
381 pg/ml before treatment. Among the patients who acquired
resistance to EGFR-TKIs, 16 pairs of plasma before treatment with
EGFR-TKIs and after acquired resistance were obtained (Table III). Since the clinicopathological
characteristics with high HGF level in plasma were similar to that
in lung cancer tissues, it is possible that HGF in plasma would
reflect the local HGF level in cancer tissue. Therefore, we next
investigated whether HGF levels were elevated at the time point of
acquired resistance to EGFR-TKIs compared to those before
treatment. EGFR T790M mutation with plasma DNA was also
examined. Plasma HGF levels ranged from 90 to 680 and 79 to 1,235
pg/ml before treatment of EGFR-TKI and after acquired resistance,
respectively. The ratio of HGF level after acquired resistance to
that before treatment ranged from 0.52 to 7.3 and 6 patients showed
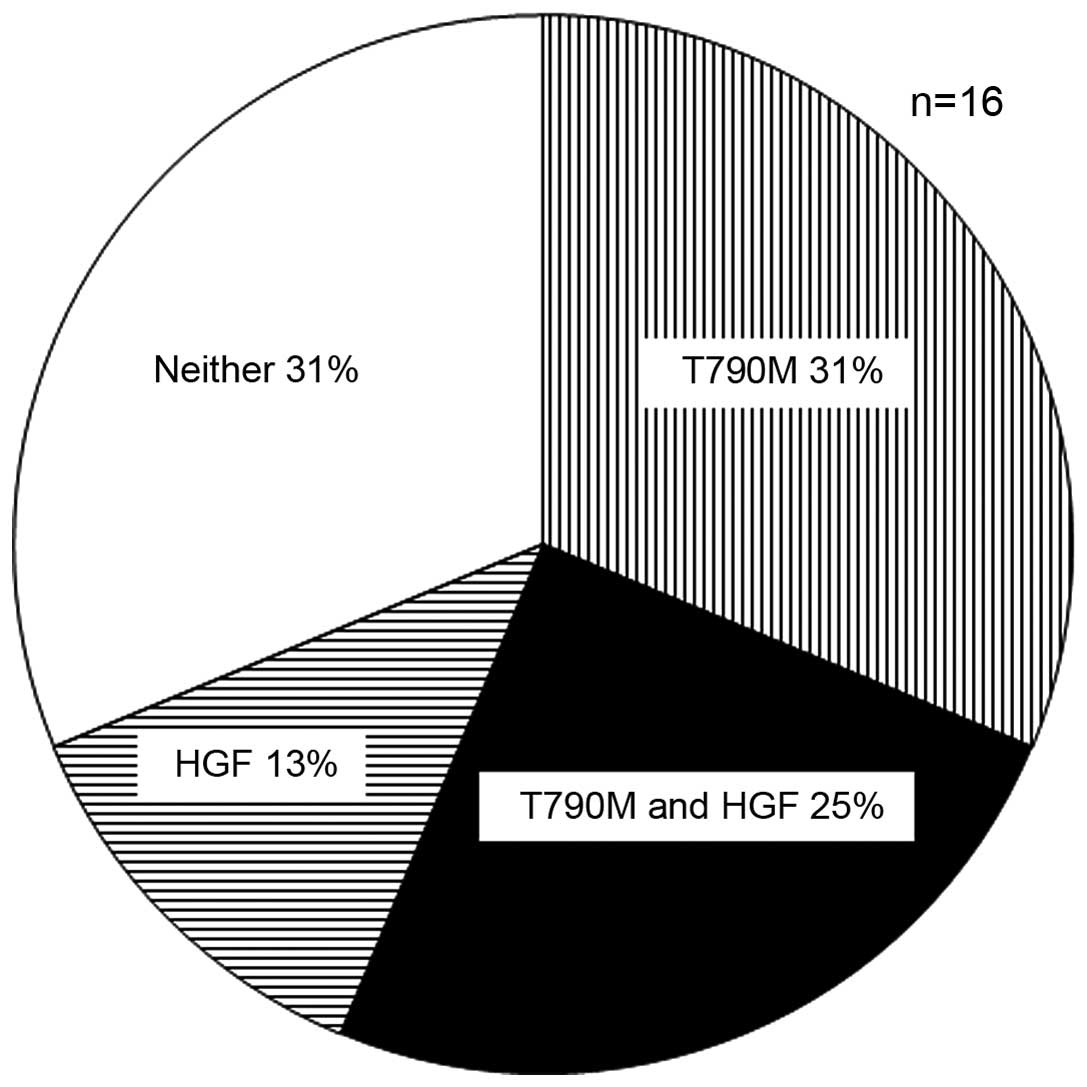
a >1.5-fold elevation in the HGF. T790M was detected with plasma
DNA in 9 patients after acquired resistance to EGFR-TKIs. Eleven of
the 16 patients (69%) showed either an HGF elevation (≤1.5-fold) or
T790M with plasma samples and elevations in both were observed in 4
patients (25%) (Fig. 5).
 | Table IIIPlasma HGF levels and T790M in plasma
DNA from the lung cancer patients who acquired resistance to
EGFR-TKIs. |
Table III
Plasma HGF levels and T790M in plasma
DNA from the lung cancer patients who acquired resistance to
EGFR-TKIs.
| HGF (pg/ml) | T790M |
|---|
|
|
|
|---|
| Before | After | Ratio | Before | After |
|---|
| 1 | 160 | 200 | 1.3 | Negative | Negative |
| 2 | 381 | 510 | 1.3 | Negative | Positive |
| 3 | 170 | 1235 | 7.3 | Negative | Positive |
| 4 | 130 | 166 | 1.3 | Negative | Positive |
| 5 | 120 | 340 | 2.8 | Negative | Positive |
| 6 | 680 | 725 | 1.1 | Negative | Positive |
| 7 | 175 | 270 | 1.5 | Negative | Negative |
| 8 | 245 | 145 | 0.59 | Negative | Negative |
| 9 | 145 | 325 | 2.2 | Negative | Negative |
| 10 | 90 | 140 | 1.6 | Negative | Positive |
| 11 | 301 | 157 | 0.52 | Negative | Positive |
| 12 | 277 | 160 | 0.58 | Negative | Negative |
| 13 | 130 | 265 | 2.0 | Negative | Positive |
| 14 | 105 | 79 | 0.76 | Negative | Negative |
| 15 | 185 | 190 | 1.0 | Negative | Negative |
| 16 | 202 | 240 | 1.2 | Negative | Positive |
Discussion
HGF plays a central role in cancer progression,
including proliferation of cancer cells, invasion, angiogenesis and
metastasis (17–19). From the viewpoint of these
biological activities, HGF has been investigated as a candidate
prognostic marker for various cancers including colon, stomach,
prostate and multiple myeloma (24–27).
In lung cancer, HGF overexpression has been reported to be a
prognostic factor using tissue samples (28,29).
As an alternative to using cancer tissue specimens, we showed that
the HGF levels in plasma were higher in advanced stage and among
smokers and it is a prognostic marker for lung cancer, which is
independent of other clinicopathological factors.
The mechanisms of acquired resistance to EGFR-TKIs
have been reported to be EGFR secondary mutation, T790M,
MET or HER2 amplification and small cell
transformation (4–6,15,16).
In addition to those mechanisms, HGF induced gefitinib, EGFR-TKI,
resistance of lung adenocarcinoma cell lines carrying EGFR
activating mutations and HGF overexpression was observed in cancer
tissues from patients who acquired resistance to EGFR-TKIs
(7). According to the results using
re-biopsy specimens in other laboratories, T790M was detected in
51–69% and HGF overexpression examined by immunohistochemistry was
observed in 61% of cases (15,16,30).
Although re-biopsy would be the most reliable method for
determination of the mechanisms of acquired resistance to
EGFR-TKIs, it is associated with various issues. One is that
re-biopsy is invasive, since most lung cancer recurrence occurs as
distant metastases in liver, bone, brain, adrenal gland and
intrapulmonary regions. The other is that the molecular
characteristics sometimes vary among the metastatic lesions,
suggesting that a biopsy specimen in one lesion would not reflect
the entire body (31). Considering
these issues, peripheral blood could be an appropriate sample for
determining the dominant molecular alterations in the entire body.
Since collecting peripheral blood is non-invasive, it is suitable
for monitoring acquired resistance, which requires repeated
examinations. We showed that T790M was detected in 56% of cases and
elevation of HGF was observed in 38% of cases. Although the
frequency of HGF elevation was lower than that using re-biopsy, the
T790M detection rate was equivalent. The possible reason for the
lower frequency of HGF elevation in plasma could be that cells
expressing HGF exist not only in cancer tissue but also in
peripheral blood, resulting in a high background level of HGF in
plasma.
These mechanisms of acquired resistance sometimes
co-exist such as T790M and MET amplification, small cell
transformation and T790M and HGF overexpression and T790M (15,16).
This phenomenon may cause primary resistance to second generation
EGFR-TKIs and therefore it is critical for making decisions whether
combination therapy is needed or not. Our results revealed that
co-existence of T790M and HGF elevation was observed in 25% (4/16)
of the patients who acquired resistance to EGFR-TKIs, which is also
equivalent to that using re-biopsy. Combining detection systems for
HGF and T790M using plasma samples enabled us to detect the
mechanisms of acquired resistance to EGFR-TKIs in 69% of patients
who acquired resistance to EGFR-TKIs. A prospective study to
investigate the utility of these detection systems for predicting
the anticancer effects of next generation EGFR-TKIs and/or MET
inhibitors is worthy of investigation.
Acknowledgements
This study was supported in part by a Grant-in-Aid
for Young Scientists (B) 25870518. The authors are grateful to Dr
John B. Cologne for suggestions regarding the statistical
analysis.
References
|
1
|
Maemondo M, Inoue A, Kobayashi K, et al:
Gefitinib or chemotherapy for non-small-cell lung cancer with
mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mitsudomi T, Morita S, Yatabe Y, et al:
Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer harbouring mutations of the epidermal
growth factor receptor (WJTOG3405): an open label, randomised phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar
|
|
3
|
Mok TS, Wu YL, Thongprasert S, et al:
Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N
Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kobayashi S, Boggon TJ, Dayaram T, et al:
EGFR mutation and resistance of non-small-cell lung cancer to
gefitinib. N Engl J Med. 352:786–792. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Engelman JA, Zejnullahu K, Mitsudomi T, et
al: MET amplification leads to gefitinib resistance in lung cancer
by activating ERBB3 signaling. Science. 316:1039–1043. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zakowski MF, Ladanyi M and Kris MG:
Memorial sloan-kettering cancer center lung cancer oncogenome
group. EGFR mutations in small-cell lung cancers in patients who
have never smoked. N Engl J Med. 355:213–215. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yano S, Wang W, Li Q, et al: Hepatocyte
growth factor induces gefitinib resistance of lung adenocarcinoma
with epidermal growth factor receptor-activating mutations. Cancer
Res. 68:9479–9487. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li D, Ambrogio L, Shimamura T, et al:
BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in
preclinical lung cancer models. Oncogene. 27:4702–4711. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang W, Li Q, Takeuchi S, et al: Met
kinase inhibitor E7050 reverses three different mechanisms of
hepatocyte growth factor-induced tyrosine kinase inhibitor
resistance in EGFR mutant lung cancer. Clin Cancer Res.
18:1663–1671. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nakagawa T, Takeuchi S, Yamada T, et al:
Combined therapy with mutant-selective EGFR inhibitor and Met
kinase inhibitor for overcoming erlotinib resistance in EGFR-mutant
lung cancer. Mol Cancer Ther. 11:2149–2157. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miller VA, Hirsh V, Cadranel J, et al:
Afatinib versus placebo for patients with advanced, metastatic
non-small-cell lung cancer after failure of erlotinib, gefitinib,
or both and one or two lines of chemotherapy (LUX-Lung 1): a phase
2b/3 randomised trial. Lancet Oncol. 13:528–538. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Katakami N, Atagi S, Goto K, et al:
LUX-Lung 4: a phase II trial of afatinib in patients with advanced
non-small-cell lung cancer who progressed during prior treatment
with erlotinib, gefitinib, or both. J Clin Oncol. 31:3335–3341.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sequist LV, Pawel J, Garmey EG, et al:
Randomised phase II study of erlotinib plus tivantinib versus
erlotinib plus placebo in previously treated non-small-cell lung
cancer. J Clin Oncol. 29:3307–3315. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nakamura T, Sueoka-Aragane N, Iwanaga K,
et al: A noninvasive system for monitoring resistance to epidermal
growth factor receptor tyrosine kinase inhibitors with plasma DNA.
J Thorac Oncol. 6:1639–1648. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yano S, Yamada T, Takeuchi S, et al:
Hepatocyte growth factor expression in EGFR mutant lung cancer with
intrinsic and acquired resistance to tyrosine kinase inhibitors in
a Japanese cohort. J Thorac Oncol. 6:2011–2017. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu HA, Arcila ME, Rekhtman N, et al:
Analysis of tumor specimens at the time of acquired resistance to
EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers.
Clin Cancer Res. 19:2240–2247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mizuno S and Nakamura T: HGF-MET cascade,
a key target for inhibiting cancer metastasis: The impact of NK4
discovery on cancer biology and therapeutics. Int J Mol Sci.
14:888–919. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang WG, Martin TA, Parr C, et al:
Hepatocyte growth factor, its receptor and their potential value in
cancer therapies. Crit Rev Oncol Hematol. 53:35–69. 2005.
View Article : Google Scholar
|
|
19
|
Martin TA, Parr C, Davies G, et al: Growth
and angiogenesis of human breast cancer in a nude mouse tumor model
is reduced by NK4, a HGF/SF antagonist. Carcinogenesis.
24:1317–1323. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wisles M, Rabbe N, Marchal J, et al:
Hepatocyte growth factor production by neutrophils infiltrating
bronchioloalveolar subtype pulmonary adenocarcinoma: Role in tumor
progression and death. Cancer Res. 63:1405–1412. 2003.
|
|
21
|
Grenier A, Chollet-Martin S, Crestani B,
et al: Presence of a mobilizable intracellular pool of hepatocyte
growth factor in human polymorphonuclear neutrophils. Blood.
99:2997–3004. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Goldstrow P, Crowley J, Chansky K, et al:
The IASLC Lung Cancer Staging Project: Proposals for the revision
of the TNM stage groupings in the forthcoming (seventh) edition of
the TNM classification of malignant tumours. J Thorac Oncol.
2:706–714. 2007. View Article : Google Scholar
|
|
23
|
Jackman D, Pao W, Riely GJ, et al:
Clinical definition of acquired resistance to epidermal growth
factor receptor tyrosine kinase inhibitors in non-small-cell lung
cancer. J Clin Oncol. 28:357–360. 2010. View Article : Google Scholar
|
|
24
|
Takanami I, Tanana F, Hashizume T, et al:
Hepatocyte growth factor and c-Met/hepatocyte growth factor
receptor in pulmonary adenocarcinomas: An evaluation of their
expression as prognostic markers. Oncology. 53:392–397. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Siegfried JM, Weissfeld LA, Luketich JD,
et al: The clinical significance of hepatocyte growth factor for
non-small cell lung cancer. Ann Thorac Surg. 66:1915–1918. 1998.
View Article : Google Scholar
|
|
26
|
Toiyama Y, Miki C, Inoue Y, et al: Serum
hepatocyte growth factor as a prognostic marker for stage I or II
colorectal cancer patients. Int J Cancer. 125:1657–1662. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tanaka K, Miki C, Wakuda R, et al:
Circulating level of hepatocyte growth factor as a useful tumor
marker in patients with early-stage gastric carcinoma. Scand J
Gastroenterol. 8:754–760. 2004. View Article : Google Scholar
|
|
28
|
Gupta A, Karakiewicz PI, Roehrborn CG, et
al: Predictive value of plasma hepatocyte growth factor/scatter
factor levels in patients with clinically localized prostate
cancer. Clin Cancer Res. 14:7385–7390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Seidel C, Borset M, Turesson I, et al:
Elevated serum concentrations of hepatocyte growth factor in
patients with multiple myeloma. The Nordic Myeloma Study Group.
Blood. 91:1806–1812. 1998.
|
|
30
|
Arcila MA, Oxnard GR, Nafa K, et al:
Rebiopsy of lung cancer patients with acquired resistance to EGFR
inhibitors and enhanced detection of the T790M mutation using a
locked nucleic acid-based assay. Clin Cancer Res. 17:1169–1180.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Suda K, Murakami I, Katayama, et al:
Reciprocal and complementary role of MET amplification and EGFR
T790M mutation in acquired resistance to kinase inhibitors in lung
cancer. Clin Cancer Res. 16:5489–5498. 2010. View Article : Google Scholar : PubMed/NCBI
|