Introduction
Breast cancer is the most significant worldwide
health concern for females, accounting for the leading cause of
cancer incidence and the second mortality rate in 2008 (1–3). In
spite of advancements in early detection, treatment options, and
prevention strategies for controlling breast cancer, a high
percentage of breast cancer cases continue to progress,
metastasize, and eventually cause patients to succumb to the
disease. In this regard, identification of genes or molecular
pathways that are responsible for the development and progression
of breast cancer with understanding of their clinical significance
are necessary, and may aid in the development of novel approaches
for the effective treatment and prognostic prediction of breast
cancer. Towards this end, our study focused on Ras
GTPase-activating protein 1 (RASA1), which functions to inactivate
Ras-GTPase and inhibit mitogenic signaling to downstream protein
partners through its N-terminal SH2-SH3-SH2 domains (4–6).
Aberrant expression of RASA1 protein occurs in ~30% of all human
cancers, including breast cancer, but in up to 90% of pancreatic
cancers (7).
The Ras family members belong to a class of small
GTPase proteins that switch between the active GTP-bound and
inactive GDP-bound states to control intracellular signaling
networks; for example, the activation of Ras signaling can lead to
cell cycle progression and cell proliferation, and can alter cell
differentiation, adhesion, and migration, while reducing cell
apoptosis (4,5). Altered expression of the Ras oncogene
and Ras-related proteins promotes tumorigenesis and increases tumor
cell invasion and metastasis (4,5). As a
Ras family member, P120 Ras GTPase-activating protein (RasGAP)
encoded by the RASA1 gene is involved in the negative regulation of
the Ras oncogene (8), with an
altered expression of RASA1 protein being reported in several types
of carcinoma (9,10). A reduction of RASA1 expression or
activity can trigger the RAS-MAPK signaling pathway by activating
the GTPase activity of RAS, thereby increasing cell proliferation,
suppressing apoptosis, deregulating the cell cycle, and eventually
leading to the malignant transformation of cells (10). Thus, in this study, we assessed
RASA1 expression in breast cancer tissues and determined whether
aberrant expression was associated with clinicopathological factors
and prognosis of breast cancer patients. This allowed us to
evaluate the potential of using RASA1 expression as a biomarker for
predicting clinical outcome and prognosis of breast invasive ductal
carcinoma patients (IDC).
Materials and methods
Patients and tissue specimens
In this study, we collected two sets of tissue
specimens. The first set included 45 patients with breast IDC who
were subjected to surgical resection for breast cancer at The Third
Affiliated Hospital, Harbin Medical University between March and
August 2012. We also collected fresh breast cancer tissue samples
and corresponding non-tumor breast tissue immediately after
surgical resection, which were immediately snap-frozen in liquid
nitrogen and stored at −80°C at the Heilongjiang Breast Tumor
Biobank. The second set of breast cancer tissue specimens, 373
paired paraffin-embedded IDC and corresponding non-tumor breast
tissue blocks, were obtained from the Department of Pathology, The
Third Affiliated Hospital, Harbin Medical University between March
and December 2006. The percentage of breast cancer in the tumor
specimens was >75%. All the patients were confirmed as breast
IDC by a pathological diagnosis of breast tissues and all the
samples were collected prior to any radiotherapy or chemotherapy.
The Ethics Committee of Harbin Medical University approved our
study protocol and written informed consent was obtained from each
patient.
Clinicopathological data were collected from patient
medical records and the patients (n=373) were followed up until
December 2012. The patients were closely monitored after surgery
with a median follow-up period of 74 months (range, 3–82 months).
Overall survival (OS) and disease-free survival (DFS) were defined
as the time from the date of surgery to date of death and the first
recurrence, respectively.
Individual samples were routinely assessed for
expression of the estrogen receptor (ER), progesterone receptor
(PR), human epidermal growth factor receptor 2 (HER2) and Ki-67 by
immunohistochemistry. A tumor lesion (case) with ≥14% of
Ki-67-positive tumor cells was considered highly proliferative
(11). The intensity of anti-HER2
staining was semi-quantitatively analyzed and graded as 0–3: grade
0 or 1, negative; grade 2, weakly positive; and grade 3, positive.
Tissues with a weak positive expression (grade 2) were also
subjected to duplicate fluorescence in situ hybridization
(FISH) analysis of the HER-2 gene and scored as negative
(<2-fold change) or positive (>2-fold increase) HER-2 gene
amplification (12,13). Furthermore, each breast cancer case
was subjected to molecular subtyping according to the following
criteria: Luminal A type, ER- and/or PR-positive, HER2-negative and
Ki-67-positive cells <14%; Luminal B type, (HER2-negative) ER-
and/or PR-positive, HER2-negative and Ki-67-positive cells ≥14%,
(HER2-positive) ER- and/or PR-positive, with HER2 being
overexpressed and/or amplified; HER2 type, where ER and PR were
negative and HER2 was overexpressed and/or amplified; or
triple-negative breast cancer type where ER, PR, and HER2 were all
negative (14). The
clinicopathological characteristics of the 373 patients are listed
in Table I.
 | Table IAssociation of RASA1 expression with
clinicopathological characteristics from invasive ductal carcinoma
patients (n=373). |
Table I
Association of RASA1 expression with
clinicopathological characteristics from invasive ductal carcinoma
patients (n=373).
| | RASA1 | |
|---|
| |
| |
|---|
| Characteristics | n | Negative | Positive | P-value |
|---|
| Age (years) |
| <50 | 208 | 132 | 76 | 0.203 |
| ≥50 | 165 | 94 | 71 | |
| Tumor size (cm) |
| T1 (≤2) | 138 | 75 | 63 | 0.059 |
| T2–T3 (>2) | 235 | 151 | 84 | |
| Lymph node
metastasis |
| Negative | 167 | 83 | 84 | 0.002 |
| Positive | 206 | 143 | 63 | |
| TNM stage |
| I/II | 255 | 144 | 111 | 0.017 |
| III | 118 | 82 | 36 | |
| ER |
| Negative | 159 | 111 | 48 | 0.002 |
| Positive | 214 | 115 | 99 | |
| PR |
| Negative | 126 | 83 | 43 | 0.136 |
| Positive | 247 | 143 | 104 | |
| HER-2 |
| Negative | 240 | 148 | 92 | 0.568 |
| Positive | 133 | 78 | 55 | |
| Ki-67 |
| Negative | 162 | 86 | 76 | 0.009 |
| Positive | 211 | 140 | 71 | |
| Histological
grade |
| G1–G2 | 237 | 160 | 77 | <0.001 |
| G3 | 136 | 66 | 70 | |
| Molecular
subtype |
|
Triple-negative | 67 | 48 | 19 | 0.041 |
| Others | 306 | 178 | 128 | |
| Recurrence |
| Yes | 139 | 92 | 47 | 0.088 |
| No | 234 | 134 | 100 | |
| Death |
| Yes | 76 | 60 | 16 | <0.001 |
| No | 297 | 166 | 131 | |
Quantitative RT-PCR (RT-qPCR)
Total RNA was isolated from frozen tissue samples
using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to
the manufacturer’s instructions and quantified using a NanoDrop
spectrophotometer (Thermo Scientific, Waltham, MA, USA). Then, 100
ng of each RNA sample was reverse transcribed into cDNA using a
High-Capacity cDNA reverse transcriptase kit (Applied Biosystems,
Foster City, CA, USA) and qPCR was amplified. The relative levels
of RASA1 transcripts were normalized to control GAPDH mRNA using
the 2−ΔΔCt method. The primer sequences for RASA1 were
forward, 5′-CCAACGCCAAAC AATCAG-3′ and reverse,
5′-ATTTCCTTGCCATCCACT-3′; and GAPDH forward, 5′-GCCAGCCGAGCCACAT-3′
and reverse, 5′-CTTTACCAGAGTTAAAAGCAGCCC-3′. The qPCR amplification
was performed in triplicate at 95°C for 10 min and 40 cycles of
95°C for 15 sec and 60°C for 30 sec. Negative controls consisted of
distilled H2O.
Tissue microarray and
immunohistochemistry
Immunohistochemical staining was carried out using
the two-step plus poly-HRP method as described previously (15). Briefly, tissue blocks were cut for 4
μm sections and placed on poly-L-lysine coated slides. The slides
were deparaffinized, dehydrated, immersed in 10 mM sodium citrate
buffer (pH 6.0) and pretreated in a microwave oven for 10 min. This
was followed by a 10-min rinse with PBS. After blocking with 3%
hydrogen peroxide for 10 min at room temperature, the slides were
incubated at 4°C overnight with primary anti-RasGAP antibody (1:150
mouse mAb; Abcam, Cambridge, MA, USA). Afterwards, the slides were
stained with the two-step plus poly-HRP anti-mouse IgG detection
system (ZSGB-Bio, Beijing, China). After visualization of the
reaction with DAB chromogen, the slides were counterstained with
hematoxylin and covered with a glycerin gel. For the negative
controls, the primary antibody was substituted with PBS to confirm
the specificity of the primary antibody.
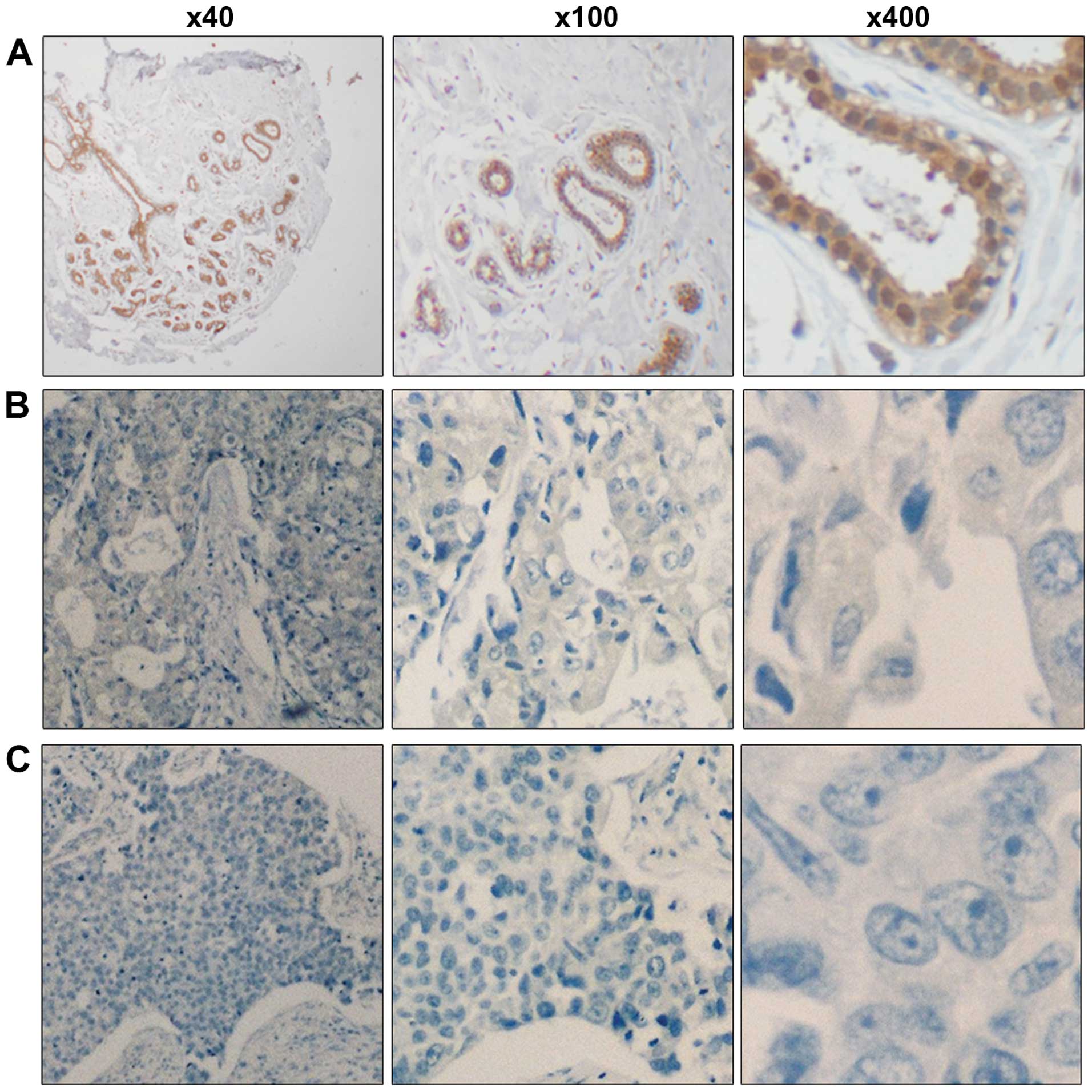
The relative levels of RasGAP expression were
evaluated semi-quantitatively as described in a previous study
(16). Briefly, the percentage and
intensity of positive anti-RasGAP staining was evaluated and scored
in randomly selected 10 high-power fields (magnification, ×400) by
two investigators in a blinded manner. The percentage of positively
stained tumor cells was scored as 0, zero; 1, <10%; 2, 10–50%;
and 3, >50%. The staining intensity was scored as 0, no
staining; 1, weak (appearing as light yellow); 2, moderate staining
(appearing as yellowish-brown); and 3, strong staining (appearing
as brown). The staining index (SI) of individual sections was then
calculated as: (averaged staining intensity score) × (proportion
score). The cut-off value for anti-RasGAP staining was determined
by measuring heterogeneity accordingly. An SI score of 4 as the
cut-off value was used to distinguish low (<4) vs. high (≥4)
RasGAP expression. The cases with discrepancies in the staining
scores were re-reviewed by the two pathologists together with a
senior pathologist to reach consensus. Ultimately, the staining
assessment and allocation of tumors by two investigators were
similar with perfect inter-rater reliability.
Statistical analysis
Data were expressed as either real-case numbers or
percentages. The difference between groups was analyzed by the
χ2 test or Fisher’s exact test. The OS and DFS of the
patients were estimated by the Kaplan-Meier survival curve and
analyzed by the log-rank test. The potential factors affecting
survival were analyzed by risk ratios (RRs), 95% confidence
intervals (CI), and univariate and multivariate regression analyses
using the Cox proportional hazards model. Statistical analyses were
performed using SPSS, ver. 11.0. (SPSS, Chicago, IL, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Differential expression of RASA1 mRNA and
protein in breast IDC tissues
In this study, we first assessed the expression of
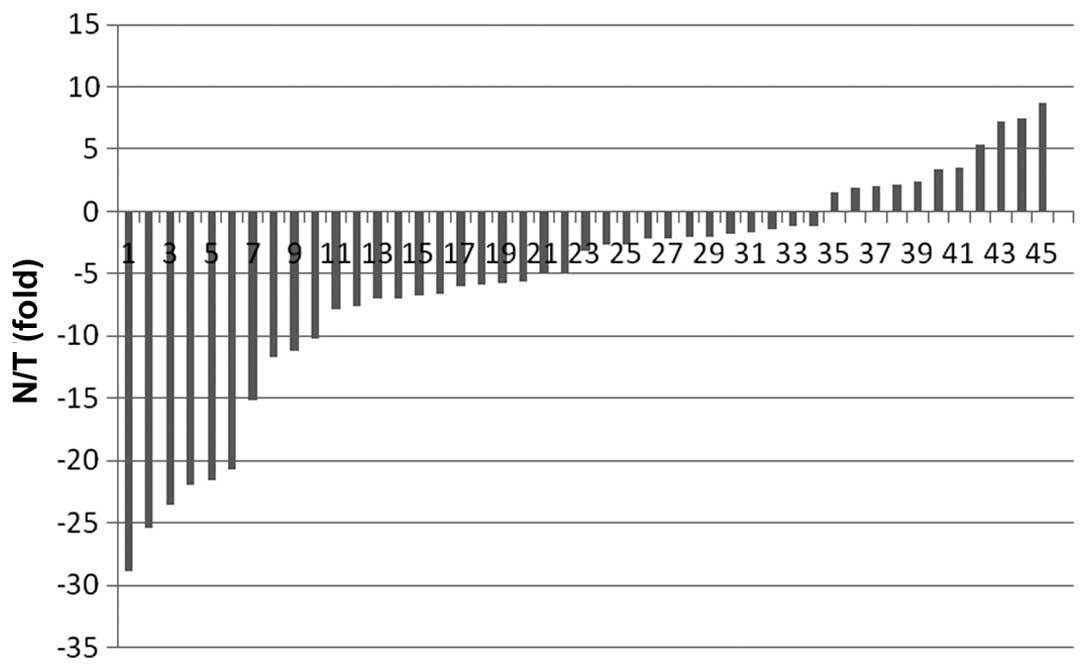
RASA1 mRNA in 45 samples of breast IDC tissue and found that the
mean expression value of RASA1 mRNA in the tumor tissues was
2.93±17.98 (normalized to the GAPDH level), which was significantly
lower than that of the corresponding normal tissue (20.16±125.98;
P<0.001 by a Wilcoxon rank test). We then defined 2-fold changes
as a reduced expression of RASA1 mRNA between the tumor and
corresponding normal tissue specimens (Fig. 1). The results revealed that 64.4%
(29/45) of breast cancer tissue expressed a low level of RASA1
compared to the paired normal specimens, indicating that the
downregulation of RASA1 expression occurs at the mRNA level. To
verify this finding, we used another set of tissue samples to
assess RASA1 protein expression and then determined whether the
levels of RASA1 expression were associated with clinicopathological
data and prognosis of the patients.
We assessed the expression of RASA1 protein in 373
cases of IDC and the paired normal cases using
immunohistochemistry. Our results showed that 226 (60.59%) out of
373 breast IDC tissues had a reduced expression of the RasGAP
protein, whereas 147 (39.41%) of the 373 breast IDC tissues
expressed relatively high levels of RasGAP protein compared to the
paired normal breast tissue (Fig.
2). The results regarding the association between RasGAP
protein expression and clinicopathological data are shown in
Table I. Specifically, the
stratification analysis showed that there was no statistically
significant difference between RASA1 expression and patient age,
tumor size, progesterone receptor (PR) expression, or HER-2
expression. By contrast, the reduced RASA1 expression was
significantly associated with lymph-node metastasis (P=0.002),
advanced TNM stage (P=0.017), ER expression (P=0.002), Ki-67
expression (P=0.009) and higher histopathological grades
(P<0.001). A reduction in RASA1 expression was also frequently
observed in triple-negative breast cancers (P=0.041).
Association between RASA1 expression and
survival of breast IDC patients
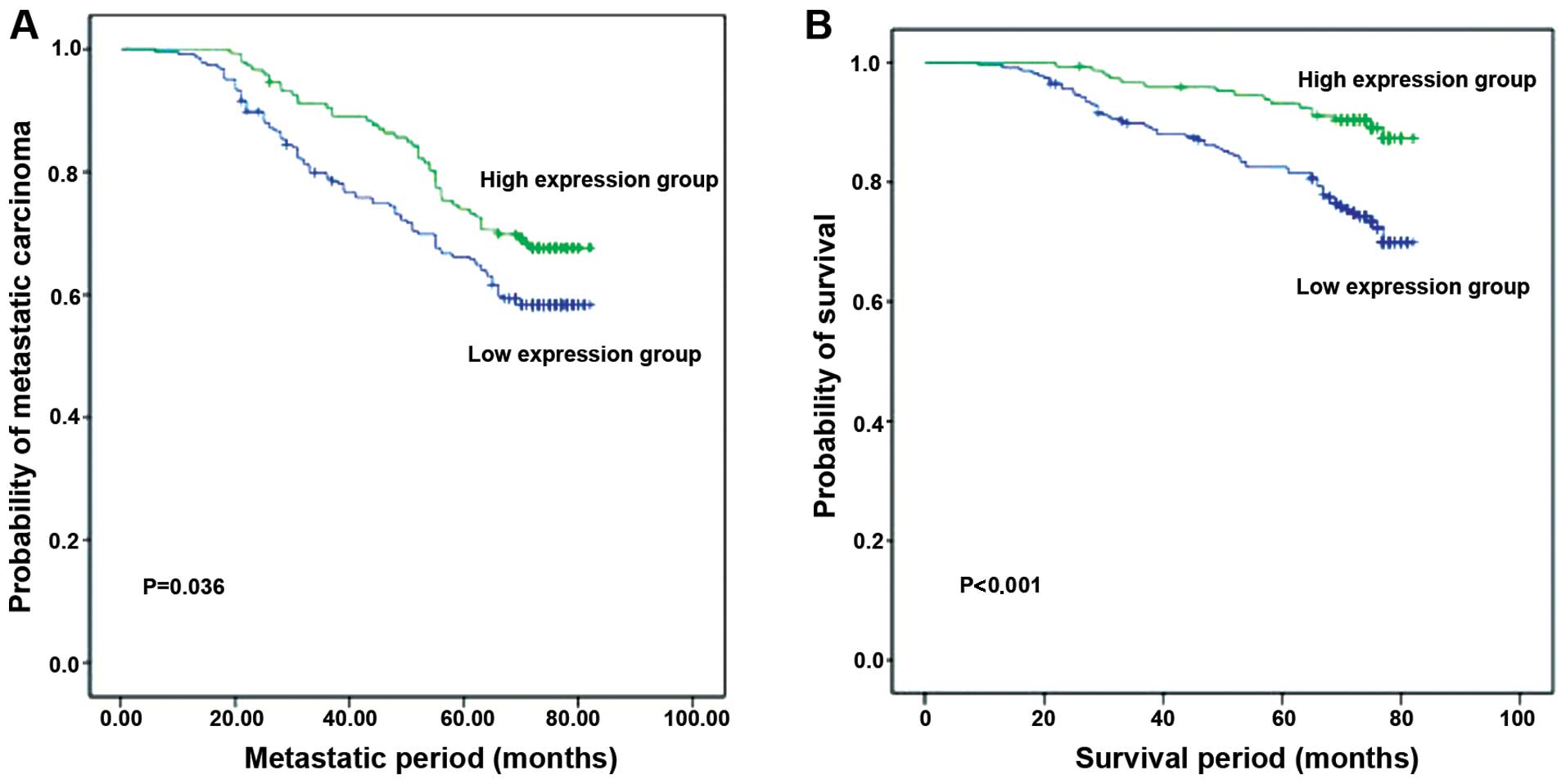
We determined whether levels of RASA1 expression
were associated with survival of breast IDC patients (Fig. 3 and Table II). All of the 373 patients were
followed up after surgery with a median of 74 months of the
follow-up (ranging between 3 and 82 months). Our results showed
that a reduced RASA1 expression was significantly associated with
poorer DFS compared to when tumors show high levels of RASA1
expression (P=0.036). Similarly, patients with low levels of RASA1
expression in tumors had a shorter OS rate than patients with
tumors showing a high RASA1 expression (P<0.001).
 | Table IIUnivariate and multivariate analyses
for overall survival in 373 invasive ductal carcinoma patients. |
Table II
Univariate and multivariate analyses
for overall survival in 373 invasive ductal carcinoma patients.
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
|
Characteristics | RR | 95% CI | P-value | RR | 95% CI | P-value |
|---|
| Age (≤50/>50
years) | 1.281 | 0.82–2.01 | 0.281 | - | - | - |
| Tumor size
(T1/T2+T3) | 1.604 | 0.97–2.65 | 0.036 | - | - | - |
| Lymph node
metastasis (−/+) | 4.625 | 2.54–8.41 | <0.001 | 2.540 | 1.23–5.22 | 0.011 |
| TNM stage
(I–II/III) | 4.140 | 2.61–6.57 | <0.001 | 2.365 | 1.36–4.10 | 0.002 |
| ER (−/+) | 0.335 | 0.21–0.54 | <0.001 | - | - | 0.188 |
| PR (−/+) | 0.376 | 0.24–0.59 | <0.001 | - | - | 0.727 |
| Her-2 (−/+) | 1.664 | 1.06–2.61 | 0.027 | 3.796 | 2.07–6.96 | <0.001 |
| Ki-67 | 0.639 | 0.41–1.00 | 0.051 | - | | |
| Histopathological
grade (1+2/3) | 0.795 | 0.49–1.29 | 0.353 | - | - | - |
| Molecular subtype
(triple-negative/others) | 1.470 | 1.25–1.73 | <0.001 | 1.806 | 1.46–2.24 | <0.001 |
| RASA1 expression
(high/low) | 0.362 | 0.21–0.63 | <0.001 | 0.485 | 0.28–0.85 | 0.012 |
Univariate analysis revealed that lymph-node
metastasis (P<0.001), advanced TNM stage (P<0.001),
expression of ER (P<0.001) and Her-2 (P=0.027), triple-negative
breast cancer (P<0.001) and reduced RASA1 expression
(P<0.001), all contributed to poor patient survival. In
addition, multivariate analysis showed that lymph node metastasis
(P=0.011), advanced TNM stage (P=0.002), Her-2 expression
(P<0.001), triple-negative breast cancer (P<0.001) and
reduced RASA1 expression (P=0.012) were all independent factors in
predicting the survival of these breast cancer patients.
Discussion
Breast cancer is the most frequently diagnosed
malignancy in women in western countries and is the most prevalent
malignant disease in almost all countries (17). Hampering the development of
effective treatment methods, breast cancer is a complex and
heterogeneous disease (18). On a
molecular level, breast cancer can be classified using ER, PR and
Her-2 expression as either ER- and PR-positive or as negative for
all three markers (i.e., triple-negative breast cancer). Hormone
blocking therapy can effectively control and treat ER- and
PR-positive tumors, however, triple-negative breast cancers prove a
greater challenge (19,20). Thus, development of novel strategies
for the treatment and prognostic prediction of breast cancer,
especially triple-negative breast cancer, may aid in the reduction
of breast cancer mortality. For example, identification of genes in
breast cancer may be used as novel adjuvant diagnostic and
prognostic biomarkers to improve treatment decisions in combination
with these parameters (21).
In this study, we have demonstrated that the
expression of RASA1 is significantly reduced in breast IDC tissues
compared to the corresponding normal tissues. Functionally, the
RASA1 gene encodes a p120-RasGTPase-activating protein,
which switches the active GTP-bound Ras to the inactive GDP-bound
form. In RASA1 knockout mice, the embryos exhibit abnormal
vascular development (22,23). Subsequent studies have demonstrated
that RASA1 is an important member of the RAS pathway and a putative
tumor suppressor (24). On a
molecular level, the p120-RasGAP protein contains two SH2, SH3, PH
(pleckstrin homology) and CaLB/C2 (calcium-dependent
phospholipid-binding domain) domains at the N-terminal, which
function to regulate cell proliferation, migration and apoptosis
depending on their downstream binding partners (25–29).
The p120-RasGAP protein interacts with other proteins such as Ras,
Akt, Aurora and RhoGAP to regulate cell functions in angiogenesis
and cancer development. For example, RASA1 acts as a suppressor of
RAS function by enhancing the weak intrinsic GTPase activity of RAS
proteins (24), resulting in an
increase in the inactive GDP-bound form of RAS, thereby leading to
aberrant intracellular signaling such as inhibition of the RAF-
MEKERK and PI3K-Akt pathways (30–32).
In addition, RASA1 is suggested to play a role in the regulation of
angiogenesis and tumor progression (33). Our current ex vivo
experiments have demonstrated that the reduced RASA1 expression was
significantly associated with lymph node metastasis, advanced TNM
stage, ER expression, Ki-67 expression, higher histopathological
grades and triple-negative breast cancers. These findings indicate
that a reduced RASA1 expression is likely to contribute to breast
cancer progression. A previous study also showed that a reduction
in RASA1 expression induced human colorectal cancer cell growth
in vitro and stimulated tumorigenesis in nude mice (24). Another recent study has shown that
the selective suppression of RASA1 promoted the unrestrained
activation of Ras signaling in wild-type RAS-expressed
hepatocellular carcinoma cells, resulting in increased tumor cell
proliferation and resistance to apoptosis (34). Our current study did not assess the
cause of the reduced RASA1 expression in breast cancer. However, a
previous study identified alterations in chromosome copy number in
association with different subtypes, chromosomes that contain
different candidate oncogenes and tumor suppressors including RASA1
(9). This suggests that our
findings of a reduced RASA1 expression in breast cancer may occur
at the genomic level. Future studies are needed to establish
whether the loss of heterozygosity or RASA1 mutations contribute to
a reduced RASA1 expression in breast cancer.
Our current study has shown that a low RASA1
expression frequently occurs in triple-negative breast cancers,
which is a novel finding. A previous study demonstrated that
triple-negative breast cancer was associated with a higher
expression of Ki-67 (21). Future
studies are needed to confirm our current finding and to
investigate whether targeting RASA1 is a novel therapeutic strategy
for controlling triple-negative breast cancer. In addition, this
study revealed that a reduced RASA1 expression is associated with
poor OS and DFS of breast cancer patients, which has not been
previously reported. We have also found that RASA1 expression,
together with tumor lymph node metastasis, TNM stage, Her-2
expression, and triple-negative breast cancer were all independent
factors in predicting the survival of breast cancer patients.
Acknowledgements
This study was supported in part by grants from The
Scientific Research Project of Heilongjiang Provincial Health
Department (2013155) and from The Scientific Research Foundation of
The Third Affiliated Hospital, Harbin Medical University
(JJ2011-14).
References
|
1
|
Al-Mubarak M, Sacher AG, Ocana A,
Vera-Badillo F, Seruga B and Amir E: Fulvestrant for advanced
breast cancer: a meta-analysis. Cancer Treat Rev. 39:753–758. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lianidou ES, Mavroudis D and Georgoulias
V: Clinical challenges in the molecular characterization of
circulating tumour cells in breast cancer. Br J Cancer.
108:2426–2432. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lavigne E, Holowaty EJ, Pan SY, et al:
Breast cancer detection and survival among women with cosmetic
breast implants: systematic review and meta-analysis of
observational studies. BMJ. 346:Apr 29–2013.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stites EC and Ravichandran KS: A systems
perspective of ras signaling in cancer. Clin Cancer Res.
15:1510–1513. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Saxena N, Lahiri SS, Hambarde S and
Tripathi RP: RAS: target for cancer therapy. Cancer Invest.
26:948–955. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu YF, Deth RC and Devys D: SH3
domain-dependent association of huntingtin with epidermal growth
factor receptor signaling complexes. J Biol Chem. 272:8121–8124.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vigil D, Cherfils J, Rossman KL and Der
CJ: Ras superfamily GEFs and GAPs: validated and tractable targets
for cancer therapy? Nat Rev Cancer. 10:842–857. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pamonsinlapatham P, Hadj-Slimane R,
Lepelletier Y, et al: p120-Ras GTPase activating protein (RasGAP):
a multi-interacting protein in downstream signaling. Biochimie.
91:320–328. 2009. View Article : Google Scholar
|
|
9
|
Hu X, Stern HM, Ge L, et al: Genetic
alterations and oncogenic pathways associated with breast cancer
subtypes. Mol Cancer Res. 7:511–522. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fang JY and Richardson BC: The MAPK
signalling pathways and colorectal cancer. Lancet Oncol. 6:322–327.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheang MC, Chia SK, Voduc D, et al: Ki67
index, HER2 status, and prognosis of patients with luminal B breast
cancer. J Natl Cancer Inst. 101:736–750. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Aksoy S, Dizdar O, Harputluoglu H and
Altundag K: Demographic, clinical, and pathological characteristics
of Turkish triple-negative breast cancer patients: single center
experience. Ann Oncol. 18:1904–1906. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bauer KR, Brown M, Cress RD, Parise CA and
Caggiano V: Descriptive analysis of estrogen receptor
(ER)-negative, progesterone receptor (PR)-negative, and
HER2-negative invasive breast cancer, the so-called triple-negative
phenotype: a population-based study from the California cancer
Registry. Cancer. 109:1721–1728. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xue C, Wang X, Peng R, et al:
Distribution, clinicopathologic features and survival of breast
cancer subtypes in Southern China. Cancer Sci. 103:1679–1687. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Cheng S, Zhang G, et al: Low
expression of RECK indicates a shorter survival for patients with
invasive breast cancer. Cancer Sci. 103:1084–1089. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu T, Zhang X, Shang M, et al:
Dysregulated expression of Slug, vimentin, and E-cadherin
correlates with poor clinical outcome in patients with basal-like
breast cancer. J Surg Oncol. 107:188–194. 2013. View Article : Google Scholar
|
|
17
|
Adam Maciejczyk A: New prognostic factors
in breast cancer. Adv Clin Exp Med. 22:5–15. 2013.PubMed/NCBI
|
|
18
|
do Carmo França-Botelho A, Ferreira MC,
França JL, França EL and Honorio-França AC: Breastfeeding and its
relationship with reduction of breast cancer: a review. Asian Pac J
Cancer Prev. 13:5327–5332. 2012. View Article : Google Scholar
|
|
19
|
Liedtke C, Mazouni C, Hess KR, et al:
Response to neoadjuvant therapy and long-term survival in patients
with triple-negative breast cancer. J Clin Oncol. 26:1275–1281.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Carey LA, Dees EC, Sawyer L, et al: The
triple negative paradox: primary tumor chemosensitivity of breast
cancer subtypes. Clin Cancer Res. 13:2329–2334. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cadoo KA, McArdle O, O’Shea AM, Power CP
and Hennessy BT: Management of unusual histological types of breast
cancer. Oncologist. 17:1135–1145. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Henkemeyer M, Rossi DJ, Holmyard DP, et
al: Vascular system defects and neuronal apoptosis in mice lacking
ras GTPase-activating protein. Nature. 377:695–701. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lapinski PE, Bauler TJ, Brown EJ, Hughes
ED, Saunders TL and King PD: Generation of mice with a conditional
allele of the p120 Ras GTPase-activating protein. Genesis.
45:762–767. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun D, Yu F, Ma Y, et al: MicroRNA-31
activates the RAS pathway and functions as an oncogenic MicroRNA in
human colorectal cancer by repressing RAS p21 GTPase activating
protein 1 (RASA1). J Biol Chem. 288:9508–9518. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Trahey M, Wong G, Halenbeck R, et al:
Molecular cloning of two types of GAP complementary DNA from human
placenta. Science. 242:1697–1700. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moran MF, Koch CA, Anderson D, et al: Src
homology region 2 domains direct protein-protein interactions in
signal transduction. Proc Natl Acad Sci USA. 87:8622–8626. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Musacchio A, Gibson T, Rice P, Thompson J
and Saraste M: The PH domain: a common piece in the structural
patchwork of signalling proteins. Trends Biochem Sci. 18:343–348.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Clark JD, Lin LL, Kriz RW, et al: A novel
arachidonic acid-selective cytosolic PLA2 contains a
Ca(2+)-dependent translocation domain with homology to PKC and GAP.
Cell. 65:1043–1051. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gawler DJ, Zhang LJ, Reedijk M, Tung PS
and Moran MF: CaLB: a 43 amino acid calcium-dependent
membrane/phospholipid binding domain in p120 Ras GTPase-activating
protein. Oncogene. 10:817–825. 1995.PubMed/NCBI
|
|
30
|
Glennon TM, Villa J and Warshel A: How
does GAP catalyze the GTPase reaction of Ras? A computer simulation
study. Biochemistry. 39:9641–9651. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shurki A and Warshel A: Why does the Ras
switch ‘break’ by oncogenic mutations? Proteins. 55:1–10. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Denhardt DT: Signal-transducing protein
phosphorylation cascades mediated by Ras/Rho proteins in the
mammalian cell: the potential for multiplex signalling. Biochem J.
318:729–747. 1996.PubMed/NCBI
|
|
33
|
Cailliau K, Browaeys-Poly E and Vilain JP:
RasGAP is involved in signal transduction triggered by FGF1 in
Xenopus oocytes expressing FGFR1. FEBS Lett. 496:161–165. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Calvisi DF, Ladu S, Conner EA, et al:
Inactivation of Ras GTPase-activating proteins promotes
unrestrained activity of wild-type Ras in human liver cancer. J
Hepatol. 54:311–319. 2011. View Article : Google Scholar :
|