Introduction
Human aspartyl-(asparaginyl)-β-hydroxylase (HAAH) is
a type 2 transmembrane protein and an α-ketoglutarate-dependent
dioxygenase that can stereospecifically catalyze the
post-translational hydroxylation reaction of β-carbon atoms of
aspartic acid and asparagine residues present in epidermal growth
factor-like domains of certain specific proteins (1). The HAAH gene was first reported
by Korioth et al (2). It is
positioned on q12 of chromosome No. 8, and the full length of mRNA
is 2449 bp. The alternative splicing of the HAAH gene
generates at least five homeotic subfamily mRNA transcripts a, b,
c, d and e, which encode the proteins AAH, Junctate,
Humbug, Junctin and Junctin-1, respectively
(3–5). AAH is a unique protein with enzymatic
activity, and Junctate, Junctin and Junctin-1, which are mainly
expressed on the endoplasmic reticulum membrane and the sarcoplasm
membrane of the myocardium diaphragm, are calcium-binding proteins
connected with the instantaneous release of calcium ions, and with
protein modification in the process of protein synthesis.
Humbug is a truncated isoform of HAAH that lacks the
catalytic domain. Humbug also participates in calcium
regulation and increases intracellular calcium levels by promoting
calcium release from intracellular stores. In recent years, using
immunohistochemistry, gene expression analysis, in vivo
binding and enzyme-linked immunosorbent assay, it has been
confirmed that HAAH/humbug is overexpressed in a broad range
of malignant tumor tissues and cell lines and it is not expressed
or has a low expression in normal, non-cancer inflammatory lesion
and paracancerous tissues. Over 1,000 cases of malignant tumor
specimens including cholangiocarcinoma; hepatocellular carcinoma
(HCC); prostate, breast, colon, lung, gastric cancer and ovarian
cancer; pancreatic carcinoma; glioblastoma; oligodendroglioma and
primitive neuroectodermal tumors were previously detected,
exhibiting a positive rate of >95% or even >99% (6–10).
Previous study results showed that humbug has been
associated with a variety of human cancers, and although
humbug lacks enzymatic activity, it is expressed at levels
comparable with that of HAAH in various cancer cell lines.
Humbug was also useful in the evaluation of HCC prediction
and prognosis (5,11). In addition, as a transmembrane
protein, HAAH/humbug can be dissociated from the surface of
cancer cells and is readily shed into the blood of cancer patients.
Free HAAH in the blood serum of 857 cases of lung, breast,
prostatic and colon cancer and non-tumor serum samples, was
quantitatively determined. The sensitivity of the test was 94.7%
(n=857), specificity was 94.3% (n=211) and overall accuracy was
94.6%. The average values of free HAAH in the blood serum were 0
ng/ml (non-cancer), 25.7 ng/ml (prostate cancer), 34.6 ng/ml (lung
cancer), 17.6 ng/ml (breast cancer) and 30.0 ng/ml (colon cancer)
(12). The present study showed
that HAAH is a special carcinoembryonic antigen and an important
marker in the formation of malignant cells. HAAH is overexpressed
in a variety of malignant tumor tissues and transformed cell lines.
As a new tumor marker, HAAH has a broad spectrum and specificity
(13). In the preliminary study, we
conducted a qualitative analysis on the distribution and expression
of HAAH in seven types of tumor cell lines and 20 tumor tissues
including liver cancer, cholangiocarcinoma, kidney and breast
cancer. The present study indicates that HAAH mRNA was
overexpressed in all seven (100%) tumor cell lines examined,
although the levels of the gene expression varied and the positive
rate was 90.4% (94/104) in 20 tumor tissues (14).
RT-PCR (mRNA detection) and northern blotting
(protein detection) are frequently used to detect the expression
level of HAAH mRNA and protein. However, these methods have certain
drawbacks, such as a long operation time, are prone to
false-positive or -negative results, deliver low diagnostic yields
and are only used for qualitative detection. The molecular beacon
(MB) method, however, is simple, rapid and highly specific, and has
been used for the quantitative detection of various types of
tumor-marker genes.
In the present study, we examined the expression
levels of the HAAH/humbug gene and proteins in tissue
specimens of patients with HCC and adjacent HCC-free liver tissues,
to determine whether the HAAH/humbug gene and protein
overexpression levels are directly associated with HCC
clinicopathologically and to evaluate the overexpression of the
HAAH gene and protein as a diagnostic and prognostic
biomarker in HCC.
Materials and methods
Patients and tissue specimens
A total of 120 surgically resected tissue specimens
from patients with HCC and 40 adjacent HCC-free liver tissue were
obtained from The First Affiliated Hospital of Medical College of
Xi’an Jiaotong University and the Xijing Hospital of Digestive
Diseases, Fourth Military Medical University, respectively. None of
these patients underwent radiotherapy or chemotherapy prior or
subsequent to surgical resection. The study included 84 males and
36 females, with an age range of 35–70 years (median 55.1±8.2
years). The samples were obtained in accordance with the Ethics
requirements for conducting medical experiments. These samples were
tested and diagnosed at the Department of Pathology, Xijing
Hospital.
Main reagents
TRIzol, reverse transcriptase M-MLV, RNase
inhibitor, quantitative PCR kit (SuperMix-UDG) were purchased from
Invitrogen Life Technologies, Carlsbad, CA, USA, and anti-HAAH mAb
was prepared in our laboratory (15).
RT-PCR primer and HAAH/humbug MB
Three pairs of RT-PCR primers, FP1/RP1, FP2/FRP2 and
FP/RP, were designed using Oligo 6.0 analysis software. Primer FP1
contained the restriction endonuclease sites XhoI, Kozak
sequence and His-tag sequence. Primer RP1 contained stop code TAA
and the SpeI restriction endonuclease sites. Products from
the primer pair FP1/RP1 were applied to amplify HAAH/humbug
regions that were used for the construction of recombinant plasmid
carrying the humbug gene fragment. Primer pair FP2/RP2 was
utilized for the detection of the HAAH/humbug gene in the MB
quantitative RT-PCR (RT-qPCR) reaction. Primer pair FP/RP was used
to amplify the housekeeping gene β-actin. Humbug
molecular beacon probes (humbug MB, a sequence of 27
nucleotides, which can recognize a target molecule) and β-actin MB
(a sequence of 30 nucleotides, which can recognize a target
molecule) were designed using Beacon Designer 7.0 software. Their
target specificity was verified using BioEdit ver. 7.0.9.0
software. The primers and MB probe above were synthesized by Sangon
Co. (Shanghai, China) (Table
I).
 | Table ISequences of molecular beacon and
primers. |
Table I
Sequences of molecular beacon and
primers.
| Name of
primer/MB | Sequence
(5′-3′) | Gene position
(bp) | Amplified fragments
(bp) |
|---|
| Humbug
FP1 |
CCATCGATGCCACCATGTGGGCCATCAT
CATCATCATCATATGGTGATTGCATTGCTGGGC | 175–195 | 766 |
| Humbug
FP2 |
GGACTAGTTTATGTTTCTGGTGGTAC | 922–939 | |
| Humbug
FP2 |
ATGGTGATTGCATTGCTGGGC | 186–206 | 267 |
| Humbug
RP2 |
CAGAATATCGAAGATGAAGCA | 432–452 | |
| Humbug
MB |
FAM-AGCAGGTTCCTGTGGAGGCAGAACCC
CAGAATACCTGCT-DABCYL | 413–440 | |
| β-actin FP |
TGGCATTGCTGACAGGATG | 227–245 | 114 |
| β-actin RP |
AAGTACTCCGTGTGGATCG | 331–350 | |
| β-actin MB |
FAM-CCGCATTGGCTCCCAGCACCATGAAG
ATCAAGATCAATGCGG-TAMRA | 266–294 | |
Specimen collection and RNA
extraction
Fresh surgically resected HCC tissue samples and
paracancerous HCC-free liver tissue specimens were collected. These
fresh samples were rapidly transferred and stored in liquid
nitrogen prior to RNA purification. Total RNA from snap-frozen
tissue was extracted using TRIzol reagent (Invitrogen Life
Technologies) according to the manufacturer’s instructions. The
integrity of the total RNA was validated using UV spectrophotometry
and 20 g/l agarose gel electrophoresis. Only high-quality RNA with
intact 18S and 28S RNA was used for subsequent experiments, and
then stored at −80°C until use.
Preparation of HAAH/humbug cDNA
RNA samples were reverse-transcribed using the M-MLV
First Strand cDNA synthesis kit (Roche, Basel, Switzerland).
Oligo-dT18 was used for primers of the first chain synthesis. cDNA
was reverse-transcribed from 2 μg of the extracted RNA in a 20 μl
reaction system containing 2 μl of 5X reaction buffer, 1 μl of
M-MLV reverse transcriptase, 1 μl of RNAsin, 2 μl of 0.1 M DTT, 1
μl of Oligo-dT 18 primers (50 μg/ml), 6 μl of dNTP mix (10 mM) and
DEPC-treated water. The samples were heated to 70°C for 5 min,
followed by incubation at 42°C for 1 h. Reactions were terminated
by heating at 95°C for 5 min. β-actin mRNA levels measured in
parallel reactions were used to calculate the relative abundance of
each mRNA transcript.
Construction of the recombinant plasmid
carrying an HAAH/humbug-encoding gene
cDNA obtained by reverse transcription was used as a
template for the polymerase chain reaction (PCR). The primer pair
humbug FP1/RP1 was used for the amplification reaction. The
total volume of 50 μl contained 5 μl of cDNA template, 5 μl 10X PCR
Buffer (Mg2+ Plus), 1 μl primer humbug FP1 and
RP1 each, 4 μl 10 mM dNTPs, 0.5 μl rTaq DNA polymerase (Dalian
Takara Co.), and 33.5 μl ultrapure water. The initial denaturation
step at 95°C for 5 min was followed by 30 cycles at 94°C for 1 min,
60°C for 1 min, 72°C for 1 min, followed by 72°C for 10 min
(Mastercycler Gradient, Eppendorf Co., Hamburg, Germany). PCR
products were then separated on a 1.2% agarose gel containing 5
μl/100 ml Goldview using DL2000 ladder (Shanghai Shinegene Co.,
Shanghai, China) as a size marker. The PCR products were cloned
into pMD18-T vector using a TA cloning kit (Promega Co., Madison,
WI, USA) and sequenced. The recombinant plasmid was identified
using the restriction enzyme digestion method. The concentration
and purity of the recombinant plasmid DNA were estimated by UV. DNA
was diluted in 0.9% NaCl at a concentration of 1 mg/ml. The ratio
of OD260/280 nm ranged 1.8–2.0. The plasmid DNA was stored at −20°C
in 0.5-ml aliquots.
Sensitivity experiment and linear
standard curve
According to the manufacturer’s instructions,
AccuBlue™ dsDNA quantification kits were used to have a precise
quantitative detection on the pMD-Humbug plasmid DNA that
had the HAAH/humbug genes (uQuant Microplate Scanning
spectrophotometer; Bio-Tek, Winooski, VT, USA). The precise
quantitative plasmid DNA was used as the template. Sterile
deionized water was used to achieve gradient dilution, and the
concentration gradients were 105, 104,
103, 102, 101 and 100
(copies/μl). The control was pMD empty plasmid diluted with sterile
deionized water. A 25 μl reaction system was utilized with 2.5 μl
of 10X PCR buffer, 0.25 μl of dNTP (10 mmol/l), 6 μl of
MgCl2 (25 mmol/l), 0.5 μl for Primer humbug FP2
and RP2 (10 μmol/l), respectively, 0.5 μl of Taq DNA polymerase (5
U/μl, Dalian Takara Co.), 1 μl of pMD-Humbug plasmid
template and 0.25 μl of Humbug MB (10 μmol/l). The reaction
conditions were 95°C for 5 min, 95°C for 45 sec, 55°C for 60 sec,
72°C for 60 sec for 45 cycles. Experiments were performed in
triplicate and repeated twice. Following the reaction, the
fluorescent quantitative PCR analysis software was used to set the
baseline and cycle threshold (Ct) and quantitative amplification
curves were obtained. The Ct value was used as the abscissa and
ordinate, with the fluorescence intensity value as ordinate, the
linear standard curve cycle threshold (Ct) as abscissa, and the
logarithms of DNA concentration as the ordinate.
Immunohistochemical staining
Cryostat sections of frozen tissue were cut at 6 μm
(freezing microtome; Leica CM1950, Nussloch, Germany), placed on
glass slides, air dried and fixed in a 1:1 solution of
alcohol:acetone. The slides were then washed gently with PBS 2–3
times and were submerged in methanol containing 10%
H2O2 to quench endogenous peroxidase
activities in tissues. Following washing and air drying, the
sections were stored at −20°C until use. Immediately before
commencement of immunostaining, the sections were washed in PBS
buffer for 5 min and treated with 5% skimmed milk to block
non-specific bindings on the sections. The slides were rinsed with
PBS buffer to remove the blocking solution and air dried again. The
anti-HAAH MAbs (also recognizing humbug) were then added and
incubated in a moist chamber at 37°C for 45 min. The slides were
rinsed again in PBS buffer. HRP-conjugated anti-mouse antibodies
diluted in PBS were added, and incubated in a moist chamber for 45
min at 37°C. The slides were washed gently with PBS 3–5 times. TMB
blotting substrate was added and developed for 5 min. The sections
were gently counterstained with hematoxylin, and then viewed under
an inverted microscope.
MB quantitative RT-PCR
As mentioned above, in the cDNA preparation, total
RNA was extracted from tumor tissues, Oligo(dT)18 was regarded as
the reverse transcription primer, and single-stranded cDNA was
synthesized under M-MLV reverse transcriptase. The PCR reaction was
conducted with this template, and the primer pair FP2 (upstream
primer)/RP2 (downstream primer) and FP (upstream primer)/RT
(downstream primer) was used to amplify the HAAH/humbug and
β-actin gene fragments, respectively. The MB RT-qPCR reaction was
performed on Chromo4™ four-color real-time detector (MJ Research
Inc., Waltham, MA, USA) to detect the expression levels of the
HAAH/humbug genes and housekeeping gene β-actin in
the specimens. The PCR reaction conditions were the same as those
for the cDNA preparation. β-actin RNA levels measured in parallel
reactions were used to calculate the relative abundance of each
mRNA transcript. After the reaction ended, 5 μl of PCR product were
used to conduct agarose gel electrophoresis at a concentration of
2.0%. Images were captured on a Gel Doc XR gel-imaging system, and
gray was analyzed to the result of electrophoresis.
Results
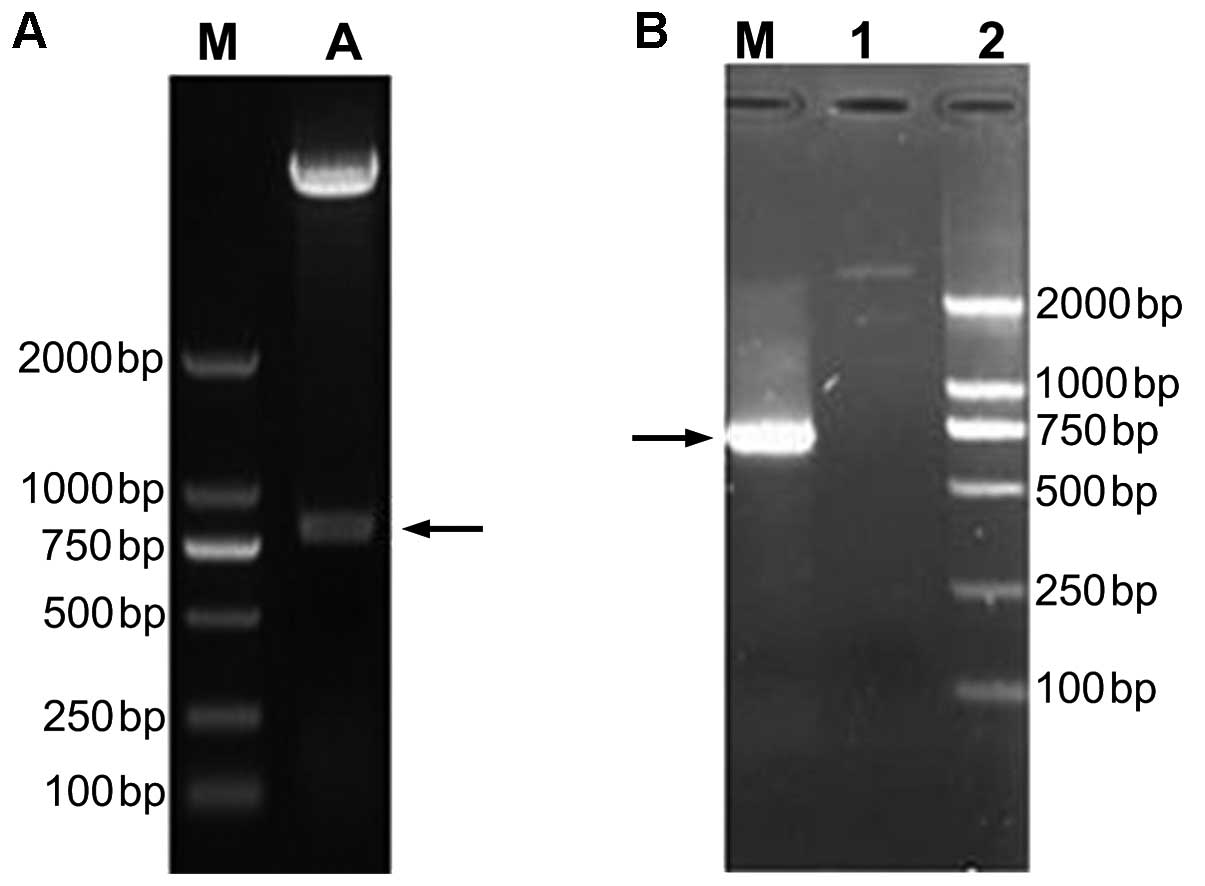
Amplification of the HAAH/humbug gene and
restriction analysis of the recombinant plasmid pMD-humbug
The complete humbug-encoding gene segment
with a molecular weight of ~760 bp was amplified using RNA
extracted from HCC tumor tissue as a template, with FP1/RP1 being
used as a RT-qPCR primer (Fig. 1B).
However, no DNA fragments of the same size were amplified using RNA
isolated from the adjacent tumor-free liver tissues as a template
and the same primer pair. The PCR product was gel-purified and then
ligated into a pMD18-T vector. The ligation product was incubated
overnight at 16°C and used for transformation of the E. coli
strain DH5α. The transformants were plated on LB agar plates
containing 100 μg/ml ampicillin and incubated at 37°C for 24 h.
Recombinant plasmid was extracted from DH5α using an EasyExtraction
kit (Biofuture Group Inc., Beijing, China). The recombinant plasmid
pMD-humbug was identified by XhoI/SpeI double
restriction enzyme digestion. Restriction digestion analyses
clearly showed that there was a specific digestion strip at ~760 bp
in agarose gel electrophoresis and the molecular weight of the
digested fragment was consistent with the expected size (Fig. 1A). The positive recombinant plasmid
was selected and sequenced (Genscript Co., Nanjing, China). The
sequencing result was compared with the HAAH/humbug sequence
of the NCBI Genbank database. It was highly homologous to the
HAAH/Humbug sequence of the database, except that No. 346
had a G to A mutation, which was the nonsense mutation (data not
shown). The positive recombinant plasmid carrying the
HAAH/humbug-encoding gene fragments was designated as
pMD-humbug.
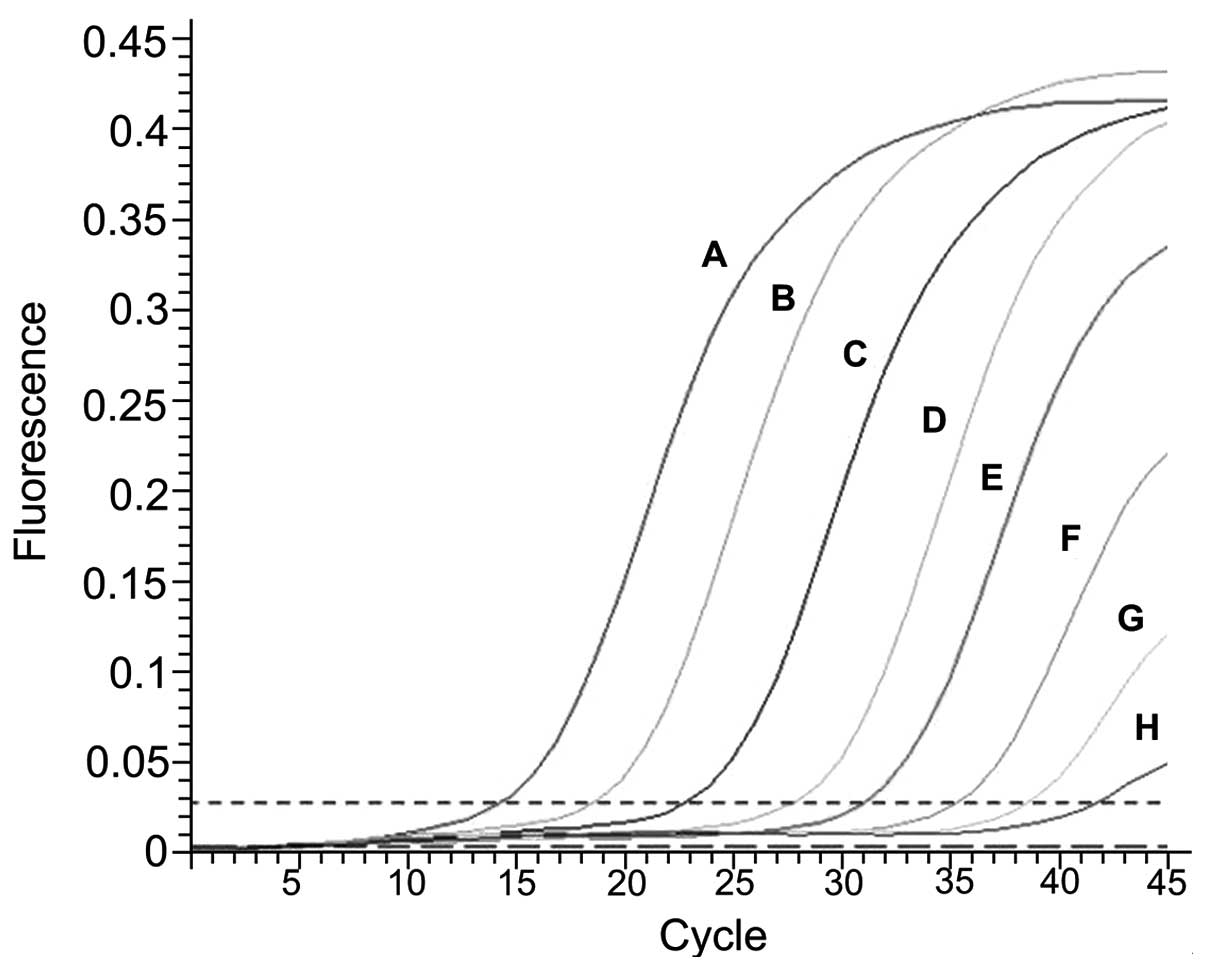
Sensitivity and specificity of the MB
RT-qPCR system
In the 1×100–1×106 copies/μl
range, with an increase in the number of cycles (Ct), the
fluorescence intensity of each sample increased at varying degrees.
The amplification curve was S-shaped. The values of Ct and the
fluorescence intensity were positively correlated with the initial
concentrations of the plasmid DNA templates. In the 25-μl reaction
system, the limit of detection was 1 copy/reaction system (Fig. 2 and Table II).
 | Table IICt value of MB real-time qRT-PCR
standard curve. |
Table II
Ct value of MB real-time qRT-PCR
standard curve.
| Log(C) | 106 | 105 | 104 | 103 | 102 | 101 | 100 |
|---|
| Run 1 | 14.0 | 20.1 | 21.4 | 26.9 | 29.8 | 33.9 | 41 |
| Run 2 | 14.6 | 17.8 | 25.1 | 28 | 30.7 | 37 | 39.3 |
| Run 3 | 16.4 | 19.1 | 22.5 | 29.1 | 32.5 | 34.1 | 36.7 |
| Mean ± SD | 15±1.3 | 19±1.2 | 23±1.9 | 28±1.1 | 31±1.3 | 35±1.7 | 39±2.2 |
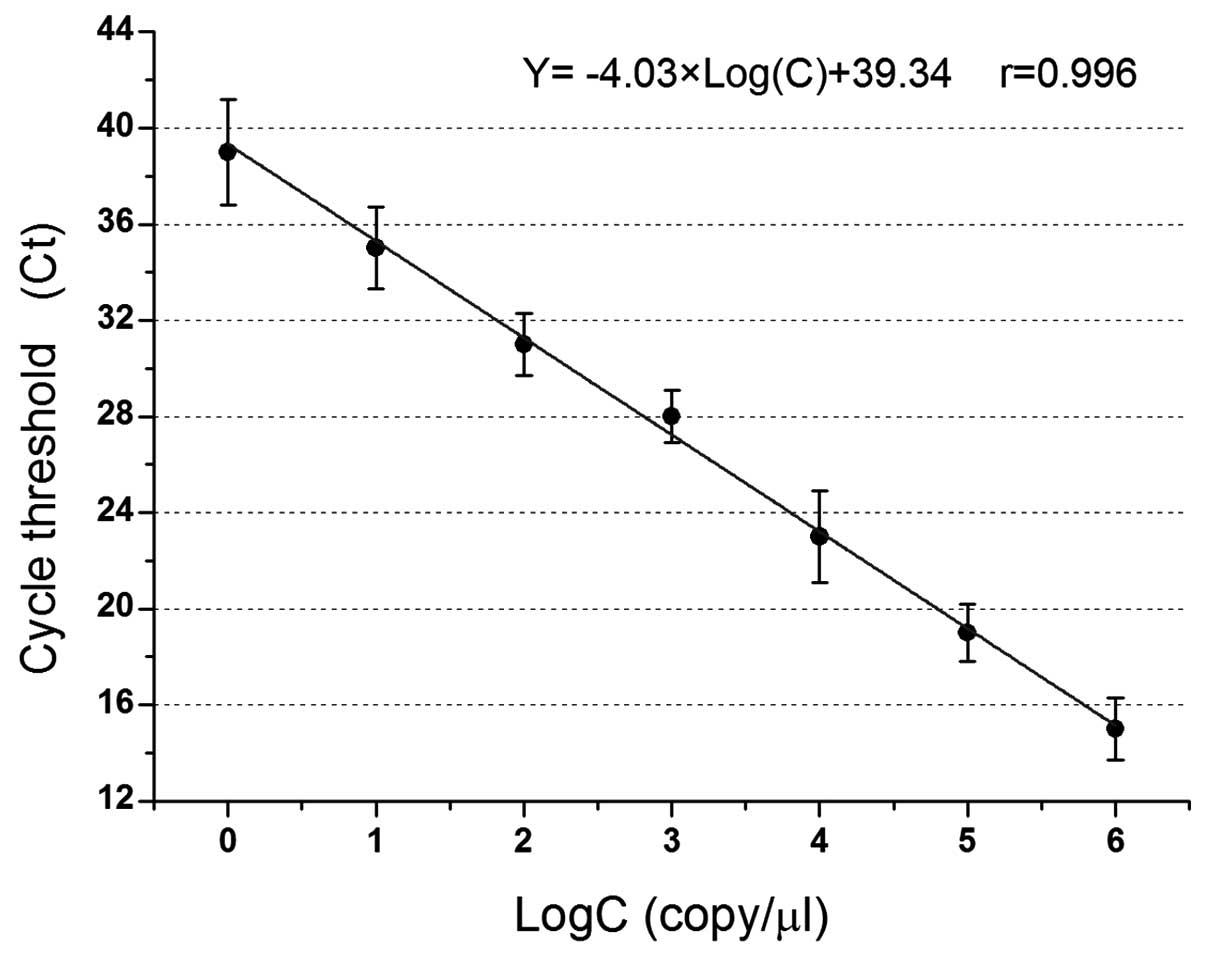
MB qPCR standard curve of pMD-humbug
plasmid
The canonical plotting of MB quantitative RT-PCR of
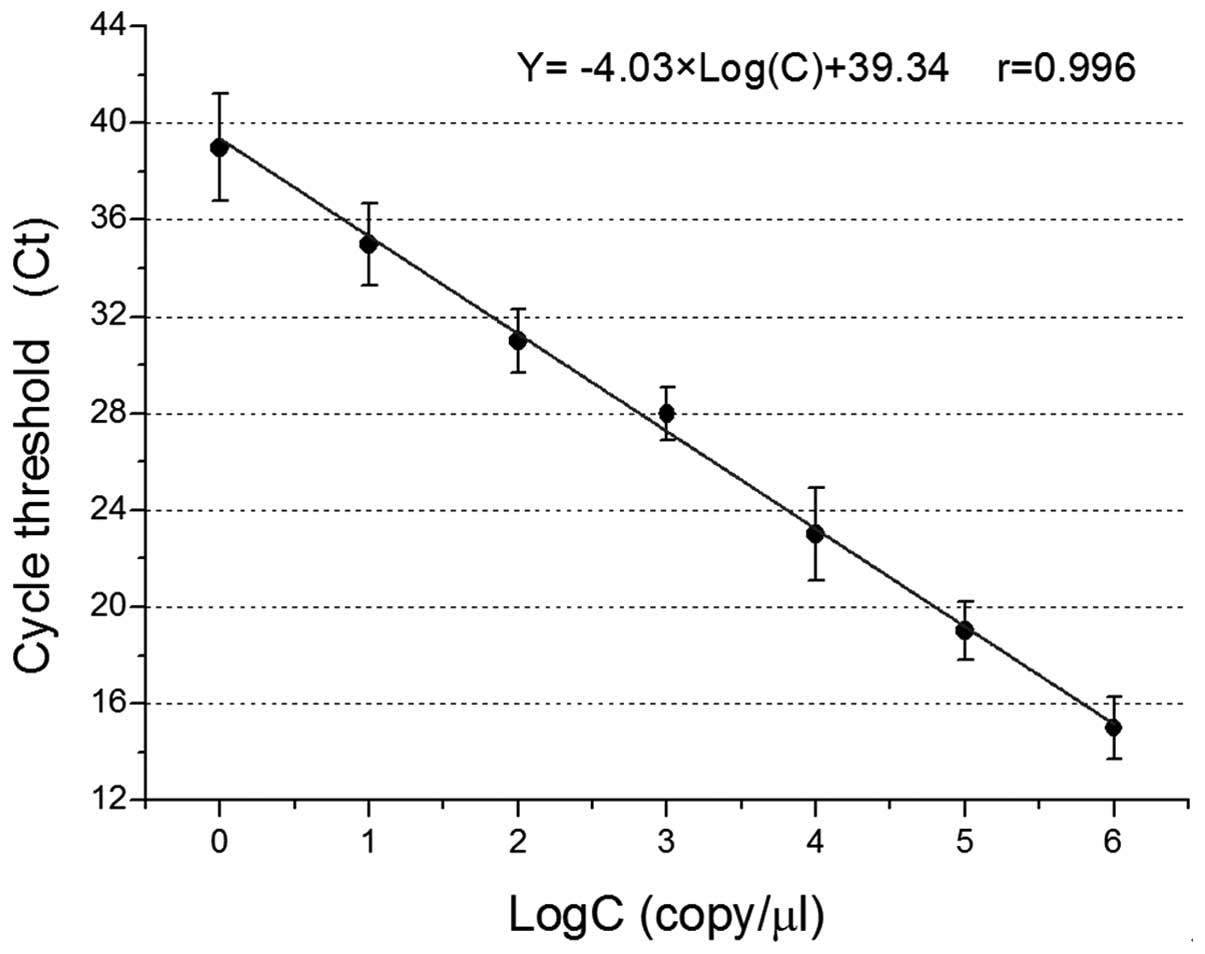
plasmid pMD-humbug is shown in Fig. 3. The logarithm of the standard copy
number was regarded as the x-coordinate, and the Ct value that
passed through when fluorescent signal intensity reached the preset
threshold was the y-coordinate. A straight standard curve was
obtained. In the range of 1×101–1×106
copies/μl, alterations in the number of Ct indicated a good linear
relationship with the concentration of the initial plasmid
pMD-humbug DNA, and the calibration curve equation obtained
was Y = −4.03xLog(C) + 39.34, r=0.996. The DNA samples of unknown
concentration were amplified, the Ct values were entered into the
calibration curve equation, and the logarithm of the copy number of
DNA samples was calculated. The copy number of DNA samples was
subsequently obtained through this standard curve.
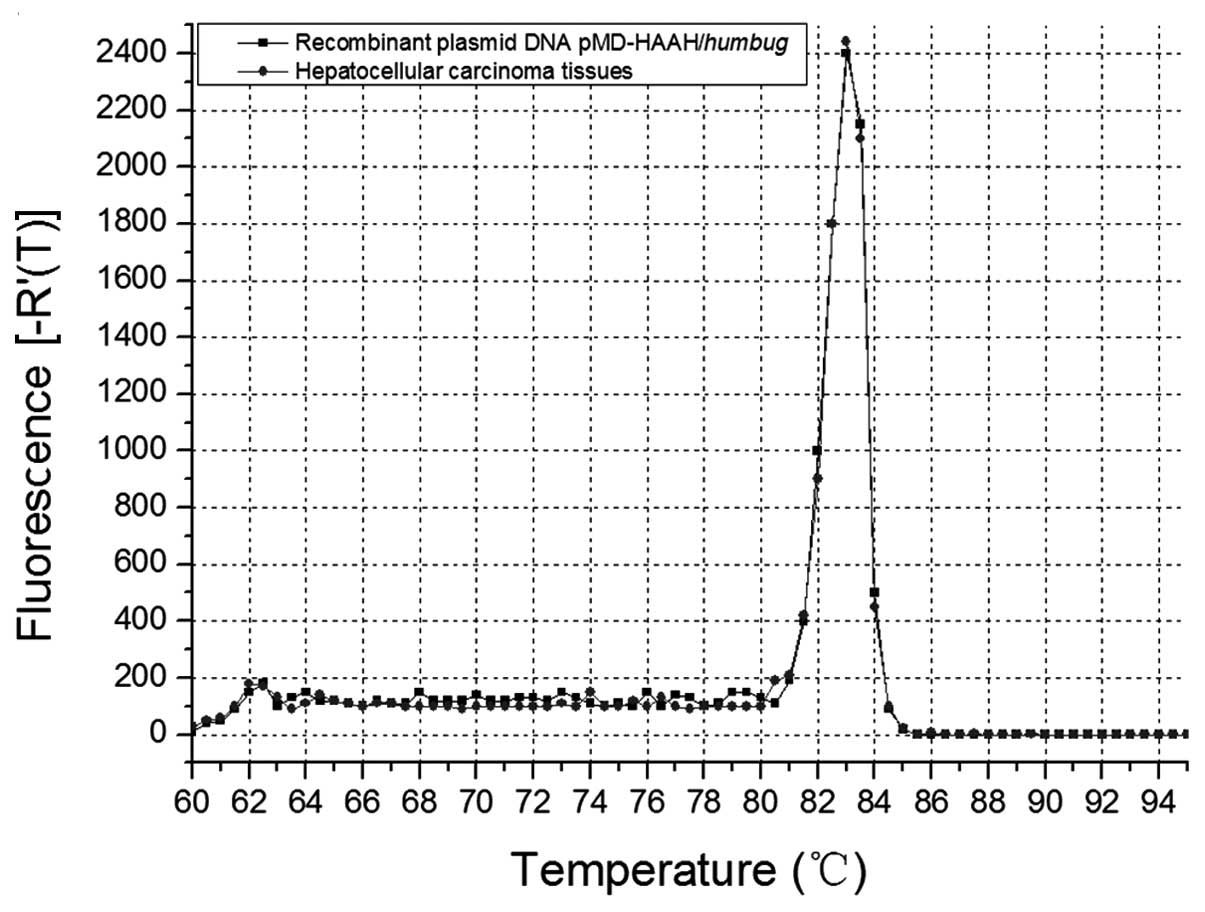
Analysis of melting curve
Melting curve (Tm) analysis allows for different PCR
amplification products and identification of primer dimers
according to the difference of melting points in order to
differentiate non-specific amplification. Therefore, the influence
of non-specific products can be decreased. The ideal melting curve
should be a single-peak curve. Two or more peaks indicates that
non-specific amplification products, such as primer dimers, are
produced in the reaction system. Therefore, Tm analysis was
constantly employed to determine PCR primers, PCR products and the
specificity of an amplification system. Fig. 4 shows the Tm peak of MB RT-qPCR
products of HCC carcinoma tissues highly coincides with that of the
products of recombinant plasmid pMD-humbug MB RT-qPCR
(positive control), which indicates that the two peaks are
identical, and that the MB and primer we designed are of a high
specificity.
Expression of the HAAH/humbug gene in HCC
tissues
Paired samples of HCC tissues and adjacent HCC-free
liver tissues were used to measure HAAH/humbug mRNA levels
by RT-qPCR. The β-actin mRNA levels measured in the same samples
were used to calculate relative HAAH/humbug mRNA abundance
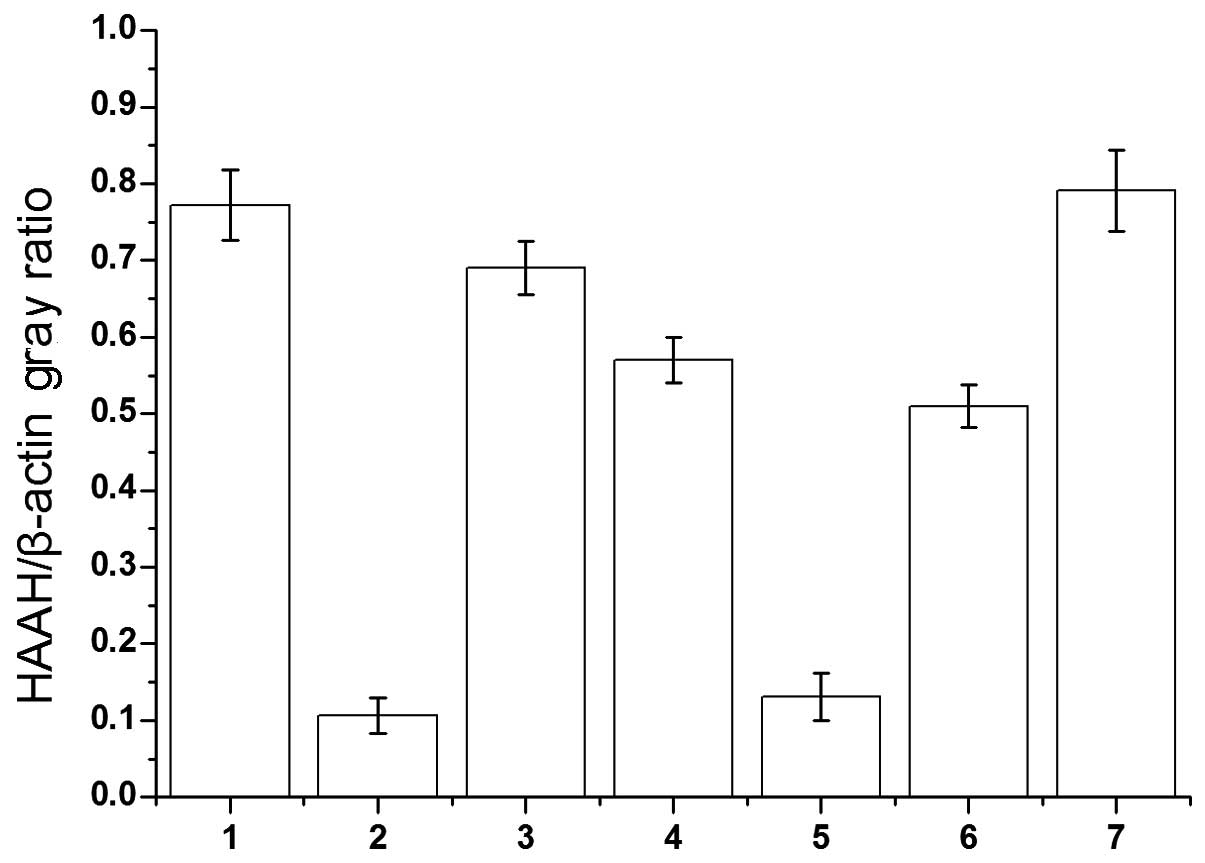
(Fig. 5). The intra-group gray
value of agarose gel electrophoresis comparisons were performed
using Quantity One software (Gel Doc EQ System; Bio-Rad Co.,
Hercules, CA, USA). MB qPCR studies demonstrated that the
expression levels of the HAAH/humbug gene in the HCC tissues
and adjacent non-tumor liver tissues were 0.789±0.287 and
0.121±0.098. The mean level of humbug mRNA was ~5- to 8-fold
higher in the cancer samples, compared to the adjacent cancer-free
tissues, respectively (Fig. 6).
Immunohistochemical staining analysis of
HAAH/humbug in HCC tissues
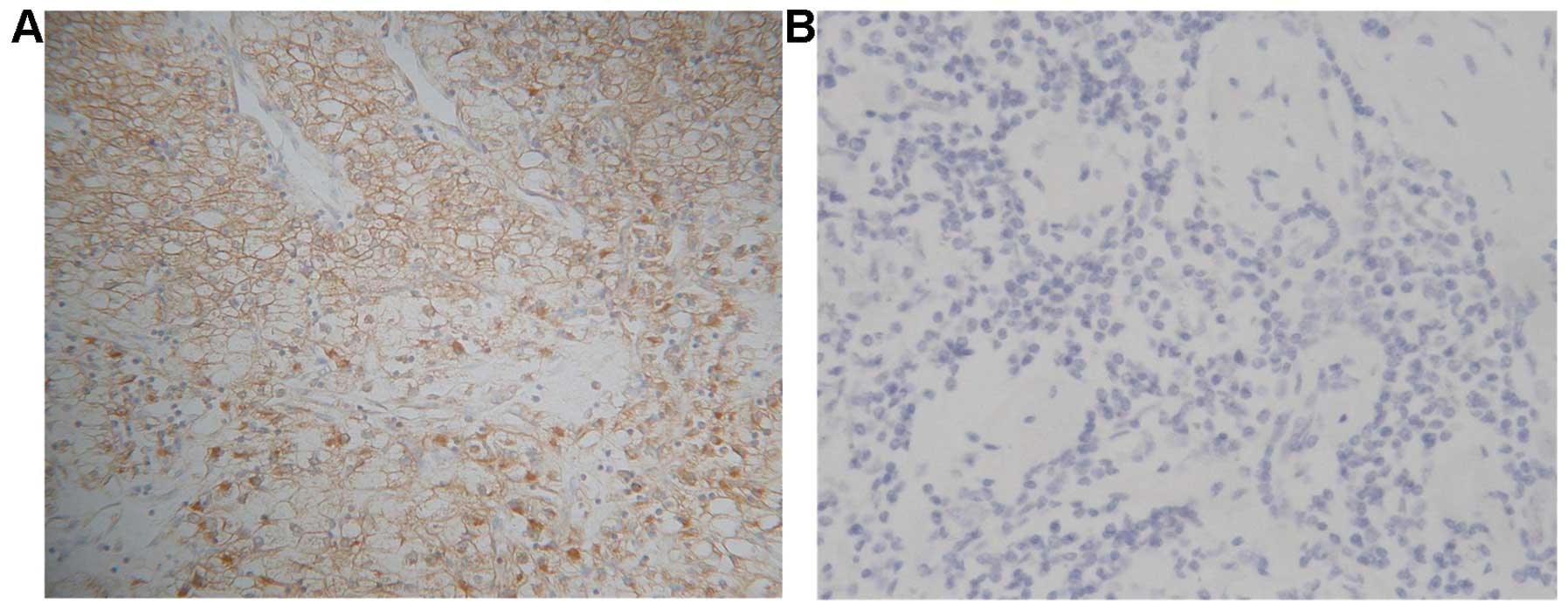
The results of immunohistochemical (IHC) staining
showed that 117 (97.5%) of the 120 frozen sections of patients with
HCCs exhibited HAAH/humbug-positive immunoreactivity,
whereas the 40 adjacent non-tumor liver tissues exhibited no
staining. The staining pattern for HAAH/humbug in the tumor
cells was primarily cytoplasmic with a distinct perinuclear
accentuation and plasmalemmal (Fig.
7).
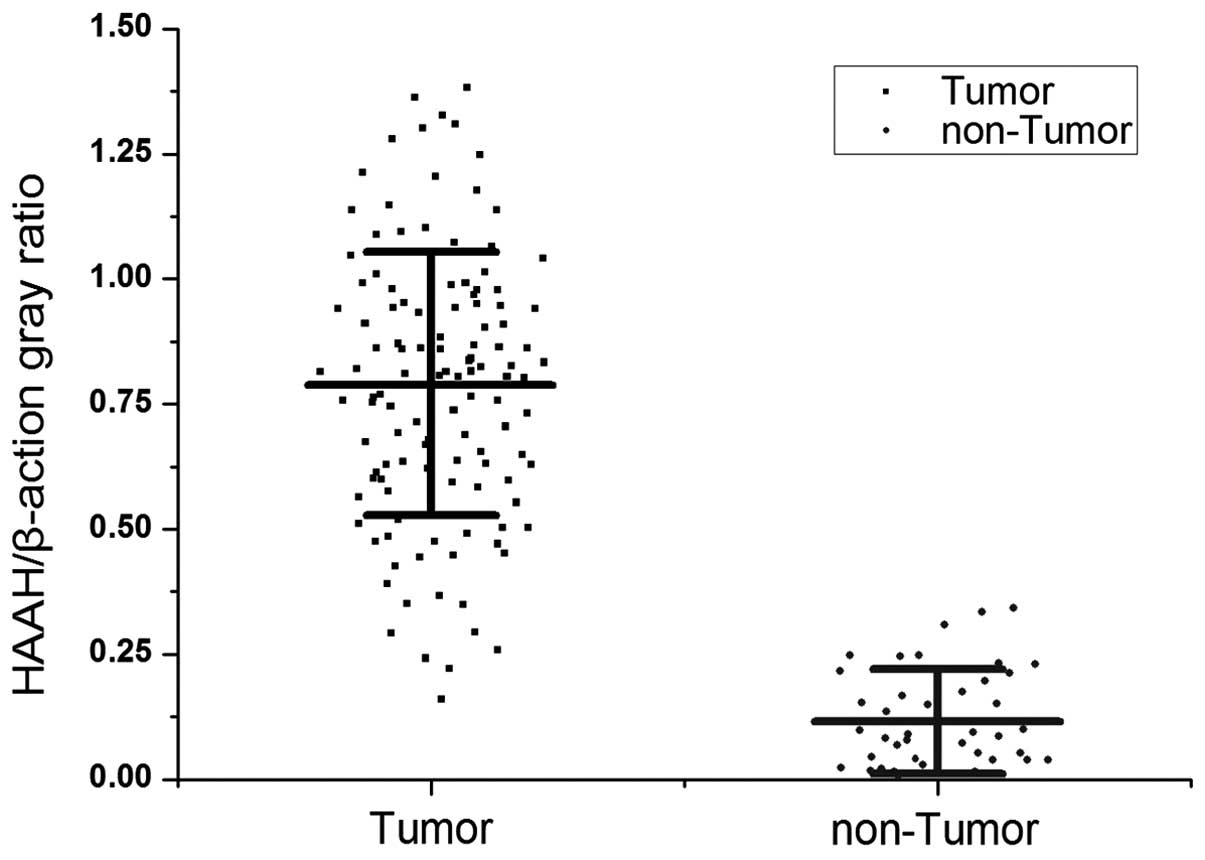
MB qPCR detection of HAAH/humbug
The MB RT-qPCR studies demonstrated higher levels of
HAAH/humbug mRNA in 113 (94.2%) of the 120 cases of the HCC
tissues relative to the adjacent cancer-free tissue. The ratio of
HAAH/β-actin abundance was used as the cut-off point (0.315).
Comparatively, only 1 of 40 cases were weakly positive in the
adjacent non-tumor liver tissues specimens and the false-positive
rate was 2.5%. While the copy number of the HAAH/humbug gene
was used as the cut-off point (77.35 μg), the positive rate was 114
(95%) of 120 in HCC tissues, and the false-positive rate was also
5% in the adjacent non-tumor liver tissues. Briefly, the
HAAH/humbug expression level was upregulated in almost all
the HCC tissue cells relative to normal liver cells, irrespective
of the cut-off point used (P<0.01; Fig. 8).
ROC curve analysis
ROC analysis was performed based on the expression
levels of the HAAH/humbug gene in 120 cases of HCC tissues
and 40 cases of adjacent non-tumor liver tissues using SPSS 19.0
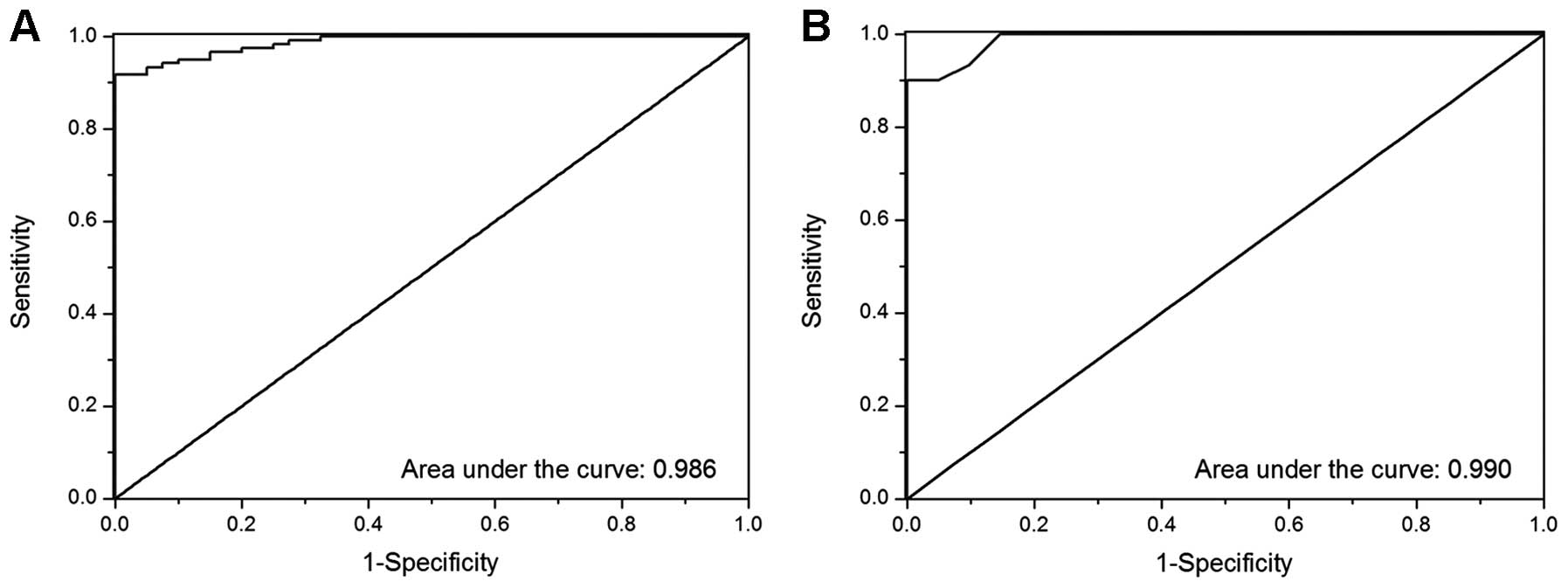
software (IBM SPSS Statistics Co., Armonk, NY, USA). Fig. 9A shows the result of ROC curve
analysis of the ratio of HAAH/β-actin abundance. Sensitivity was
90.1%, specificity 97.6% and ROC AUC was 0.986 with 0.315 copies/μl
as the cut-off point. Fig. 9B shows
the gene copy number identified by ROC curve analysis. Sensitivity
was 99.2%, specificity 96.7% and ROC AUC was 0.990 when the cut-off
point was 77.35 copies/μl. No statistically significant difference
was noted for the two factors. The results showed that the
overexpression of HAAH/humbug has a potential diagnostic and
prognostic value for HCC.
Discussion
HCC is a highly invasive neoplasm with multicentric
occurrence, and is also one of the five most prevalent lethal human
malignant cancer-types. At present, the mortality rate for HCC
ranks third worldwide. In recent years, the HCC incidence rate has
been on the increase. The incidence of HCC in North America and
Europe is <10 in 10 million individual, while its incidence in
Asia and Africa is 50–150 in 10 million individuals. China is one
of the regions most seriously affected by HCC (16). The annual incidence of 17.83%
accounts for >40% of the annual total number of patients with
HCC worldwide. HCC leads to a high mortality rate and is the third
leading cause of cancer-related death in China (24,25).
It has become the second largest cause of cancer-related mortality
in rural areas. Primary HCC is characterised by high invasiveness,
insidious onset, no obvious symptoms at the early stage and poor
prognosis. Therefore, the early diagnosis, prevention and timely
treatment of HCC are particularly important. Numerous approaches,
such as liver biopsy puncture, ultrasonography, helical computed
tomography, or magnetic resonance imaging, may be applied to detect
HCC. As the ‘golden standard’, α-fetoprotein (AFP) is the most
common tumor marker in the clinical diagnosis of HCC, but its low
sensitivity (70%), even lower specificity (64%) when utilized alone
in HCC diagnosis easily lead to misdiagnosis and missed diagnosis
(17). Therefore, identification of
new biomarkers is of great significance for the early diagnosis and
early prevention of HCC.
A series of reports (9,26) have
documented that the expression levels of HAAH/humbug in HCC
tissues is significantly higher than adjacent non-cancerous liver
tissues, irrespective of the mRNA or protein levels.
The overexpression of HAAH/humbug increases
motility, invasiveness, malignant transformation and the
infiltrative proliferation of HCC cells. It has been confirmed that
antisense constructs produced against the AUG start codon inhibited
gene expression and tumor cell motility and migration (18) and correspondingly, HAAH siRNA
exhibited significantly lower mean directional motility indices
relative to the control cells. Previous studies have shown that the
overexpression of HAAH in transfected normal cells was sufficient
to induce cell transformation and suppression of HAAH expression
(siRNA) (9,19) or neutralized activity (mAb) that
returns cancer cells to a normal phenotype (20,21).
The present results indicate that the overexpression of HAAH is
closely associated with poor prognosis, tumor recurrence and
patient survival (22).
The HAAH gene encodes the proteins AAH,
Junctate, Humbug, Junctin and Junctin-1. The N-terminus of
HAAH-related proteins has a role in calcium homeostasis, whereas
the C-terminal region, which is essential for catalytic activity,
is only present in HAAH. As one of the HAAH protein
post-translational splicing isoforms, humbug is a truncated
homolog of HAAH. It is locked in the domain with the catalytic
activity of the enzyme in the C-terminal fragment of the HAAH
protein. The molecular weight of the humbug gene is 2.9 kb,
encoding 50–55 kDa humbug protein. Although the
humbug protein lacks enzymatic activity, it is expressed at
levels comparable with that of HAAH in a variety of human cancer
types (18).
To assess the value of HAAH/humbug in the
diagnosis and prediction of HCC, we examined the expressed levels
of the HAAH/humbug gene using MB RT-qPCR and the expressed
levels of HAAH protein using immunohistochemical staining in 120
cases of HCC patients and 40 cases of adjacent HCC-free liver
tissues.
In the present study, HAAH/humbug expression
levels were detected by immunohistochemistry (protein level) and MB
RT-qPCR (mRNA level) in 120 tumor tissues of patients with HCC
specimens and 40 adjacent non-tumor tissues. The results of
immunohistochemistry showed that, in 120 cases of HCC tissue
samples, 117 (97.5%) exhibited positive immunoreactivity for or
against HAAH/humbug monoclonal antibodies (mAb), whereas in
all 40 cases of adjacent non-cancerous tissues, immunoreactivity
was negative. In the experiment of MB RT-qPCR, of 120 HCCs, the
HAAH/humbug gene expression was strongly positive in 113
tumor tissues (94.2%), whereas HAAH/humbug mRNA expression
was weakly positive in 2 of 40 adjacent non-tumor liver tissues
(5%), using the specific value of HAAH/β-actin abundance as the
cut-off point (0.315). When the copy number of the
HAAH/humbug gene (77.35 μg) was used as cut-off point, the
positive rates were 114 (95%) of 120 in HCC tissues and upregulated
in the majority of HCC tissues relative to the adjacent cancer-free
tissue, irrespective of the cut-off point used (P>0.05).
The following conclusions can be drawn from the
aforementioned tests: the HAAH/humbug at the mRNA and
protein levels in Chinese HCC patients was markedly higher than
that of non-cancerous liver tissues. This finding is consistent
with the results of a previous study (20).
ROC analysis was performed based on the expression
levels of the HAAH/humbug gene in 120 cases of HCC tissues
and 40 cases of adjacent non-tumor liver tissues using SPSS 19.0
software. The results of the ROC curve analysis concerning the
ratio of HAAH/β-actin abundance showed that the sensitivity was
90.1%, specificity 97.6% and ROC AUC 0.986 when the cut-off point
was 0.315 (Fig. 9A). The results of
the ROC curve analysis concerning gene copy number (copies/μl)
demonstrated that the sensitivity was 99.2%, specificity 96.7% and
ROC AUC was 0.990 when the cut-off point was 77.35 copies/μl
(Fig. 9B). No significant
difference was identified between these factors. The present study
results confirm that HAAH/humbug is extremely valuable for
the diagnosis of HCC. Thus, HAAH/humbug serves as a
potential diagnostic biomarker for HCC.
In order to improve the sensitivity and specificity
of HAAH/humbug mRNA detection on tumor cells, qPCR and
hot-start PCR were utilized in previous experiments, although the
results obtained were unsatisfactory (data not shown). In this
study, the method of MB RT-qPCR was applied to quantitatively
measure HAAH/humbug expression levels.
The MB probe technique is a new nucleic acid
quantitative detection technique developed by Tyagi and Kramer
(23), based on fluorescence energy
transfer technology and the linear probe technique. MBs are
hairpin-shaped molecules, consisting of a stem loop structure with
single-stranded DNA molecules. The MB stem is formed by the
attachment, to both termini of the loop, of two short (5–7
nucleotide residues) oligonucleotides that are complementary to
each other. The 18–30 bp region of the MB loop in the middle is
complementary to the target DNA or RNA and they do not base pair
with one another. At the 5′-end of the stem, a fluorescent dye is
covalently attached. The quencher dye is covalently attached to the
3′-end of the stem. When the MB is in closed loop shape, the
quencher resides in proximity to the fluorophore, which results in
quenching the fluorescent emission of the latter (23). When the MB is bound to a target
nucleic, the acid sequence causes the separation of the stem and
thus of fluorescence quenching.
The stem of the classical MB comprises complementary
sequences unrelated to target sequences. In this study, base pairs
at each terminus of the MB stem are complementary to each other,
and to the target DNA. After the MB was complemented with the
target genes, the space interval between fluorescent dye and the
quencher was increased. This was useful for fluorescent emission
and to intensify the luminous intensity, therefore, it improves the
sensibility of detection. Since there are specific PCR primers and
MB probes in this reaction system, the false positivity of
detection can be effectively reduced. In this reaction system, the
lowest detectable limit (LOD) of HAAH/humbug mRNA was 1
copy/reaction in a 25-μl reaction system and the linear detection
range was 101–106 copies/reaction.
The strengths of this approach are that it is
unnecessary to dilute or concentrate specimens in the experiment,
and second that the procedures of detection were completed in a
closed tube. The reaction was completed in a closed tube and
real-time monitoring of the amplification occurs. Therefore,
further handling after the PCR reaction, and the drawback of easy
pollution in the conventional PCR was avoided. After MB RT-qPCR was
terminated, agarose gel electrophoresis to inspect the products of
PCR amplification was not undertaken. When the Ct value of the
unknown sample is achieved, the initial sample copy number can be
calculated from the standard curve.
Based on the findings of this study, the new MB
RT-qPCR method with advantages of high specificity, speed and
simplicity can be widely applied for HAAH/humbug mRNA
detection in different tumor tissue samples, including early-stage
tissue samples.
Acknowledgements
The authors are grateful for the support offered by
the Research Project of Shaanxi Provincial Key Laboratory of
Biotechnology (11JS085 and 14JS088) and the Development Project of
Science and Technology Research of Shaanxi Province
(2011K12-61).
References
|
1
|
Lavaissiere L, Jia S, Nishiyama M, et al:
Overexpression of human aspartyl(asparaginyl)beta-hydroxylase in
hepatocellular carcinoma and cholangiocarcinoma. J Clin Invest.
98:1313–1323. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Korioth F, Gieffers C and Frey J: Cloning
and characterization of the human gene encoding aspartyl
beta-hydroxylase. Gene. 150:395–399. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jones LR, Zhang L, Sanborn K, Jorgensen AO
and Kelley J: Purification, primary structure, and immunological
characterization of the 26-kDa calsequestrin binding protein
(junctin) from cardiac junctional sarcoplasmic reticulum. J Biol
Chem. 270:30787–30796. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Treves S, Feriotto G, Moccagatta L,
Gambari R and Zorzato F: Molecular cloning, expression, functional
characterization, chromosomal localization, and gene structure of
junctate, a novel integral calcium binding protein of
sarco(endo)plasmic reticulum membrane. J Biol Chem.
275:39555–39568. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee JH: Overexpression of humbug promotes
malignant progression in human gastric cancer cells. Oncol Rep.
19:795–800. 2008.PubMed/NCBI
|
|
6
|
Dinchuk JE, Henderson NL, Burn TC, et al:
Aspartyl beta-hydroxylase (Asph) and an evolutionarily conserved
isoform of Asph missing the catalytic domain share exons with
junctin. J Biol Chem. 275:39543–39554. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ince N, de la Monte SM and Wands JR:
Overexpression of human aspartyl (asparaginyl) beta-hydroxylase is
associated with malignant transformation. Cancer Res. 60:1261–1266.
2000.PubMed/NCBI
|
|
8
|
Maeda T, Taguchi K, Aishima S, et al:
Clinicopathological correlates of aspartyl (asparaginyl)
beta-hydroxylase overexpression in cholangiocarcinoma. Cancer
Detect Prev. 28:313–318. 2004. View Article : Google Scholar
|
|
9
|
De la Monte SM, Tamaki S, Cantarini MC, et
al: Aspartyl-(asparaginyl)-beta-hydroxylase regulates
hepatocellular carcinoma invasiveness. J Hepatol. 44:971–983. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Luu M, Sabo E, de la Monte SM, et al:
Prognostic value of aspartyl (asparaginyl)-beta-hydroxylase/humbug
expression in non-small cell lung carcinoma. Hum Pathol.
40:639–644. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J, de la Monte SM, Sabo E, et al:
Prognostic value of humbug gene overexpression in stage II colon
cancer. Hum Pathol. 38:17–25. 2007. View Article : Google Scholar
|
|
12
|
Moshiri M, Lebowitz MS and Roberts SF:
Cancer biomarker, haah (human aspartyl (asparaginyl)
beta-hydroxylase), a companion diagnostic strategy. In: Proceedings
of the Fifth Oncology Biomarker Conference; Zurich. pp. 2332012
|
|
13
|
Silbermann E, Moskal P, Bowling N, Tong M
and de la Monte SM: Role of aspartyl-(asparaginyl)-beta-hydroxylase
mediated notch signaling in cerebellar development and function.
Behav Brain Funct. 6:682010. View Article : Google Scholar
|
|
14
|
Yang H, Song K, Xue T, et al: The
distribution and expression profiles of human Aspartyl/Asparaginyl
β-hydroxylase in tumor cell lines and human tissues. Oncol Rep.
24:1257–1264. 2010.PubMed/NCBI
|
|
15
|
Xue T, Xue XP, Huang QS, Wei L and Sun K:
Monoclonal antibodies against human aspartyl (asparaginyl)
beta-hydroxylase developed by DNA immunization. Hybridoma.
28:251–257. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xian ZH, Zhang SH, Cong WM, Yan HX, Wang K
and Wu MC: Expression of aspartyl beta-hydroxylase and its
clinicopathological significance in hepatocellular carcinoma. Mod
Pathol. 19:280–286. 2006. View Article : Google Scholar
|
|
17
|
Lander ES, Linton LM, Birren B, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sepe PS, Lahousse SA, Gemelli B, et al:
Role of the aspartyl-asparaginyl-beta-hydroxylase gene in
neuroblastoma cell motility. Lab Invest. 82:881–891. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Davila JA, Morgan RO, Shaib Y, McGlynn KA
and El Serag HB: Hepatitis C infection and the increasing incidence
of hepatocellular carcinoma: a population-based study.
Gastroenterology. 127:1372–1380. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ho SP, Scully MS, Krauthauser CM, et al:
Antisense oligonucleotides selectively regulate aspartyl
beta-hydroxylase and its truncated protein isoform in vitro but
distribute poorly into A549 tumors in vivo. J Pharmacol Exp Ther.
302:795–803. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fuller S, Stewart S, Lebowitz M, et al:
Immunogenicity of a lambda phage-based anti-cancer vaccine
targeting HAAH. J Immunother Cancer. 1:P2102013. View Article : Google Scholar
|
|
22
|
Wang K, Liu J, Yan ZL, et al:
Overexpression of aspartyl-(asparaginyl)-beta-hydroxylase in
hepatocellular carcinoma is associated with worse surgical outcome.
Hepatology. 52:164–173. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tyagi S and Kramer FR: Molecular beacons:
probes that fluoresce upon hybridization. Nat Biotechnol.
14:303–308. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang Z, Zhou M and Wang L: Study on the
geographic distribution of liver cancer mortality and HBsAg carrier
rate in China. Disease Surveillance. 22:242–245. 2007.(In
Chinese).
|
|
25
|
Fan X, Zhao H and Zhang L: A 1:2 matched
case-control study on risk factors of hepatocellular carcinoma in
North Shaanxi. J Fourth Military Medical University. 23:891–895.
2002.(In Chinese).
|
|
26
|
Cantarini MC, De la Monte SM, Pang M, Tong
M, D’Errico A, Trevisani F and Wands JR: Aspartyl-asparagyl beta
hydroxylase over-expression in human hepatoma is linked to
activation of insulin-like growth factor and notch signaling
mechanisms. Hepatology. 44:446–457. 2006. View Article : Google Scholar : PubMed/NCBI
|