Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an
aggressive leukemia derived from malignant transformation of T cell
progenitors and is more common in males than in females. T-ALL
affects mainly older children and adolescents and represents 10–15%
of pediatric and 25% of young adult ALL cases (1). Hyperdiploidy (>46 chromosomes) is
found in 30% of childhood and 10% of adulthood ALL cases. Notably,
high hyperdiploidy (51–65 chromosomes) has been connected with high
survival rates and excellent outcome (2,3), while
low hyperdiploidy (47–50 chromosomes) has been associated with
worse prognosis (4). The most
commonly gained chromosomes in ALL are #4, #6, #10, #14, #17, #18,
#21 and X (5). Trisomy 4 is rarely
observed as a sole cytogenetic abnormality in T-ALL (6). However, the mechanism for chromosomal
gains in ALL and their role in leukemogenesis are still ambiguous
(7,8). In hyperdiploid karyotypes, the
t(9;22)(q34;q11), 11q23 (MLL gene) rearrangements,
t(12;21)(p13;q22), t(1;19)(q23;p13) and t(8;14)(q24;q32) are the
most common structural cytogenetic abnormalities in ALL. However,
in T-ALL, involvement of the T cell receptor (TCR) gene in
14q11 in rearrangements such as t(1;14)(p31;q11), t(10;14)(q24;q11)
or t(8;14)(q24;q11) are frequently observed; also del(6)(q15) and
del(1)(p32) have been described (3,9–11).
Still, cryptic structural chromosomal abnormalities
were and are a challenge in the cytogenetics of T-ALL. For example,
as the cryptic t(5;14)(q35;q32) is known to be present in ~20% of
childhood and in 13% of adult T-ALL cases, this aberration is
currently routinely tested by molecular (cyto)genetics, addressing
the breakpoint on the TLX3 (HOX11L2) gene in 5q35 and
to the promoter of the BCL11B gene in 14q32 (12). In addition, recent reports on newly
detected cryptic chromosomal rearrangements such as the
MLLT10 gene (previously AF10, in 10p13), and
MLL (in11q23) or PICALM (in 11q14) highlight the
necessity to further study clinical cases as detailed as possible
(13,14). The goal of these studies must be, on
the one hand, to provide the most accurate diagnosis to each
individual patient and, on the other hand, to achieve insights into
the biology and pathogenesis of T-ALL.
In the present study, an adolescent T-precursor cell
ALL case with an MLLT10 and IL3 gene rearrangement
together with trisomy 4 in complex four-way translocation is
characterized in detail retrospectively using molecular
cytogenetics and molecular genetics. This leukemia subtype would
currently be classified as early T-cell precursor ALL (15–17).
Case report
Clinical description
A 16-year-old male presented in 1998 for diagnostics
due to fever and unclear symptoms of malaise. Immunophenotypic
analysis of bone marrow cells revealed the following results:
HLA-DR+, TdT+, cyCD3+, CD5 weak,
CD7+, CD8+, CD10+,
CD13+, CD33+ and CD34+. This
supported a diagnosis of early T-ALL; at present, it would be
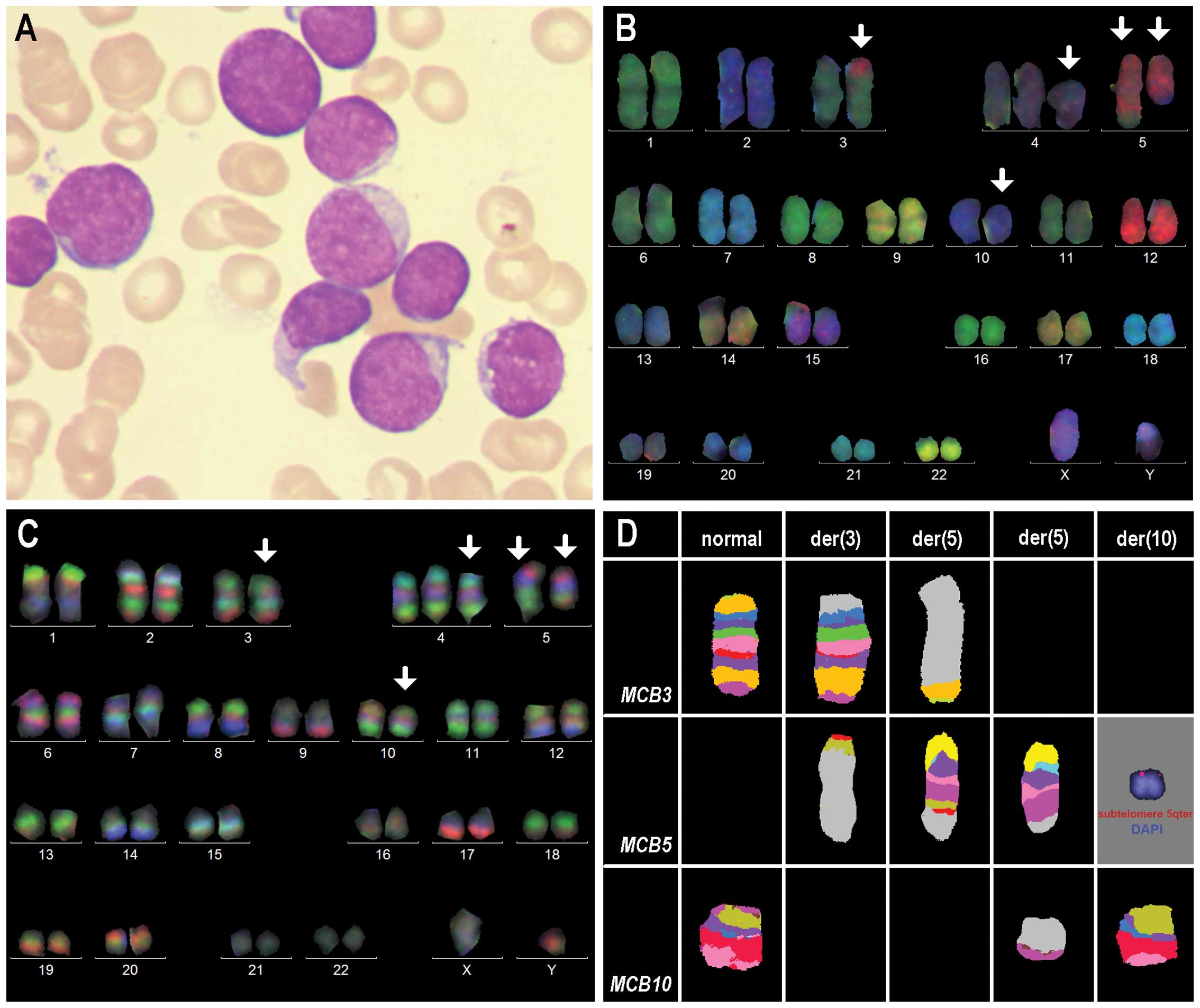
classified as early T-cell precursor ALL (Fig. 1A).
The patient was treated according to the ALL-BFM 95
protocol; the continuation therapy was completed 24 months after
the initial diagnosis. Three months later an isolated bone marrow
relapse with acute thrombocytopenia was diagnosed, and treatment
according to the ALL-REZ-BFM protocol was initiated. One month
later the patient died due to an Aspergillus sepsis and
still with 100% blasts in the bone marrow.
Tests conducted at diagnosis
Banding cytogenetic analysis was performed on an
unstimulated bone marrow aspirate according to standard procedures.
A total of 20 metaphases were available for cytogenetic evaluation
and analyzed on a banding level of 300 bands per haploid karyotype
(22). GTG-banding revealed a
normal male karyotype in our laboratory, and also a second
cytogenetic analysis on 25 metaphases performed 4 months after the
initial diagnosis in another laboratory confirmed this test result.
Molecular diagnostic PCR tests for gene fusions BCR/ABL,
MLL/AF4 and TEL/AML1 were negative (data not
shown).
Test conducted in retrospect
Molecular cytogenetics
Fluorescence in situ hybridization (FISH) was
performed according to standard procedures and/or according to the
manufacturer’s instructions.
The following homemade probes and probe sets were
used: i) 24-color-FISH using all human whole chromosome painting
(WCP) probes (19); ii)
FISH-banding probe sets as follows: genome-wide multitude
multicolor banding (mMCB) and chromosome-specific high resolution
array-proven multicolor banding (aMCB) (20–22);
iii) DNA from bacterial artificial chromosome (BAC) probes
(Table I) obtained from Resources
Center (Oakland, CA, USA) were labeled by PCR with SpectrumGreen,
SpectrumOrange or TexasRed-dUTP and applied in two- or three-color
FISH approaches.
 | Table IResults of the locus-specific probes
used for breakpoint analyses in metaphase FISH are listed. |
Table I
Results of the locus-specific probes
used for breakpoint analyses in metaphase FISH are listed.
| Cytoband | Position
[hg18] | Genes/locus | Probe | Results (signals
on…) |
|---|
| 3pter |
chr3:131,486-331,767 | D3S4559 | 3pTEL (Vysis) | der(5)t(3;5) |
| 3p24.1 |
chr3:30,275,517-30,447,565 | n.d. | RP11-69K20 | der(5)t(3;5) |
| 3p24.1 |
chr3:30,541,893-30,705,070 | STT3B | RP11-7I16 | der(5)t(3;5) |
| 3p22.3 |
chr3:32,453,732-32,650,841 | GPD1L | RP11-524O15 | der(5)t(3;5) |
| | GADL1 | | |
| | OSBPL10 | | |
| | CMTM7 | | |
| | CMTM8 | | |
| 3p22.2 |
chr3:38,928,115-39,088,251 | n.d. | RP11-159A17 | der(3)t(3;5) |
| 5q22.2 |
chr5:112,073,070-112,236,540 | n.d. | RP11-107C15 | der(5)t(5;10) |
| 5q23.1 |
chr5:117,308,035-117,479,091 | n.d. | RP11-567A12 | der(5)t(5;10) |
| 5q23.3 |
chr5:126,045,879-126,232,850 | n.d. | RP11-434D11 | der(5)t(5;10) |
| 5q23.3~q31.1 |
chr5:130,306,745-130,460,728 | 5′ of IL3 | RP11-114H7 | der(5)t(5;10) |
| 5q31.1 |
chr5:131,424,246-131,426,795 | IL3 | n.a. | n.a. |
| 5q31.1 |
chr5:131,817,004-131,977,063 | 3′ of IL3 | RP11-729C24 | der(3)t(3;5) |
| 5q31.1 |
chr5:135,739,999-135,916,051 | n.d. | RP11-114H21 | der(3)t(3;5) |
| 5q31.2 |
chr5:137,829,080-137,832,903 | EGR1 | LSI EGR1 | der(3)t(3;5) |
| 5q32.1 |
chr5:149,473,595-149,515,615 | PDGFRB | POSEIDON PDGFRB
(Kreatech) | der(3)t(3;5) |
| 5q35.1 |
chr5:170,996,421-171,159,856 | n.d. | RP11-20O22 | der(3)t(3;5) and
der(5)t(3;5) |
| 5q35.2 |
chr5:173,985,900-174,153,222 | n.d. | RP11-47J7 | der(3)t(3;5) and
der(5)t(3;5) |
| 5q35.2 |
chr5:175,502,694-175,558,904 | n.d. | RP11-844P9 | der(3)t(3;5) and
der(5)t(3;5) |
| 5q35.3 |
chr5:176,550,923-176,735,050 | n.d. | RP11-265K23 | der(3)t(3;5) and
der(5)t(3;5) |
| 5q35.3 |
chr5:178,243,600-178,455,573 | 5′ HNRNPH1 | RP11-281O15 | der(3)t(3;5) and
der(5)t(3;5) |
| 5q35.3 |
chr5:178,973,785-178,983,328 | HNRNPH1 | n.a. | n.a. |
| 5q35.3 |
chr5:179,360,362-179,524,360 | 3′ HNRNPH1 | RP11-39H3 | der(5)t(5;10) and
der(5)t(3;5) |
| 5q35.3 |
chr5:180,142,710-180,335,838 | n.d. | RP11-516K1 | der(5)t(5;10) and
der(5)t(3;5) |
| 5qter |
chr5:180,510,748-180,711,420 | D5S2907 | 5pTEL (Vysis) | der(5)t(5;10) and
der(5)t(3;5) |
| 10pter |
chr10:292,280-292,670 | Z96139 | 10pTEL (Vysis) | der(5)t(3;5) |
| 10p12.31 |
chr10:20,782,567-20,938,614 | n.d. | RP11-51E20 | der(5)t(3;5) |
| 10p12.31 |
chr10:21,321,413-21,495,264 | 5′ MLLT10 | RP11-165O3 | der(5)t(3;5) |
| 10p12.31 |
chr10:21,863,580-22,072,560 | MLLT10 | n.a. | n.a. |
| 10p12.31 |
chr10:22,399,352-22,575,929 | 3′ MLLT10 | RP11-108B14 | der(5)t(5;10) |
Additionally, the following commercially available
probes were used: LSI EGR1/D5S23, D5S721 (EGR1 in 5q31;
D5S23, D5S721 in 5p15.2; Abbott Molecular/Vysis, Mannheim,
Germany), POSEIDON PDGFRB (5q33 Break probe; Kreatech
Diagnostics, Amsterdam, The Netherlands), and subtelomeric probes
for 3p, 5p, 5q and 10p (3p in D3S4559; 5p in C84c11/T3, 5q in
D5S2907; 10p in Z96139; Abbott Molecular/Vysis).
A total of 10–15 metaphase spreads were analyzed,
using a fluorescence microscope (Axio Imager Z1 mot; Carl Zeiss AG)
equipped with appropriate filter sets to discriminate between a
maximum of five fluorochromes and the counterstain DAPI
(diaminophenylindol). Image capturing and processing were carried
out using an ISIS imaging system (MetaSystems, Altlussheim,
Germany).
DNA isolation
Genomic DNA was extracted from cells fixed in acetic
acid-methonal (1:3) using the Puregene DNA purification kit (Gentra
Systems, Inc., Minneapolis, MN, USA). DNA concentration was
determined by a NanoDrop spectrophotometer. The quality of DNA was
checked using agarose gel electrophoresis. DNA samples extracted
from fixed cells of 2 healthy males and 2 healthy females by the
same method were used as reference samples.
Multiplex ligation-dependent probe
amplification (MLPA)
The P377-A1 hematologic malignancies probemix and
SALSA reagents were used for the present study (MRC-Holland,
Amsterdam, The Netherlands). Amplified probes and GeneScan 500 ROX
standard were separated by capillary electrophoresis using a
4-capillary ABI PRISM 3130XL Genetic Analyzer (Applied Biosystems,
Foster City, CA, USA). Sizing of peaks and quantification of peak
areas and heights were performed using the GeneMarker v1.9 software
(Applied Biosystems). A minimum of 4 healthy control samples were
included in each run.
High resolution array-comparative
genomic hybridization (aCGH)
aCGH was performed using the Agilent SurePrint G3
Human Genome Microarray 180K (Agilent Technologies, Santa Clara,
CA, USA), an oligonucleotide microarray containing ~180,000 probes
60-mer with a 17-kb average probe spacing. Genomic DNA of the
patient was co-hybridized with a male control DNA (Agilent
Technologies). Labeling was performed using the Agilent Genomic DNA
Enzymatic Labeling kit (Agilent Technologies) according to the
manufacturer’s instructions. After hybridization, the aCGH slide
was scanned on an Agilent scanner, processed with the Feature
Extraction software (v.10.7) and results were analyzed using
Cytogenomics (v2.9.1.3) using ADM2 as aberration algorithm.
Results of the retrospective
analyses
Genome-wide 24-color FISH using all human WCP probes
and FISH-banding analysis using the mMCB probe set were applied as
initial tests in this retrospective case. Thereby, a previously
unrecognized numerical aberration, trisomy 4, and balanced
translocations were identified between one chromosome 3 and 10,
each, and both chromosomes 5. Overall, an abnormal karyotype was
characterized as 47,XY,+4,der(3)t(3;5)(p23;q31.1),der(5)
t(3;5)(p23;q35.3), der(5)t(5;10)(q31.1;p12.3),der(10)t(5;10)
(q35.3;p12.3)[8]/46,XY[13] (Fig. 1B and
C).
Chromosome-specific aMCB confirmed these results
(Fig. 1D) and locus-specific probes
narrowed down the breakpoints according to NCBI36/hg18 as follows
(Table I). i) The breakpoint in
3p23 was determined between the positions 30,705,070 and
32,453,732; 6 OMIM genes are located there: STT3B,
GPD1L, GADL1, OSBPL10, CMTM7 and
CMTM8. ii) The breakpoint 5q31.1 locates between positions
130,460,728 and 131,817,004 and those flank the gene IL3
(interleukin 3 precursor) in 131,424,246-131,426,795. iii) The
second breakpoint on chromosome 5 in subband q35.3 was mapped to
positions 178,455,573 to 179,360,362; here the HNRNPH1
(heterogeneous nuclear ribonucleoprotein H1) gene is included in
178,973,785-178,983,328. iv) Finally, the breakpoint in 10p12.3 was
narrowed down to localize between positions 21,495,264 and
22,399,352, where the MLLT10 (myeloid/lymphoid or
mixed-lineage leukemia) gene has been mapped to
21,863,580-22,072,560.
No submicroscopic changes were detected by MLPA and
aCGH; only the trisomy 4 was observed in aCGH (data not shown).
Discussion
Chromosomal translocations in ALL may be missed in
banding karyotyping due to several reasons. They may be cryptic, as
they are not resolvable due to a similar or identical GTG-banding
pattern; an example is the t(12;21)(p13;q22) in childhood ALL
(23). In addition, known
aberrations may be masked in a complex karyotype (24). Finally, it may simply be difficult
to obtain evaluable metaphases where chromosomes are well-spread
and not clumsy or appearing as fuzzy with indistinct margins
(25). In the present case the
latter was the major problem. In the reanalyses, all well-spread
metaphases were normal and all aberrant metaphases were clumsy and
not evaluable in standard GTG-banding. Thus, cytogenetic analyses
in two different laboratories missed the aberrations present in
this case. Otherwise gross structural and a numerical aberration
would not have been overlooked like in this case which were
detected in retrospect by molecular cytogenetics.
Trisomy 4 as a sole abnormality is rare in acute
myeloid leukemia (AML) (26) but is
scarce in ALL and is not associated with a clear prognosis
(6,27,28).
In pediatric ALL, trisomy 4 has been reported to be associated with
a favorable outcome suggesting that children who have trisomies of
both chromosomes 4 and 10 may have a particularly low risk of
treatment failure (3,5). Here, trisomy 4 was observed together
with additional structural chromosomal aberrations. Most likely the
oncogene MLLT10 in 10p12.31 was activated by the strong
promoter of HNRNPH1 in 5q35.3. In addition, the
translocation of 5q31.1 to 3p23 brought in close proximity the gene
IL3, which has been shown to have an oncogenic effect on
hematopoietic cells (29), to 6
OMIM genes listed in Table I, which
could also potentially lead to overexpression of IL3.
MLLT10 gene
Rearrangements have previously been identified in
both child and adulthood acute leukemia (30). The t(10;11) is a recurrent
reciprocal translocation present in two common variants:
t(10;11)(p12;q23) and t(10;11)(p12;q21); the latter tending to be
more frequent in T-ALL patients (31). In addition, the t(10;11)(p12;q23)
mainly found in childhood AML is rarely observed in B-ALL and T-ALL
(32). The MLLT10 gene
encodes a leucine zipper protein that functions as a transcription
factor. MLLT10 gene rearrangements are associated with a
poor outcome due to the poor response to therapy (33,34).
HNRNPH1 gene
While unbalanced structural aberration of chromosome
5 are common in myelodysplastic syndrome or AML (35,36),
they are less common in ALL. Still Brandimarte et al
(14) previously identified the
HNRNPH1 gene as a new MLLT10 fusion partner in
pediatric T-ALL, as we observed in our case of T-precursor cell
ALL.
IL3 gene
Located in 5q31.1, the IL3 gene is a
multipotent hematopoietic growth factor produced by activated T
cells (37). Its involvement in
malignancies was previously reported in B-ALL cases due to a
t(5;14)(q31;q32). Overexpression of IL3 was associated with
unfavorable outcome in such cases (38).
3p23 region
Six OMIM genes are located in the breakpoint region
of chromosome 3 in subband p23. These include: STT3B (source of
immunodominant MHC-associated), GPD1L (glycerol-3-phosphate
dehydrogenase 1-like), GADL1, (glutamate decarboxylase-like
1), OSBPL10 (oxysterol-binding protein-like protein 10),
CMTM7 (CKLF-like MARVEL transmembrane domain containing 7)
and CMTM8 (CKLF-like MARVEL transmembrane domain containing
8). It is difficult to determine which one might have provided a
strong promoter for IL3 gene expression.
In conclusion, the study in particular of ALL cases
with unexpectedly adverse outcome in retrospect and in detail by
high resolution molecular approaches is warranted. In the present
case the combination of FISH-banding, FISH with locus-specific
probes and aCGH revealed trisomy 4 but apart from that a balanced
aberrant karyotype, explaining the severe course of the disease in
this case with adverse outcome. Even if this complex karyotype
would have been identified at the time of diagnosis most likely no
additional therapy other than the applied protocol (ALL-BFM 95)
would have been used. Yet, the recurrence may have been detected
much earlier in the case of available cytogenetic markers. Thus,
the most comprehensive molecular (cyto)genetic analyses should be
offered to each individual ALL case. Even though aCGH would not
have detected the balanced translocations, the detectable trisomy 4
would have hinted at the malignant clone missed by banding
cytogenetics. In conclusion, the present case is the first one
presenting with combined trisomy 4 with a four-way translocation
activating IL3 together with MLLT10.
Acknowledgements
The present study was supported in part by the DAAD
and KAAD.
References
|
1
|
Han X and Bueso-Ramos CE: Precursor T-cell
acute lymphoblastic leukemia/lymphoblastic lymphoma and acute
biphenotypic leukemias. Am J Clin Pathol. 127:528–544. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Moorman AV, Richards SM, Martineau M, et
al: Outcome heterogeneity in childhood high-hyperdiploid acute
lymphoblastic leukemia. Blood. 102:2756–2762. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chilton L, Buck G, Harrison CJ, et al:
High hyperdiploidy among adolescents and adults with acute
lymphoblastic leukaemia (ALL): cytogenetic features, clinical
characteristics and outcome. Leukemia. 28:1511–1518. 2014.
View Article : Google Scholar
|
|
4
|
Heerema NA, Sather HN, Sensel MG, et al:
Prognostic impact of trisomies of chromosomes 10, 17, and 5 among
children with acute lymphoblastic leukemia and high hyperdiploidy
(>50 chromosomes). J Clin Oncol. 18:1876–1887. 2000.PubMed/NCBI
|
|
5
|
Paulsson K, Forestier E, Andersen MK, et
al: High modal number and triple trisomies are highly correlated
favorable factors in childhood B-cell precursor high hyperdiploid
acute lymphoblastic leukemia treated according to the NOPHO ALL
1992/2000 protocols. Haematologica. 98:1424–1432. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gupta V and Chun K: Trisomy 4 as the sole
cytogenetic abnormality in a patient with T-cell acute
lymphoblastic leukemia. Cancer Genet Cytogenet. 152:158–162. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Paulsson K, Panagopoulos I, Knuutila S, et
al: Formation of trisomies and their parental origin in
hyperdiploid childhood acute lymphoblastic leukemia. Blood.
102:3010–3015. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gruszka-Westwood AM, Horsley SW,
Martinez-Ramirez A, et al: Comparative expressed sequence
hybridization studies of high-hyperdiploid childhood acute
lymphoblastic leukemia. Genes Chromosomes Cancer. 41:191–202. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kebriaei P, Anastasi J and Larson RA:
Acute lymphoblastic leukaemia: diagnosis and classification. Best
Pract Res Clin Haematol. 15:597–621. 2002. View Article : Google Scholar
|
|
10
|
Cauwelier B, Dastugue N, Cools J, et al:
Molecular cytogenetic study of 126 unselected T-ALL cases reveals
high incidence of TCRbeta locus rearrangements and putative new
T-cell oncogenes. Leukemia. 20:1238–1244. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Inaba H, Greaves M and Mullighan CG: Acute
lymphoblastic leukaemia. Lancet. 381:1943–1955. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Berger R, Dastugue N, Busson M, et al:
t(5;14)/HOX11L2-positive T-cell acute lymphoblastic leukemia. A
collaborative study of the Groupe Français de Cytogénétique
Hématologique (GFCH). Leukemia. 17:1851–1857. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Borel C, Dastugue N, Cances-Lauwers V, et
al: PICALM-MLLT10 acute myeloid leukemia: a French cohort of 18
patients. Leuk Res. 36:1365–1369. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brandimarte L, Pierini V, Di Giacomo D, et
al: New MLLT10 gene recombinations in pediatric T-acute
lymphoblastic leukemia. Blood. 121:5064–5067. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Coustan-Smith E, Mullighan CG, Onciu M, et
al: Early T-cell precursor leukaemia: a subtype of very high-risk
acute lymphoblastic leukaemia. Lancet Oncol. 10:147–156. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Inukai T, Kiyokawa N, Campana D, et al:
Clinical significance of early T-cell precursor acute lymphoblastic
leukaemia: results of the Tokyo Children’s Cancer Study Group Study
L99-15. Br J Haematol. 156:358–365. 2012. View Article : Google Scholar
|
|
17
|
Zhang J, Ding L, Holmfeldt L, et al: The
genetic basis of early T-cell precursor acute lymphoblastic
leukaemia. Nature. 481:157–163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Claussen U, Michel S, Mühlig P, et al:
Demystifying chromosome preparation and the implications for the
concept of chromosome condensation during mitosis. Cytogenet Genome
Res. 98:136–146. 2002. View Article : Google Scholar
|
|
19
|
Liehr T and Claussen U: Current
developments in human molecular cytogenetic techniques. Curr Mol
Med. 2:283–297. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liehr T, Heller A, Starke H, et al:
Microdissection based high resolution multicolor banding for all 24
human chromosomes. Int J Mol Med. 9:335–339. 2002.PubMed/NCBI
|
|
21
|
Weise A, Heller A, Starke H, et al:
Multitude multicolor chromosome banding (mMCB) - a comprehensive
one-step multicolor FISH banding method. Cytogenet Genome Res.
103:34–39. 2003. View Article : Google Scholar
|
|
22
|
Weise A, Mrasek K, Fickelscher I, et al:
Molecular definition of high-resolution multicolor banding probes:
first within the human DNA sequence anchored FISH banding probe
set. J Histochem Cytochem. 56:487–493. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bernard OA, Romana SP, Poirel H and Berger
R: Molecular cytogenetics of t(12;21) (p13;q22). Leuk Lymphoma.
23:459–465. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Usvasalo A, Räty R, Harila-Saari A, et al:
Acute lymphoblastic leukemias with normal karyotypes are not
without genomic aberrations. Cancer Genet Cytogenet. 192:10–17.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mkrtchyan H, Glaser M, Gross M, et al:
Multicolor-FISH applied to resolve complex chromosomal changes in a
case of T-ALL (FAB L2). Cytogenet Genome Res. 114:270–273. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bains A, Lu G, Yao H, et al: Molecular and
clinicopathologic characterization of AML with isolated trisomy 4.
Am J Clin Pathol. 137:387–394. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Moreau P, Talmant P, Milpied N, et al:
Trisomy 4 associated with acute lymphocytic leukaemia. Br J
Haematol. 78:5761991. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yip SF, Wan TS, Chan LC and Chan GC:
Trisomy 4 as sole karyotypic abnormality in acute lymphoblastic
leukemia: different clinical features and treatment response
between B and T phenotypes? Cancer Genet Cytogenet. 164:94–95.
2006. View Article : Google Scholar
|
|
29
|
Steelman LS, Algate PA, Blalock WL, et al:
Oncogenic effects of overexpression of the interleukin-3 receptor
on hematopoietic cells. Leukemia. 10:528–542. 1996.PubMed/NCBI
|
|
30
|
DiMartino JF, Ayton PM, Chen EH, et al:
The AF10 leucine zipper is required for leukemic transformation of
myeloid progenitors by MLL-AF10. Blood. 99:3780–3785. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Asnafi V, Radford-Weiss I, Dastugue N, et
al: CALM-AF10 is a common fusion transcript in T-ALL and is
specific to the TCRgammadelta lineage. Blood. 102:1000–1006. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Coenen EA, Raimondi SC, Harbott J, et al:
Prognostic significance of additional cytogenetic aberrations in
733 de novo pediatric 11q23/MLL-rearranged AML patients: results of
an international study. Blood. 117:7102–7111. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dreyling MH, Schrader K, Fonatsch C, et
al: MLL and CALM are fused to AF10 in morphologically distinct
subsets of acute leukemia with translocation t(10;11): both
rearrangements are associated with a poor prognosis. Blood.
91:4662–4667. 1998.PubMed/NCBI
|
|
34
|
Caudell D and Aplan PD: The role of
CALM-AF10 gene fusion in acute leukemia. Leukemia. 22:678–685.
2008. View Article : Google Scholar
|
|
35
|
Crescenzi B, La Starza R, Romoli S, et al:
Submicroscopic deletions in 5q-associated malignancies.
Haematologica. 89:281–285. 2004.PubMed/NCBI
|
|
36
|
Kayser S, Zucknick M, Döhner K, et al:
Monosomal karyotype in adult acute myeloid leukemia: prognostic
impact and outcome after different treatment strategies. Blood.
119:551–558. 2012. View Article : Google Scholar
|
|
37
|
Mangi MH and Newland AC: Interleukin-3 in
hematology and oncology: current state of knowledge and future
directions. Cytokines Cell Mol Ther. 5:87–95. 1999.PubMed/NCBI
|
|
38
|
Gallego M, Coccé M, Felice M, et al: A new
case of t(5;14) (q31;q32) in a pediatric acute lymphoblastic
leukemia presenting with hypereosinophilia. Atlas Genet Cytogenet
Oncol Haematol. 16:183–184. 2012.
|