Introduction
Epithelial ovarian cancer (EOC) is the most lethal
gynecologic malignancy (1–3) which caused an estimated 15,520 deaths
in the USA in 2008 (2). The
standard treatment protocol used in the initial management of
advanced-stage ovarian cancer is cytoreductive surgery, followed by
primary chemotherapy with a platinum-based regimen that usually
includes cisplatin. Although the management of ovarian cancer has
improved over the last 20 years due to more effective surgical
techniques and optimized combinational chemotherapy, the overall
cure rate is only 30%. One major limitation to the successful
treatment of ovarian cancer is the resistance to the currently
preferred chemotherapeutic regimen of platinum plus paclitaxel.
Empiric-based treatment strategies are used and result in many
patients with chemotherapy-resistant disease receiving multiple
cycles of often toxic therapy without success before the lack of
efficacy is identified. Therefore, the ability to predict the
response to and the mechanism involved in platinum-based primary
therapies is a major step towards improving the outcome and
individualizing treatments for women with ovarian cancer.
In the past few years, research has focused on the
discovery of a new class of genomic regulators which are now
universally recognized as central players in virtually the
development and progression of all neoplasms, named microRNAs
(miRNAs). miRNAs are short noncoding RNAs that act as
post-transcriptional regulators. They have wide-ranging roles in
diverse biological processes, such as development, differentiation,
growth and apoptosis (4). miRNAs
may also exert a role either as an oncogene or a tumor suppressor
by affecting the response to various therapeutic interventions
(5,6). In recent years, studies have
highlighted the relationship between miRNAs and the resistance to
chemotherapy (5,7,8).
Based on miRNA microarray data in our previous
research, miR-136 was found to be significantly downregulated in
chemoresistant ovarian cancer specimens. To understand the
mechanism of miR-136 in regulating chemoresistance, we applied this
approach to identify miRNA expression patterns within primary EOC
that may predict the response to primary platinum-based
chemotherapy. Coupled with this, we analyzed the bio-functional
variations that reflect and identify the regulation of various
oncogenic-miRNA signaling pathways to identify unique mechanisms of
platinum-resistance that can guide the use of chemotherapy drugs in
platinum-resistant EOC patients.
Materials and methods
Patient samples and cell lines
We evaluated a total of 34 EOC tissue samples that
were resected at the time of primary surgery from patients who went
on to receive platinum-based chemotherapy. The clinicopathological
characteristics of the patients who contributed the OVC samples are
listed in Table I. All tissue
samples were collected at the Obstetrics and Gynecology Hospital,
Dalian, China between July 2004 and November 2010. The study was
approved by the Institutional Review Board of the Ministry of
Science and Technology of China [China Human Resource Management
Office (2009) 015]. Two independent pathologists with no knowledge
of the patient clinical data reviewed the pathological specimens
before they were formalin-fixed and paraffin-embedded (FFPE). The
cases were classified according to the FIGO staging system. A
complete response (CR) was classified as the complete disappearance
of all measurable and assessable disease or, in the absence of
measurable lesions, a normalization of CA-125 levels following
platinum-based therapy. An incomplete response (IR) was defined as
only a partial response, no response, or progression during primary
therapy (9). CA-125 response
criteria were based on established guidelines and were used only in
the cases with the absence of a measurable lesion (9–11).
 | Table IClinicopathological characteristics of
the ovarian cancer patients. |
Table I
Clinicopathological characteristics of
the ovarian cancer patients.
| Clinical
response |
|---|
|
|
|---|
| Characteristics | Complete (n=19) | Incomplete
(n=15) |
|---|
| Mean age (years) | 52.7 | 55.6 |
| Mean serum CA-125
(m/ml) |
| Before platinum
treatment | 796.5 |
1,533.8b |
| After platinum
treatment | 22.7 |
416.5b |
| Mean survival time
(months) | 57.2 | 35.8a |
| Mortality rate
(%) | 31.6 | 62.3b |
Experiments were performed with a group of human EOC
cell lines, including one cisplatin-sensitive parental cell line
(OV2008) and its cisplatin-resistant variant (C13) (1,12,13).
Cell culture, transfection and
treatment
The ovarian cancer cell lines were cultured in
RPMI-1640 medium (Invitrogen, Burlington, ON, Canada) supplemented
with 10% fetal bovine serum (FBS) and maintained at 37°C, in 5%
CO2, as reported previously (1,12,13).
All tissue culture reagents were obtained from Sigma-Aldrich (St.
Louis, MO, USA). The miR-136 mimics and inhibitors were designed
and chemically synthesized by Ambion (38422-01; Life Technologies
Corporation, Denmark).
Lipofectamine 2000 (Invitrogen) was incubated with
pre-miR-136 (miRNA mimic), anti-miR-136 (inhibitor) or their
scrambled negative controls (Ambion, mock) at a concentration of 90
nmol/l and incubated in serum-free RPMI-1640 for 20 min before
being added to C13 or OV2008 cells, respectively. Cells were
incubated at 37°C for 4 h before 10% FBS was replaced. For
cisplatin treatment, cells were maintained in medium with the
desired doses of cisplatin (P4394; Sigma, USA).
miRNA microarray and data analysis
A microarray platform optimized for the analysis of
a panel of 768 human miRNAs was used to analyze and compare the
pattern of miRNA expression between CR and IR (n=7 for all) to
platinum-based chemotherapy in EOC patients. Individual real-time
quantitative polymerase chain reaction assays were formatted into a
TaqMan low-density array (TLDA; Applied Biosystems), which was
performed at the Shannon McCormack Advanced Molecular Diagnostics
Laboratory Research Services, Dana Farber Cancer Institute, Harvard
Clinic and Translational Science Center, Boston, MA, USA. The
normalized microarray data were managed and analyzed by StatMiner
ver. 3.0 (Integromics™).
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed
using the TaqMan MicroRNA Reverse Transcription kit (Applied
Biosystems, Foster City CA, USA) on an Agilent Technologies
Stratagent Mx3000P (USA). A 500 ng mass of total RNA in 1 μl of
RNase-free water was used in 20 μl of the RT mix. The following
primer pairs were used: hsa-miR-136 (ABI miRNA-specific primers,
Cat no. 4427012). The products were detected with SYBR-Green I, and
their relative miRNA levels were calculated using the comparative
Ct (cycle threshold, 2−ΔΔCt) method with U6 as the
endogenous control.
Cell growth and viability assays
Manual cell counting using trypan blue staining was
used to determine the effect of cisplatin on the different cell
lines. Briefly, cells were cultured in 96-well plates at a density
of 1×103/well for 48 h after transfection, and were then
treated with 0, 10, 15, 20, 25, 30, 35, 60, 80 and 120 μmol/l
cisplatin (P4394; Sigma) for 48 h. Subsequently, live-cell and
dead-cell activities were detected by trypan blue staining. Cell
surviving percentage was assessed as the ratio of live to total
(live + dead) cells. Each point represents the mean ± SD of
triplicates.
Migration and invasion assays
For migration and invasion assays, 48 h after
transfection, the cells were serum starved for 12 h before
performing the assays. The assays were performed with BD Bio-Coat
Control Insert Chambers, 24-well plates with an 8-μm pore size, and
BD Bio Coat Matrigel Invasion Chambers, respectively. For the
wound-healing assay, 3×103 cells were plated in the top
chamber in 0.5 ml MEM with 0.5% FBS. The cells that did not migrate
through the membrane were removed with a cotton swab. A wound was
created using an aerosol P200 pipette tip after 12 h of incubation,
and then microscopic images were captured at 0, 8, 16 and 24 h. In
the Boyden chamber invasion assay, 3×103 cells were
seeded into 8-μm pore inserts coated with 50 μl Matrigel
(Becton-Dickinson). The bottom chamber, which contained 50% DMEM
with 10% FBS and L-glutamine, was used as a chemoattractant. After
24 h, the cells that had penetrated the Matrigel to invade the
lower surface of the membrane were fixed in methanol and stained
with crystal violet (1%), as was described for the migration assay.
Migrating or invading cells on the surface of the membranes were
counted manually using Image-Pro Plus version 6.0 (Media
Cybernetics, Inc.).
Cellular apoptosis assay with flow
cytometry
Annexin V binding with propidium iodide (PI)
staining was performed using the Annexin V-FITC Apoptosis Detection
kit (KGA106; KeyGen Biotech, Nanjing, China). Ovarian cancer cells
were initially seeded at a concentration of 1×105
cells/ml, in 6-well plates, and incubated at 37°C in a humidified
atmosphere with 5% CO2 for 48 h after transfection. The
cells were then treated with 50 and 25 μmol/l cisplatin,
respectively. After exposure to the drugs for 48 h, the cells were
dissociated using 0.05% EDTA-free trypsin and washed with cold PBS.
Approximately 1×106 cells were suspended in 100 μl of
Annexin V incubation reagent. After incubation in the dark for 20
min at room temperature, cellular fluorescence was analyzed by the
BD FACSCalibur Flow Cytometer (BD Biosciences, USA) within 30 min.
Control tubes containing binding buffer only and cells containing
Annexin V alone and PI alone were initially used to calibrate the
instrument.
Comet assay
The single cell gel electrophoresis assay (also
known as the Comet assay) is a sensitive technique for measuring
DNA strand breaks at the level of the individual cell. This is a
standard technique for evaluation of DNA biomonitoring,
genotoxicity testing and damage/repair (14,15).
The assay was performed as described by Pérez et al
(14) using a DNA damage detection
kit (KeyGen Biotech), with some modifications. Exponentially
growing cells were exposed to cisplatin 50 μmol/l (based on the
IC50 assay) for 1 h. The cells were washed and incubated
at 37°C in 5% CO2 for 48 h. Next, the cells were
trypsinized and resuspended in 1 ml culture medium. After that, 10
μl of the cells (1×104) was encapsulated in 75 μl of
0.7% low-melting-point agarose at 37°C. This mixture was layered
onto precoated slides with 0.5% standard agarose and covered with a
coverslip. The agarose was allowed to solidify for 10 min at 4°C,
and then the coverslip was gently removed. The slides were then
immersed in a precooled lysis buffer (cat. no. KGA240) for at least
90 min at 4°C and then incubated in an alkaline buffer (1 mmol/l
EDTA and 300 mmol/l NaOH; pH>13.0) for 60 min at room
temperature to allow for DNA unwinding and alkali-labile site
expression. Electrophoresis was conducted in the same alkaline
buffer for 30 min at 25 V (0.86 V/cm) and 300 mA. After
electrophoresis, the slides were washed at least three times with
0.4 mmol/l Tris-HCl (pH 7.5) prior to staining with propidium
iodide (PI) (20 μl) (KeyGen) for 10 min. Comet tails were
visualized using a FITC filtered fluorescence microscope. In these
experiments, cells with high amounts of cisplatin-induced
cross-links have shorter tails compared to cells in which
cross-links have been repaired effectively. This can be explained
by the fact that unrepaired platinum-induced DNA cross-links will
stay together after DNA degradation, resulting in larger DNA
fragments (16). The tail length is
proportional to the DNA repair. This is followed by visual analysis
with staining of DNA and calculating the percentage of DNA in comet
tails, tail length (in μm), tail moment (TM) and olive tail moment
(OTM) to determine the extent of DNA damage/repair (CASP ver.
1.2.3beta2, Krzysztof Konca; casplab.com).
Statistical analysis
SPSS 17.0 software (Chicago, IL, USA) was applied
for all quantitative analyses in the study, except for the
microarray data. Data are expressed as arithmetic means ± SD of the
number (n) of experiments. Samples were analyzed with repeated
measures analysis of variance, and differences in the incidences
were analyzed using ANOVA. The images of comet assay were analyzed
using Image-Pro Plus version 6.0. P<0.05 was considered to
indicate a statistically significant difference.
Results
Patient characteristics
Nineteen EOC patient samples were classified as CR
and 15 as IR to primary platinum-based therapy following surgery,
and FFPE blocks were obtained after surgery. The
clinicopathological characteristics of the patients who provided
the OPSC samples are listed in Table
I. The mean serum CA-125 level in the IR patients was
significantly higher than that in the patients with CR (P<0.001
for all).
microRNA microarray results and qRT-PCR
validation
To further characterize the unique miRNAs in OPSC
differentiation, specimens from the CR and IR OPSC patients were
initially analyzed by miRNA microarray, respectively. Of the 768
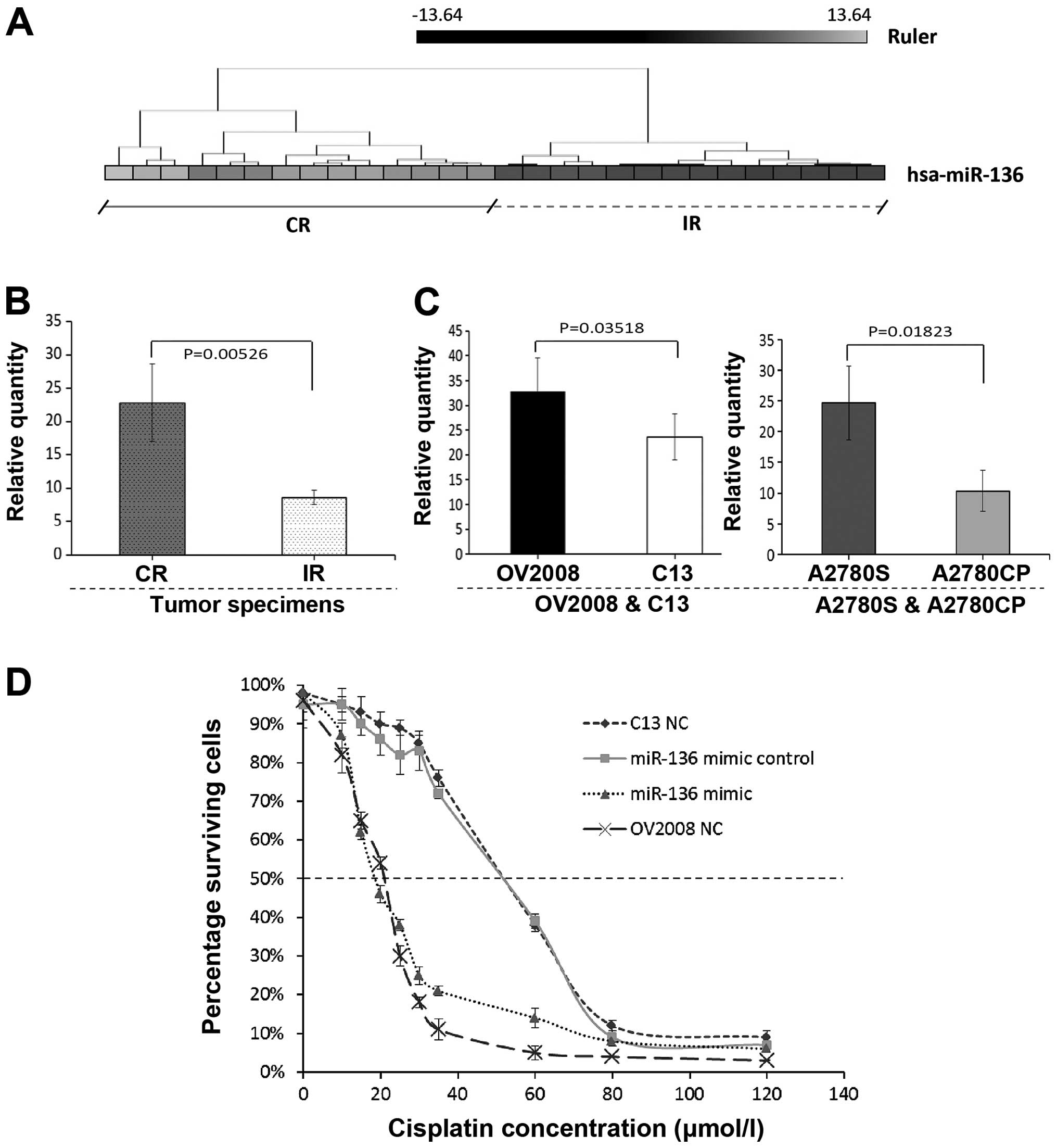
miRNAs analyzed by microarray, we identified that miR-136
expression was strongly downregulated in the IR OPSC when compared
with the CR OPSC patients (Fig.
1A). In order to confirm the microarray results, qRT-PCR
validation was performed. RNA was isolated from a new set of FFPE
tissues to increase the likelihood that the observed differences in
miRNA expression profiles represent biologically significant
changes. Consistent to the microarray results, miR-136 was weakly
expressed in the IR OPSC with significance, and representative
analysis is shown in Fig. 1B.
miR-136 expression in the different
chemoresistant ovarian cancer cell lines; overexpression of miR-136
decreases cell survival of a chemoresistant cisplatin cell
line
We demonstrated that the miR-136 expression level
was significantly higher in the CR tumor specimens than the level
in the IR tumor specimens (Fig. 1A and
B). Thus, we chose to further verify the relationship between
miR-136 expression and chemosensitivity using the human ovarian
cancer cell lines OV2008 and A2780S and the cisplatin-resistant
variants (C13 and A2780CP, respectively). In Fig. 1C, significant downregulation of
miR-136 expression was noted in the C13 and A2780CP cell lines
compared with their cisplatin-sensitive variants. These results are
consistent with those for the clinical OVC patient specimens.
Furthermore, to test whether miR-136 is causally
related to cellular sensitivity to cisplatin, we conducted
transfection experiments with cisplatin-resistant ovarian cancer
cells. Fig. 1D shows the
sensitivity to cisplatin of the C13 cells, its cisplatin-sensitive
variant OV2008 and the two transfected cell lines (C13+miR-136
mimic and C13+miR-136 mimic control). The drug concentration that
inhibited cell growth by 50% (IC50 values) in the
cisplatin-resistant ovarian cancer cells (C13) was ~2.5-fold higher
than that for its parental cell line (OV2008). C13 and C13+miR-136
mimic control showed similar IC50 values, indicating
that the transfection itself had no effect on cell growth. In
contrast, the miR-136-overexpressing cells, C13+ miR-136 mimic and
OV2008, showed substantial sensitivity to cisplatin. The
IC50 values for these cells were ~2.5-fold lower
compared to the C13 cell line (P<0.01; ANOVA test). No
difference was found in IC50 values between C13+miR-136
mimic and OV2008 cells. These results indicate that miR-136
influences the cellular sensitivity to cisplatin.
miR-136 has no effect on cell migration
and invasion but modulates cisplatin-induced apoptosis
The progression of a malignant tumor is determined
by the metastatic and invasive properties of the tumor cells, which
promote malignant cells to invade the extracellular matrix and
metastasize to distant sites. To further clarify the role of
miR-136 in the progression of ovarian cancer with primary
chemotherapy, a commercially available miR-136 mimic was applied to
the cisplatin-resistant parental cell line (C13) for a series of
migration and invasion experiments. Fig. 2A and B shows that the speed of
migration in the C13 cells had no significant change when compared
to their own mock-transfected and NC cells after overexpression of
miR-136. The slope of the general linear model and number of
migrating cells within a certain period was used to reflect the
motility potential. Using quantitative Boyden chamber analysis, the
motility of the C13 cells was not reduced after overexpression of
miR-136 (Fig. 2C and D).
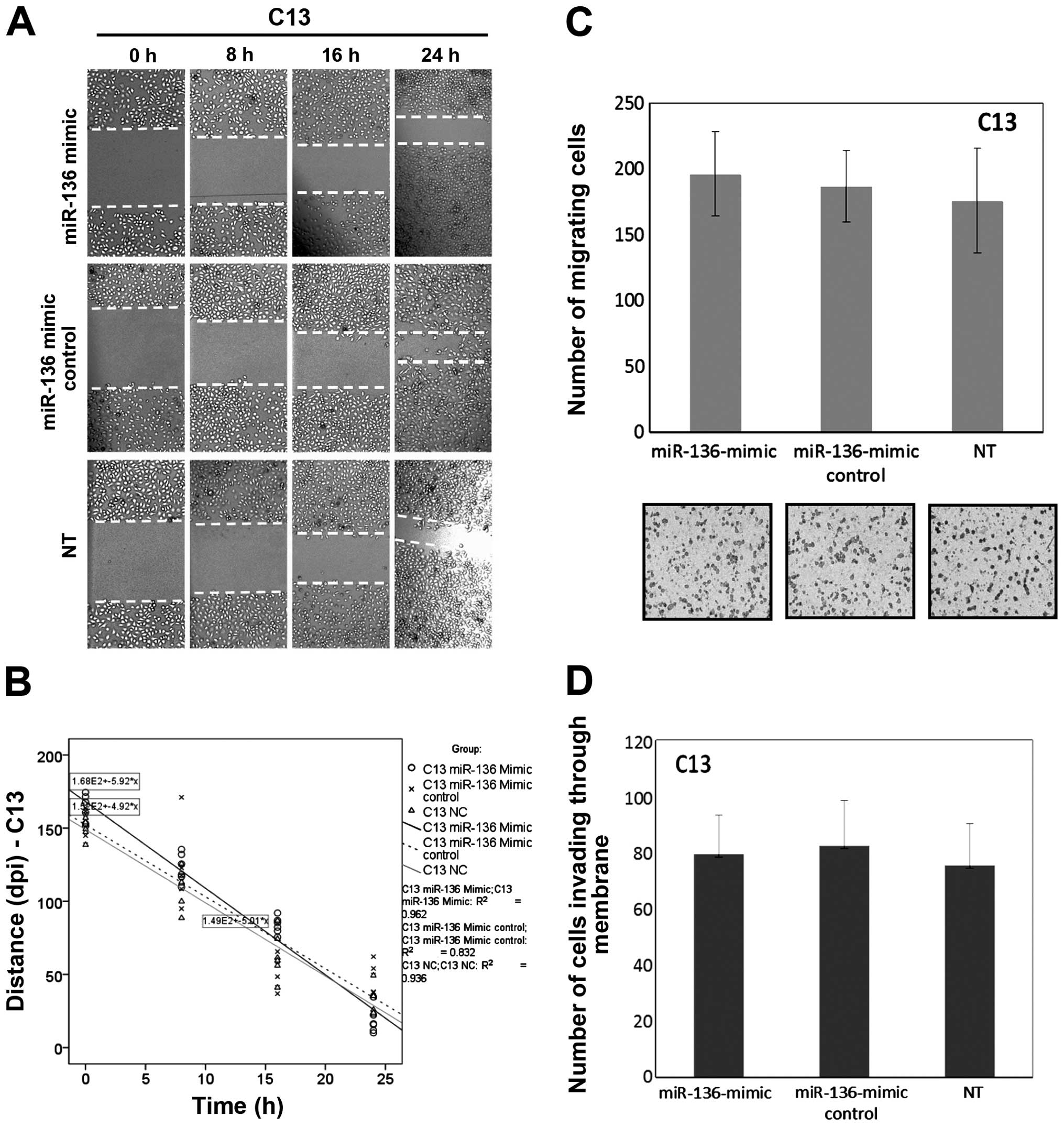 | Figure 2Overexpression of miR-136 has no
effect on cell migration and invasion in the chemoresistant OVC
cell line C13. (A) C13 cells were transfected with miR-136-mimic,
miR-136-mimic control (mock) and negative control, respectively.
After 48 h, wounds were inflicted and images were captured at 0, 8,
16 and 24 h after wounding. Lines indicate the width of the wound
at each time-point. Images shown are from one experiment
representative of six separate experiments. (B) The wound-healing
tendency (slope) of the miR-136-mimic-treated C13 cells had no
significant change when compared with their own mock-transfected
and NC cells, respectively. (C) miR-136 transfection was carried
out to assess cell motility using quantitative Boyden chamber
assay. After 48 h, the miR-136-mimic-transfected C13 cells showed
no significant decline in motility when compared to the
mock-transfected and the negative-control cells, per vision-field
at ×100. (D) The number of cells able to invade through the
Matrigel-coated Boyden chambers was also determined for each group.
Columns, mean number of cells from six replicates; bars, SD.
Compared with either negative control or mock-transfected cells
(P>0.05 for all). |
Cisplatin and other platinum-based cancer drugs bind
to double-stranded DNA and form DNA adducts, interfering with DNA
replication and RNA transcription ultimately triggering apoptosis.
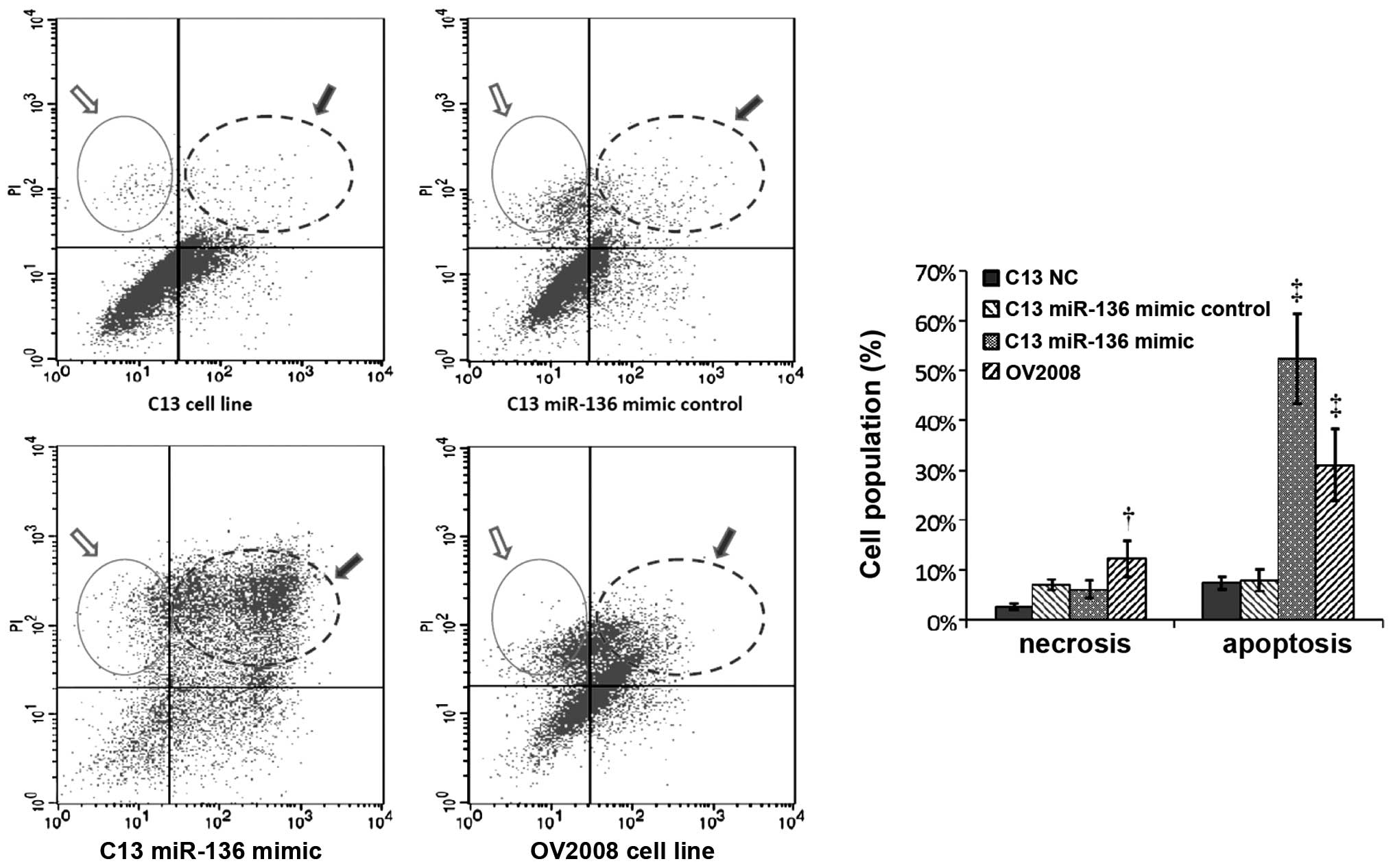
Thus, we examined whether miR-136 is involved in cisplatin-induced
chemoresistance by affecting apoptosis. As expected, the proportion
of apoptotic cells was significantly increased in the
C13+miR-136-mimic compared to its own NC- or mock-transfected
group. The cisplatin-induced apoptosis in the C13+miR-136-mimic
cells was comparable with the apoptosis of the OV2008 cells. These
results indicated that the proapoptotic activity in the
cisplatin-resistant C13 cell line was restored by introducing
miR-136 (Fig. 3).
miR-136 is essential for repair of
cisplatin-induced DNA damage in vitro
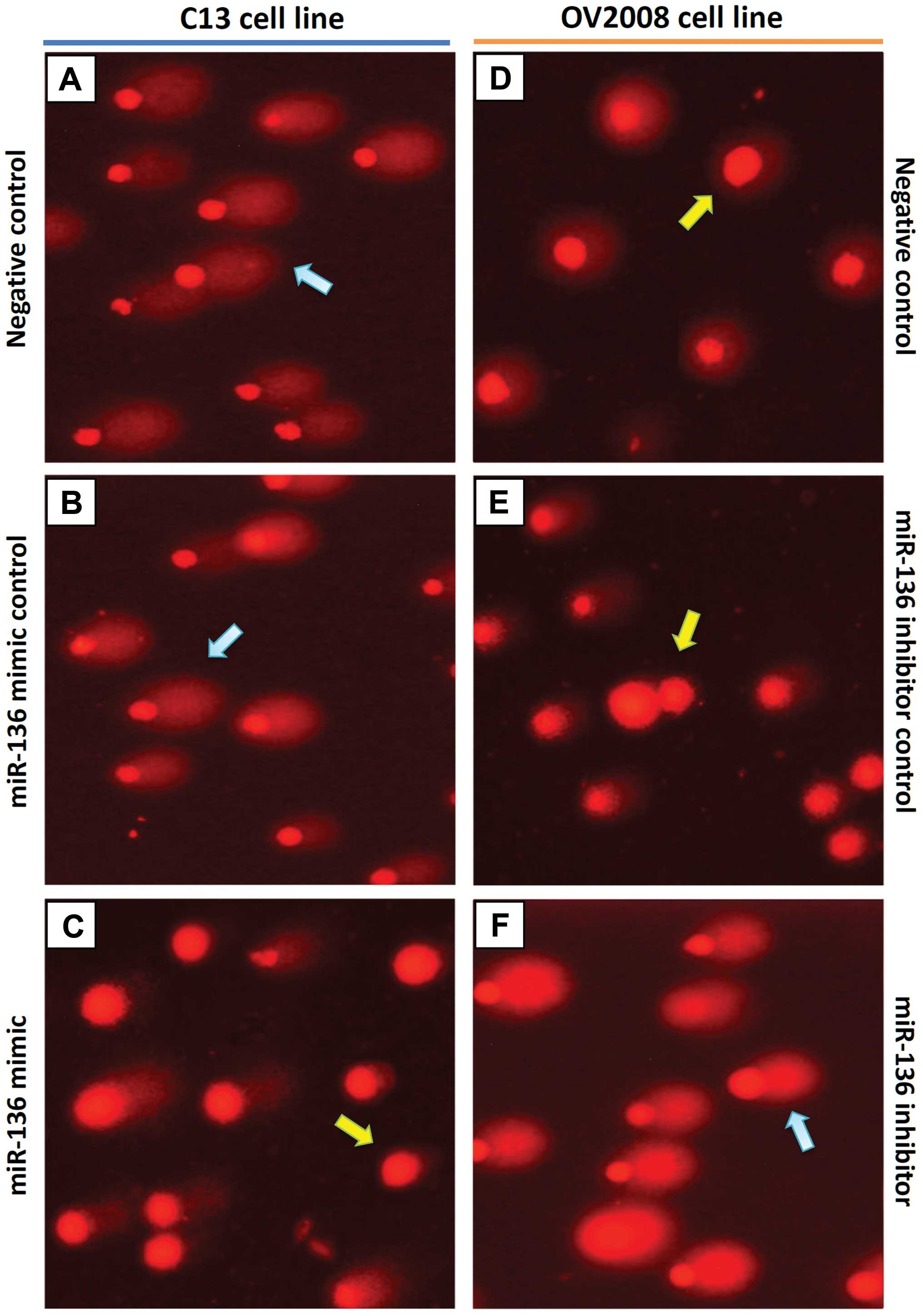
To validate whether the high DNA repair ability of
ovarian cancer cells following cisplatin treatment is caused by a
defect in miR-136, we evaluated cisplatin-induced DNA damage using
the COMET assay, in C13 cells, its cisplatin-sensitive variant
(OV2008), and the transfected variants C13+miR-136 mimic and
OV2008+miR-136 inhibitor. In order to quantify the DNA
damage/repair process, we measured the percentage of DNA in comet
tails, tail length (in μm), TM and OTM for each gel. Fig. 4A and Table II depict the decreasing trend of
DNA damage measured with CASP ver. 1.2.3beta2. The DNA damage, as
measured manually, is shown in Fig.
4B. As expected, tails of the DNA damage/repair deficient
OV2008 cell line were smaller than the C13 tails. After
transfection with miR-136, the comet tails in the C13+miR-136 mimic
were also shorter than the C13 tails. On the contrary, the miR-136
inhibitor increased the tail size compared with that of the OV2008
cells. These results indicate that DNA repair activity in the
cisplatin-resistant ovarian cancer cells was fully restored by
downregulation of miR-136 (Fig. 4).
A representative image of the modified COMET assay used for data
analysis is shown in Fig. 5.
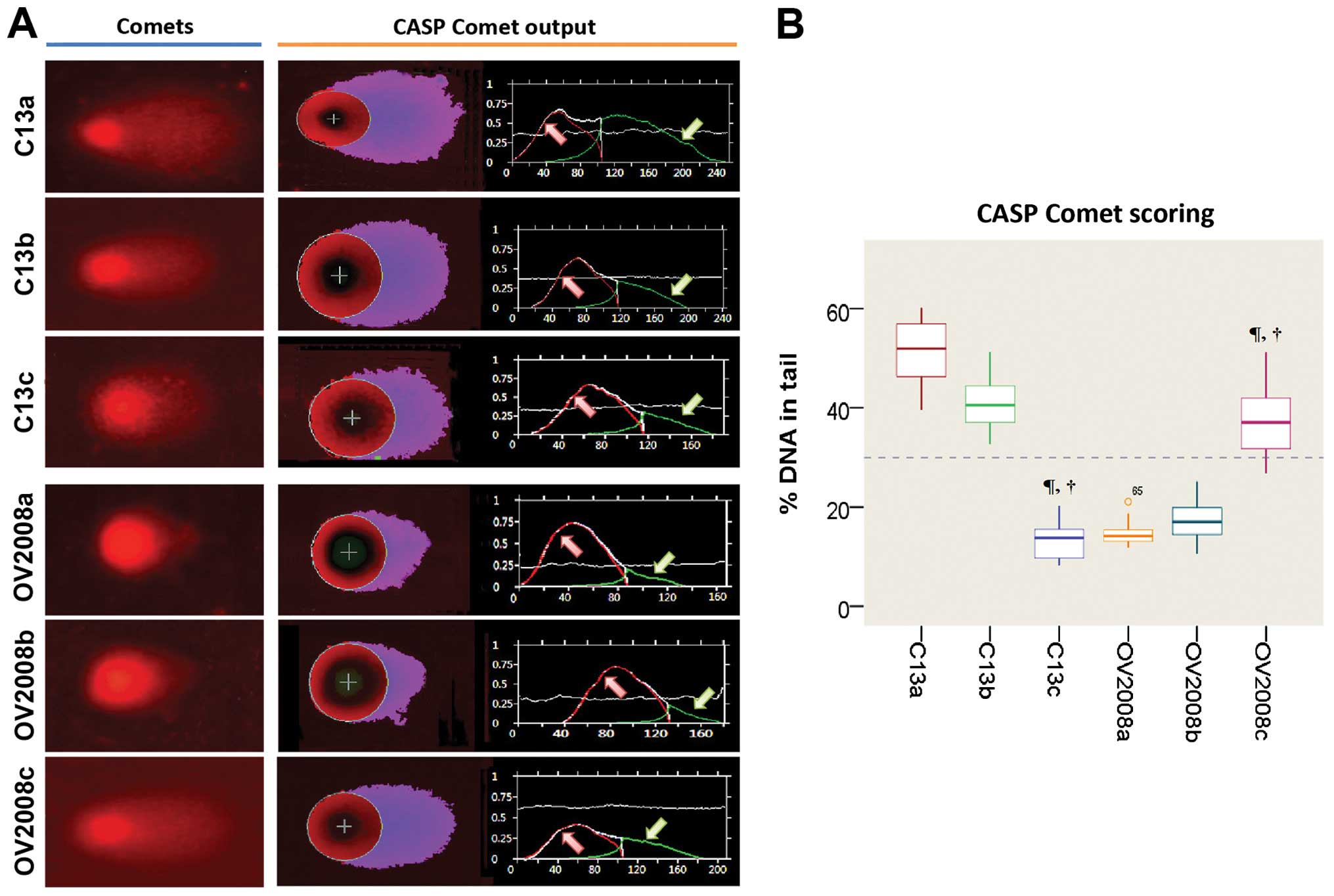 | Figure 4CASP comet analysis of alkaline comet
assay images. We used CASP comet to analyze alkaline comet assay
images of individual standard damaged cells of each group after
treatment with cisplatin. (A) CASP Comet analysis. The left column
is the alkaline comet assay image of each group, and the right
column is the output and analysis of the CASP comet. Red line (area
indicated by red arrow) represents the head of the comet, and the
green line (area indicated by green arrow) represents the area of
the tail of the comet. (B) Trend of the amount of DNA damage across
C13 and OV2008 cell lines. C13a, C13 NC; C13b, C13+miR-136 mimic
control; C13c, C13+miR-136 mimic; OV2008a, OV2008 NC; OV2008b,
OV2008+miR-136 inhibitor control; OV2008c, OV2008+miR-136
inhibitor. Compared with the normal control (NC) group of their own
cell line, ¶P<0.001; compared with their scrambled
negative controls, †P<0.001. |
 | Table IISpontaneous and cisplatin-induced mean
values and their standard deviation of four replicates of the Comet
assay parameters after treatment with cisplatin. |
Table II
Spontaneous and cisplatin-induced mean
values and their standard deviation of four replicates of the Comet
assay parameters after treatment with cisplatin.
| Ovarian cancer cell
lines | Transfection
status | % Tail DNA ± SD | Tail length ± SD | TM ± SD | OTM ± SD |
|---|
| C13 | Normal control | 51.62±5.80 | 133.95±22.43 | 70.66±11.61 | 50.10±5.36 |
| miR-136 mimic
control | 41.34±5.13 | 100.25±26.55 | 51.30±11.81 | 44.57±12.28 |
| miR-136 mimic | 13.33±3.19a,b | 46.50±7.73a,c | 9.87±2.82a,c | 10.07±3.60a,c |
| OV2008 | Normal control | 14.61±2.19 | 45.90±5.45 | 8.52±1.82 | 8.18±1.50 |
| miR-136 inhibitor
control | 17.17±3.90 | 52.95±9.76 | 10.15±2.63 | 9.77±2.60 |
| miR-136
inhibitor | 36.98±6.16a,b | 98.45±19.59a,b | 24.67±4.88a,c | 24.82±6.76a,c |
Discussion
Due to the heterogeneity and complexity of cancer
cells and the different mechanism of action of anticancer drugs,
several major mechanisms have been demonstrated to play a crucial
role in the resistance of chemotherapy, such as insensitivity to
drug-induced apoptosis, increased DNA repair, decreased drug
accumulation, and induction of drug-detoxifying mechanisms
(17,18). Most previous studies focused on
genetic and epigenetic alterations occurring during tumor formation
and progression. However, recent data have demonstrated that drug
resistance is regulated not only by genetic and epigenetic changes,
but also by miRNAs (17). In this
context the recent identification of the regulatory role of miRNAs
extended the spectrum of possible key factors involved in such a
processes.
The advent of microarray technologies offers the
opportunity to identify novel resistance mechanisms in clinically
relevant material, e.g. tumor tissue from patients before and after
chemotherapy. The discovery of a set of miRNAs strongly deregulated
in drug-resistant patients will open an avenue for innovative
cancer treatment. In a recent work, Shen et al (19) reported that miR-136 expression was
found to be deregulated in a human non-small cell lung cancer cell
line. In addition, it has been found that miR-136 is downregulated
in human glioma, and that miRNA promotes apoptosis of glioma cells
by targeting AEG-1 and Bcl-2 (19).
However, the roles of miR-136 in OVC are still largely unknown.
Here, we found that miR-136 was markedly
downregulated in cisplatin-resistant EOC patient tissues and in
cell lines. Upregulation of miR-136 inhibited EOC cell survival,
and promoted cell apoptosis. In fact, miRNAs can alter the cellular
response to a specific drug or class of drugs via many mechanisms
other than survival or apoptotic signaling, such as by interfering
with drug targets and DNA repair (17). In this study, besides regulating
cytotoxic drug-induced apoptosis, miR-136 might also alter the
cellular response to cisplatin-induced DNA excision. The levels of
adducts corrected with their extent of DNA damage/repair, in terms
of the percentage of DNA in comet tails, tail length, TM and OTM,
revealed that miR-136 is essential for repair of cisplatin-induced
DNA damage. Yet, we found no association between miR-136 and
migration or invasion potential in the ovarian cancer cell
lines.
The antitumour effect of cisplatin results from
platinum DNA cross-links, and ultimately leads to programmed cell
death. However, several cellular DNA repair mechanisms are capable
of repairing damage from platinum-DNA adducts, such as the
nucleotide excision repair (NER) system (20). In summary, in this study we
demonstrated that a specific miRNA, miR-136, is deregulated in
cisplatin-resistant ovarian cancer, and is also involved in
platinum-resistance by affecting DNA repair and cell apoptosis. We
believe that the approach described here, using microarray
technologies that predict primary chemotherapy response, coupled
with expression data that identify oncogenic pathway deregulation
to reveal mechanisms of cisplatin-resistance, represents an
important step toward the goal of personalized cancer treatment,
including EOC therapy.
Acknowledgements
This research was supported by the National Basic
Research Program of China 973 Program No. 2012CB517600 (No.
2012CB517603), and a MOST grant (2008 DFA30720) to C.S.
References
|
1
|
Zhao H, Ding Y, Tie B, et al: miRNA
expression pattern associated with prognosis in elderly patients
with advanced OPSC and OCC. Int J Oncol. 43:839–849.
2013.PubMed/NCBI
|
|
2
|
Matei DE and Nephew KP: Epigenetic
therapies for chemoresensitization of epithelial ovarian cancer.
Gynecol Oncol. 116:195–201. 2010. View Article : Google Scholar :
|
|
3
|
Yu X, Zhang X, Bi T, et al: MiRNA
expression signature for potentially predicting the prognosis of
ovarian serous carcinoma. Tumour Biol. 34:3501–3508. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNAs in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen Y, Ke G, Han D, Liang S, Yang G and
Wu X: MicroRNA-181a enhances the chemoresistance of human cervical
squamous cell carcinoma to cisplatin by targeting PRKCD. Exp Cell
Res. 320:12–20. 2014. View Article : Google Scholar
|
|
6
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Meng F, Henson R, Lang M, et al:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang H, Kong W, He L, et al: MicroRNA
expression profiling in human ovarian cancer: miR-214 induces cell
survival and cisplatin resistance by targeting PTEN. Cancer Res.
68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Passetti F, Ferreira CG and Costa FF: The
impact of microRNAs and alternative splicing in pharmacogenomics.
Pharmacogenomics J. 9:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ratner ES, Keane FK, Lindner R, et al: A
KRAS variant is a biomarker of poor outcome, platinum chemotherapy
resistance and a potential target for therapy in ovarian cancer.
Oncogene. 31:4559–4566. 2012. View Article : Google Scholar :
|
|
11
|
Rubatt JM, Darcy KM, Tian C, et al:
Pre-treatment tumor expression of ERCC1 in women with advanced
stage epithelial ovarian cancer is not predictive of clinical
outcomes: a Gynecologic Oncology Group study. Gynecol Oncol.
125:421–426. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rustin GJ, Nelstrop AE, Bentzen SM,
Piccart MJ and Bertelsen K: Use of tumour markers in monitoring the
course of ovarian cancer. Ann Oncol. 10:21–27. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rustin GJ, Nelstrop AE, McClean P, et al:
Defining response of ovarian carcinoma to initial chemotherapy
according to serum CA 125. J Clin Oncol. 14:1545–1551.
1996.PubMed/NCBI
|
|
14
|
Pérez C, Díaz-García CV, Agudo-López A, et
al: Evaluation of novel trans-sulfonamide platinum complexes
against tumor cell lines. Eur J Med Chem. 76:360–368. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gyori BM, Venkatachalam G, Thiagarajan PS,
Hsu D and Clement MV: OpenComet: An automated tool for comet assay
image analysis. Redox Biol. 2:457–465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
van Huis-Tanja LH, Kweekel DM, Lu X,
Franken K, et al: Excision Repair Cross-Complementation group 1
(ERCC1) C118T SNP does not affect cellular response to oxaliplatin.
Mutat Res Fundam Mol Mech Mutagen. 759:37–44. 2014. View Article : Google Scholar
|
|
17
|
Giovannetti E, Erozenci A, Smit J, Danesi
R and Peters GJ: Molecular mechanisms underlying the role of
microRNAs (miRNAs) in anticancer drug resistance and implications
for clinical practice. Crit Rev Oncol Hematol. 81:103–122. 2012.
View Article : Google Scholar
|
|
18
|
Li Z, Hu S, Wang J, Cai J, Xiao L, Yu L
and Wang Z: MiR-27a modulates MDR1/P-glycoprotein expression by
targeting HIPK2 in human ovarian cancer cells. Gynecol Oncol.
119:125–130. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shen S, Yue H, Li Y, Qin J, Li K, Liu Y
and Wang J: Upregulation of miR-136 in human non-small cell lung
cancer cells promotes Erk1/2 activation by targeting PPP2R2A.
Tumour Biol. 35:631–640. 2014. View Article : Google Scholar
|
|
20
|
Kweekel DM, Gelderblom H and Guchelaar HJ:
Pharmacology of oxaliplatin and the use of pharmacogenomics to
individualize therapy. Cancer Treat Rev. 31:90–105. 2005.
View Article : Google Scholar : PubMed/NCBI
|