Introduction
Forkhead transcription factors are characterized by
a winged helix DNA-binding domain and are essential for
embryogenesis (1). Some of the
members have been identified to regulate tumorigenesis and
progression, such as FoxO1 and FoxQ1 (2–4).
Decreased forkhead box F2 transcription factor (FoxF2) expression
was reported in prostate and breast cancer and was found to
correlate with early-onset metastasis and poor prognosis (5,6), which
implies the potential role of FoxF2 in tumorigenesis and
progression. Mechanistically, Ormestad et al found that
FoxF2 regulates embryonic gut development by controlling paracrine
crosstalk between the stroma and the epithelia (7). Inhibition of FoxF2 was found to
increase the mesenchymal expression of Wnt5a, which subsequently
activates the canonical Wnt signaling pathway to cause epithelial
depolarization and tissue disintegration. Notably, a quite recent
study demonstrated that mesenchymal FoxF2 restricted Lgr5-positive
stem cells of the crypt niche and reduced adenoma formation in
mouse intestines by inhibiting Wnt signaling (8). These results partly unveiled the
mechanism that underlies the initiation of intestinal carcinoma by
silencing of FoxF2 and indicate a tumor-suppressive role.
Accordingly, it would be worthy to determine the upstream
regulators and downstream effectors of the FoxF2 gene in tumor
development.
MicroRNAs (miRNAs) are a cluster of small non-coding
RNA molecules ~22 nucleotides in length. They repress the
expression of target genes by inducing mRNA degradation or by
blocking translation. Emerging evidence suggests that miRNAs are
involved in tumor development by functioning as oncogenes or tumor
suppressors, depending on their target genes and downstream
signaling pathways. miR-182 has been found to be aberrantly
overexpressed and promotes tumorigenesis in breast, ovarian and
prostate cancer (9–12). Moreover, miR-182 was also found to
facilitate metastasis in melanoma and breast cancer through
suppression of FOXO3 and MTSS1, respectively (13,14).
Importantly, Moskwa et al reported that miR-182 is
implicated in DNA damage response (15). They observed that overexpression of
miR-182 in sporadic breast cancer impeded homologous
recombination-mediated DNA repair by targeting BRCA1, but rendered
tumor cells hypersensitive to (ADP-ribose) polymerase 1 (PARP1)
inhibitor treatment. This indicates the potential of miR-182 as a
therapeutic target.
As shown in a meta-analysis, colorectal cancer (CRC)
consistently exhibits aberrant overexpression of miR-182 (16), which suggests its potential
oncogenic role in CRC. A recent study based on a comprehensive
miRNA library screen showed that aberrantly overexpressed miR-182
promotes cell proliferation and survival in CRC, and revealed a
significant inverse correlation of FoxF2 to miR-182 by employing
gene expression profiling data and target prediction tools
(17). We thus hypothesized that
aberrant expression of miR-182 in CRC promotes tumor development by
suppressing FoxF2.
Materials and methods
Tissue samples and cell lines
A panel of 33 pairs of primary CRCs and their
matched adjacent normal mucosa, and another independent panel of 49
specimens including 10 normal colon tissues and 39 CRCs classified
in different stages, were obtained from patients who underwent
surgical resections at the First People’s Hospital of Yunnan
Province, and were snap-frozen in liquid nitrogen, and then stored
at −80°C for further use. This project was approved by the Ethics
Committee of the First People’s Hospital of Yunnan Province.
Human CRC cell lines, including HT29, SW480, SW620
and HCT116, and human normal colon epithelial cell line FHC, were
purchased from the American Type Culture Collection (ATCC). CRC
cells were cultured routinely in RPMI-1640 medium (Invitrogen)
supplemented with 10% fetal bovine serum (Gibco) and cultured in a
37°C humidified atmosphere of 5% CO2. FHC cells were
cultured following the instructions of the ATCC.
Bioinformatic analyses
All analyses of microarray datasets were performed
using bioinformatic tool R2 (http://r2.amc.nl)
following the instructions.
Lentivirus packaging and stable cell line
establishment
The miR-182-knockdown or FoxF2-expressing lentivirus
particles were packaged by co-transfecting the miR-182 inhibitor
clone or the FoxF2-expressing clone with lentiviral packaging
plasmids into HEK293FT cells, using Lenti-Pac™ HIV Expression
Packaging Systems (both from GeneCopoeia) according to the
manufacturer’s instructions. For the lentivirus infection, cells
were incubated with viral supernatant in the presence of 8
μg/ml Polybrene for 24 h, followed by hygromycin or
puromycin (Invitrogen) selection until drug-resistant colonies
became visible.
RNA extraction and quantitative
RT-PCR
Total RNA and miRNA were isolated using the
RNeasy® Mini kit and miRNeasy® Mini kit
(Qiagen) respectively. Mature miR-182 expression in cells was
determined using the Hairpin-it™ miRNAs qPCR kit (GenePharma,
China). RNU6B was used as an endogenous control. mRNA expression
was determined using the SYBR-Green qPCR assay (Takara). Data were
analyzed using the 2−ΔΔCt method.
Cell growth and invasion assays
Cell growth was detected using an MTT cell
proliferation assay kit (ATCC) according to the manufacturer’s
instructions. Cell invasion ability was measured using Transwell
inserts with 8-μm pores (Corning). Briefly, the cell density
was adjusted to 106/ml in serum-free RPMI-1640 medium.
The cell suspension (200 μl) was added into each upper
insert pre-coated with Matrigel matrix (BD), while 500 μl
RPMI-1640 medium containing 10% fetal bovine serum (FBS) was added
into a matched lower chamber. After a 48-h incubation, the
non-invaded cells were removed from the upper surface of the
Transwell membrane with a cotton swab, while the invaded cells on
the lower membrane surface were fixed in methanol, stained with
0.1% crystal violet, photographed and counted. Six random fields at
×100 magnification for each insert were observed.
Western blot analysis
Cultured cells were lysed in RIPA buffer with 1%
phenylmethylsulphonyl fluoride (PMSF). Protein was loaded onto an
SDS-PAGE Minigel and transferred onto a PVDF membrane. After being
probed with 1:500 diluted anti-FoxF2 (Abcam) and 1:1,000 diluted
anti-β-catenin (BD) at 4°C overnight, the blots were subsequently
incubated with HRP-conjugated anti-IgG (Cell Signaling). Signals
were visualized using ECL substrates (Pierce). α-tubulin (Santa
Cruz) was used as an endogenous protein for normalization.
Colony formation assay
Cells were trypsinized and plated on 6-well plates
(300 cells for each well) and cultured for 10 days. The colonies
were stained with 0.1% crystal violet solution containing 80%
methanol for 5 min. The number of colonies defined as >50
cells/colony were counted.
Luciferase reporter assay
A fragment of the wild-type 3′ untranslated region
(3′UTR) of FoxF2 containing the putative miR-182 binding site was
amplified by PCR. The PCR product was subcloned into a psiCHECK-2
vector (Promega) immediately downstream to the luciferase gene
sequence. A corresponding psiCHECK-2 construct containing 3′UTR of
FoxF2 with a mutant seed sequence of miR-82 was also
synthesized.
HEK293 cells were plated in 96-well clusters, and
then co-transfected with 100 ng constructs with or without miR-182
mimics. At 48 h post-transfection, luciferase activity was detected
using a Dual-Luciferase Reporter Assay System (Promega) and
normalized to Renilla activity.
Statistical analyses
Statistical analyses were performed using the SPSS
15.0.0 and GraphPad Prism software. Comparisons between the two
groups were performed by the Student’s t-test or the Mann-Whitney U
test. Comparisons among multiple groups were performed by one-way
ANOVA. A P-value of <0.05 was considered to indicate a
statistically significant result.
Results
FoxF2 expression is downregulated in CRC
and is negatively associated with β-catenin
FoxF2 has been reported to reduce intestinal adenoma
formation, by antagonizing the activity of the Wnt/β-catenin
pathway through upregulating the expression of SFRP1 which encodes
an inhibitor of Wnt (8), yet
knowledge concerning FoxF2 in human CRC is sparse. Therefore, we
first analyzed the gene expression profiles of different human CRC
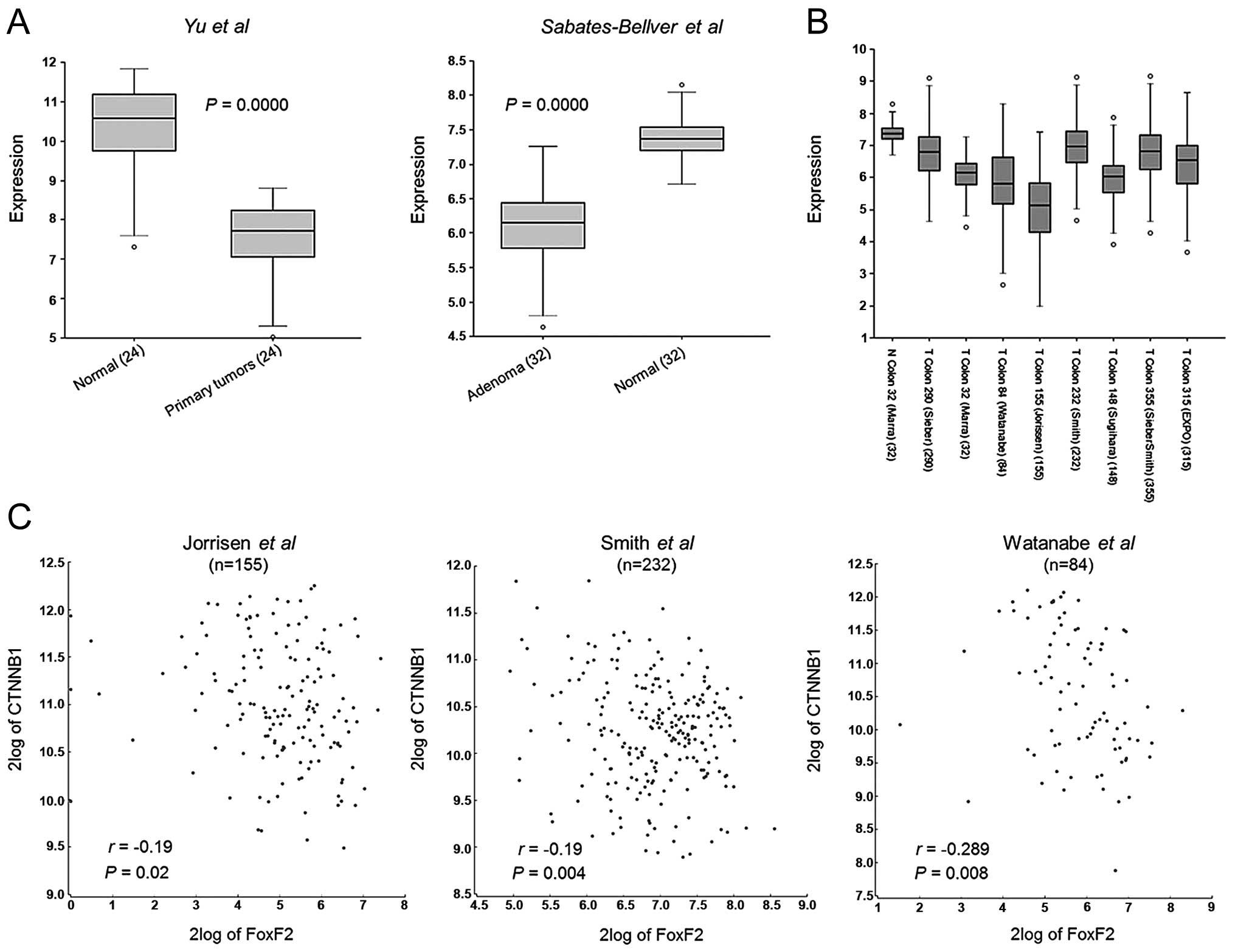
cohorts to observe the status of FoxF2 expression. As shown in
Fig. 1A, FoxF2 was significantly
downregulated in colorectal adenomas (18) and primary tumors (19) when compared with the paired normal
colon tissues. In the case of multiple dataset analysis, FoxF2
expression was commonly lower in primary CRC, compared with the
cohort of normal colon tissues [N Colon 32 (Mara) (18)] (Fig.
1B). Additionally, FoxF2 expression was observed to be
negatively associated with oncogenic β-catenin expression in
several datasets (20–23) (Fig.
1C).
Restoration of FoxF2 inhibits CRC cell
growth and invasion by antagonizing β-catenin activity
We next attempted to investigate the role of FoxF2
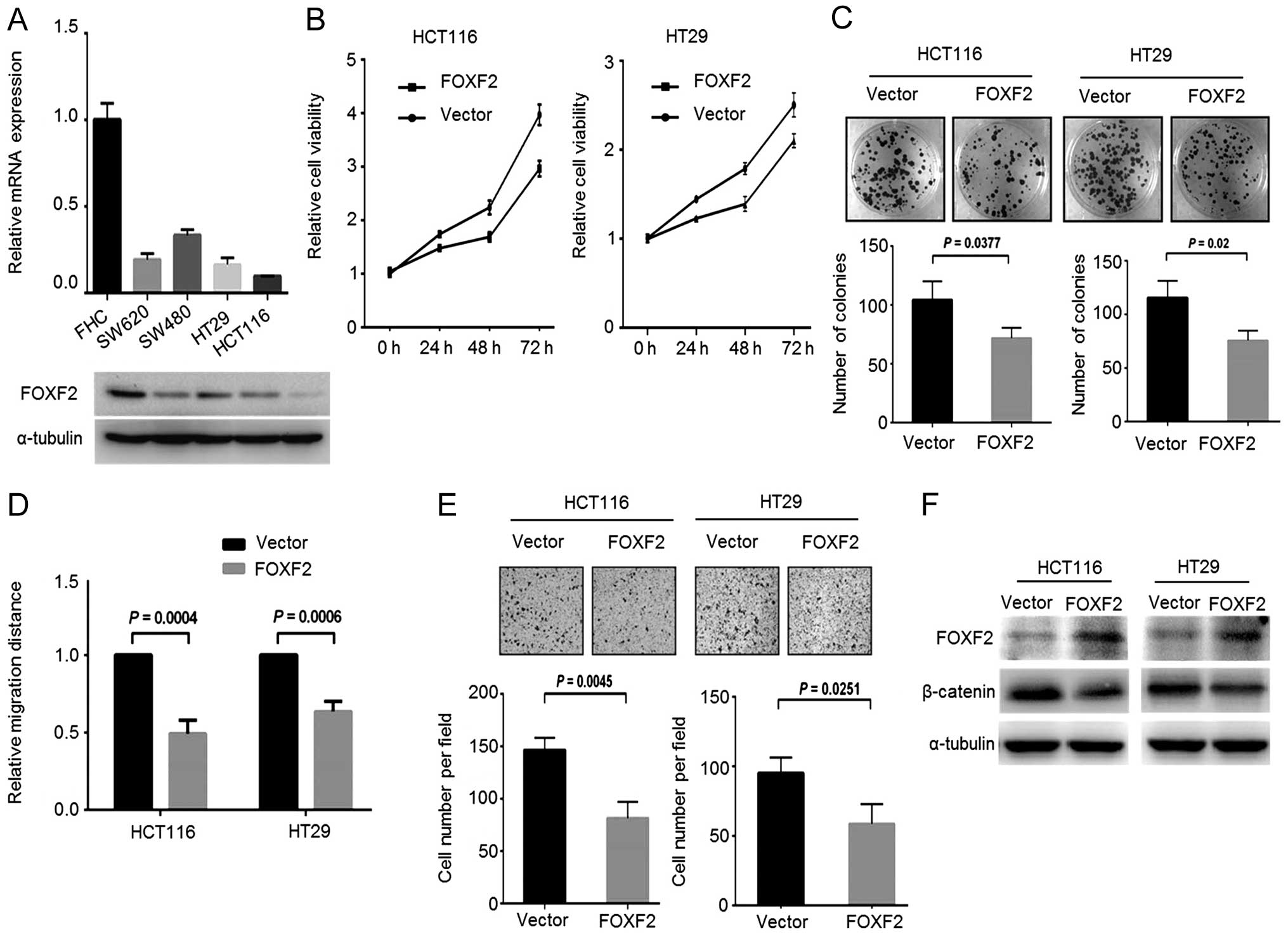
in CRC progression and the potential mechanism. We screened the
FoxF2 expression in different CRC cell lines, and observed a
notable decrease in FoxF2 expression both at the mRNA and protein
levels in CRC cells compared with the normal colon epithelial cell
line FHC (Fig. 2A). Stable
FoxF2-expressing CRC cells were generated by introducing
FoxF2-expressing lentiviral particles into HCT116 and HT29 cells
which express relatively lower FoxF2. In the gain-of-function
assays, upregulation of FoxF2 reduced cell growth, colony formation
(Fig. 2B and C) and impaired cell
motility including migration (Fig.
2D) and invasion (Fig. 2E). By
immunoblotting, in the FoxF2-expressing stable cells, β-catenin was
downregulated upon the restoration of FoxF2 (Fig. 2F), demonstrating the antagonistic
effect of FoxF2 on β-catenin activity in CRC.
miR-182 is aberrantly upregulated in CRC
tissues and cell lines
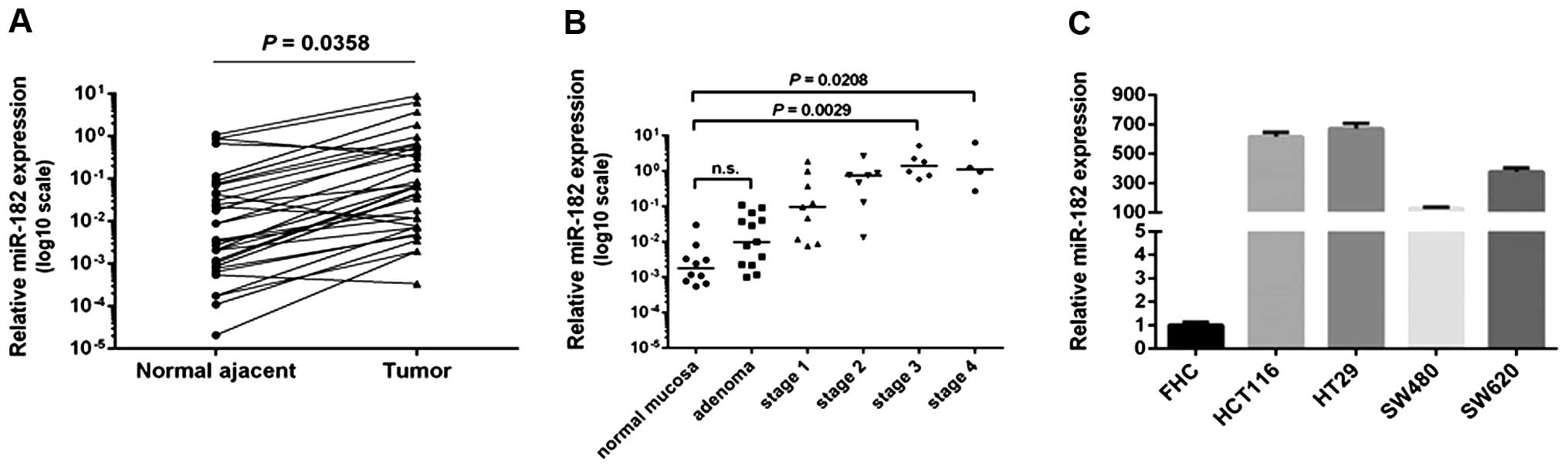
To demonstrate the expression pattern of miR-182 in
CRC, we performed quantitative PCR to detect the miR-182 level in
both clinical specimens and cell lines. In the cohort of 33 cases
of paired CRC tissues and adjacent colon epithelia, miR-182 was
significantly upregulated in 26 (78.8%) tumors compared to that in
the matched normal epithelia (Fig.
3A). In further analysis of miR-182 expression in 49 colon
specimens (10 normal colon tissues and 39 CRCs diagnosed at
different clinical stages), miR-182 in the advanced stage CRCs (III
and IV) was significantly increased compared to the normal colon
tissues and the early stage tumors, while no alteration of miR-182
level was observed in the adenomas (Fig. 3B).
We further examined the miR-182 expression level in
the CRC cell lines: HCT116, HT29, SW480 and SW620, in comparison
with that in the normal colonic epithelia cell line FHC. As
expected, miR-182 was consistently overexpressed in all of the 4
CRC cell lines (Fig. 3C). Since
HCT116 and HT29 cells expressed relatively high levels of miR-182,
they were selected for further functional assays.
FoxF2 is a direct target gene of miR-182
in CRC
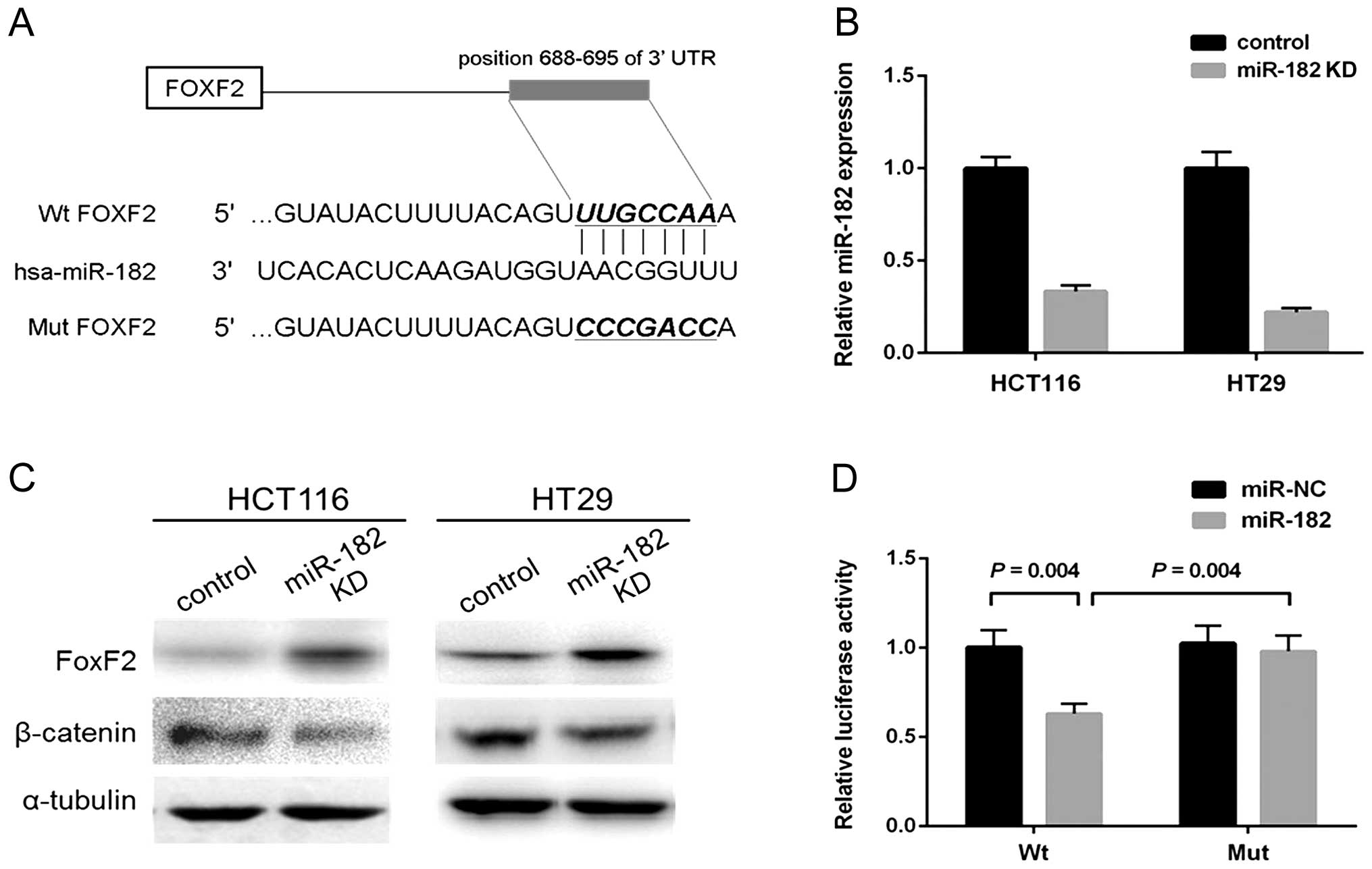
In an integrative analysis, miR-182 was predicted to
inversely correlate with FoxF2 that has a putative binding site of
miR-182 (17). A recent study on
prostate cancer further validated that miR-182 could target FoxF2
to promote cell invasion and proliferation (24). By utilizing bioinformatic algorithms
(Targetscan/Pictar/miRecords), we noted that FoxF2 was a highly
potential target gene of miR-182. The predicted binding site of
miR-182 within 3′UTR of FoxF2 mRNA is shown in Fig. 4A. Since CRC cell lines exhibited a
significant decrease in FoxF2 expression both at the mRNA and
protein levels, we thus hypothesized a link between miR-182 and
FoxF2. For further assays, we generated stable miR-182-knockdown
CRC cells by infecting HCT116 and HT29 cells with lentiviral
particles. The efficiency of knockdown of miR-182 expression was
validated by qPCR (Fig. 4B).
Immunoblotting showed that FoxF2 was notably upregulated in the
miR-182-knockdown HCT116 and HT29 cells, while reduced β-catenin
was also observed simultaneously (Fig.
4C). To further investigate whether the predicted binding site
of miR-182 to 3′UTR of FoxF2 is responsible for the upregulation,
we cloned the 3′UTR of FoxF2 into a luciferase reporter vector
(wt-FoxF2); a corresponding mutant version (mut-FoxF2) was also
constructed. We co-transfected wt-FoxF2 and miR-182 mimics or
scramble control into the HEK293 cells. The luciferase activity of
the miR-182-transfected cells was significantly reduced compared to
the scramble control cells (Fig.
4D). Moreover, miR-182-mediated repression of luciferase
activity was abolished by the mutant putative binding site
(Fig. 4D).
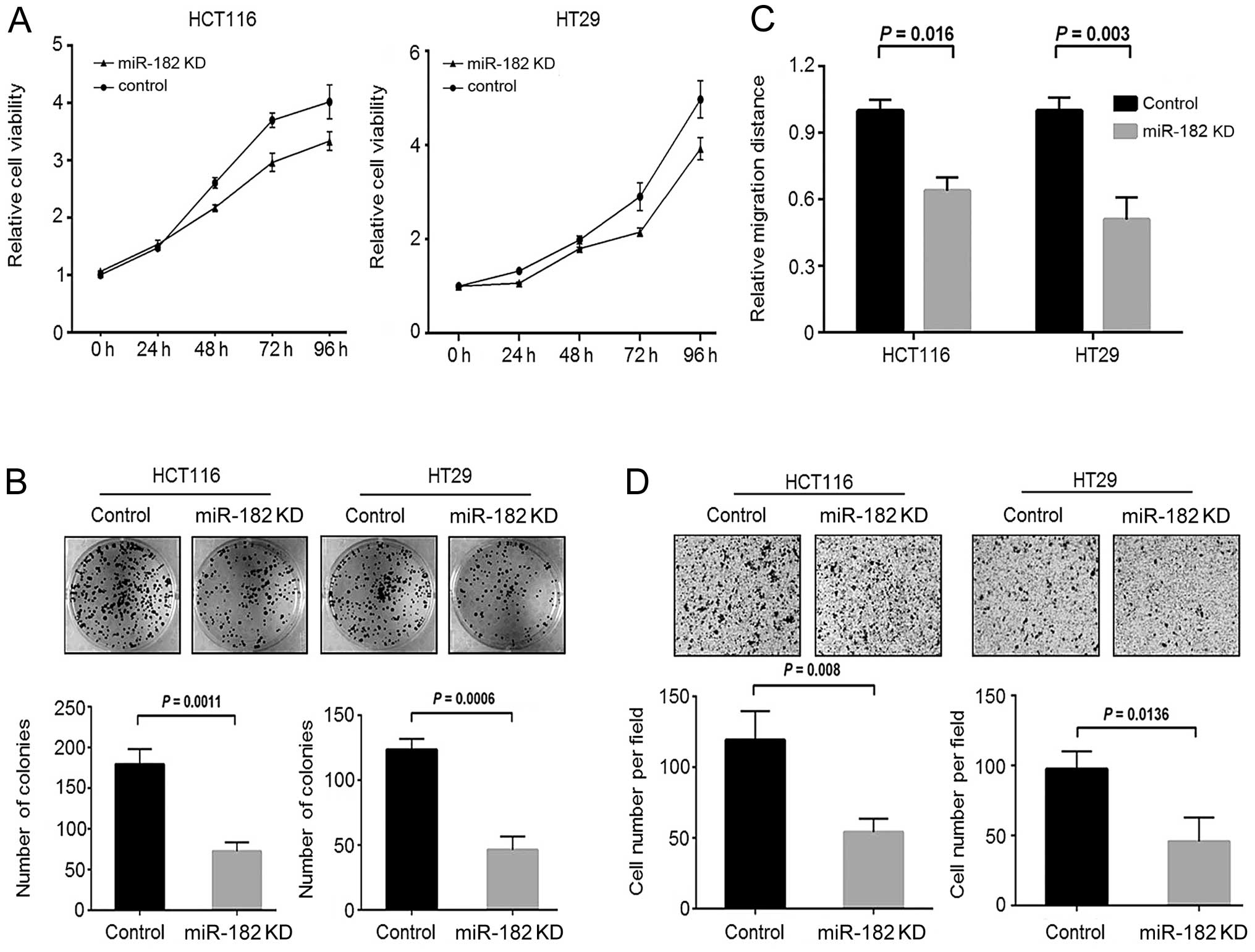
Knockdown of miR-182 suppresses CRC cell
growth and invasion in vitro
To explore whether miR-182 contributes to oncogenic
phenotypes of CRC, we performed loss-of-function assays on
miR-182-knockdown CRC cells. By an MTT assay, we observed that
downregulation of miR-182 significantly reduced the cell growth of
CRC cells compared to the scramble control cells (Fig. 5A). Similarly, we also observed a
decrease in colony formation activity in the miR-182-knockdown
cells (Fig. 5B). These findings
were similar to those presented for the stable FoxF2-expressing CRC
cells. Furthermore, we performed wound healing and Transwell
invasion assays to investigate the involvement of miR-182 in cell
motility of CRC. As shown in Fig. 5C
and D, downregulation of miR-182 significantly reduced the cell
migration and cell invasive capacity of CRC cells to migrate
through Matrigel. These findings indicate that aberrant
upregulation of miR-182 is pro-oncogenic and may confer malignant
potential to CRC cells.
Discussion
Growing evidence has linked the expression of
miR-182 to multiple types of tumors including prostate cancer,
melanoma, breast cancer and glioma (12–14,25),
although studies have reported different, even contradictory, roles
of miR-182 in tumorigenesis in different tumor models (11,14,26).
In CRC, miR-182 was shown to be aberrantly upregulated as well as
in precursor lesions (27), and its
upregulation was significantly associated with lymph node
metastasis, TNM stage and poor prognosis (28). Diep et al reported that the
miR-182 gene was amplified in 26% of primary CRC tumors and 30% of
liver metastases (29). It was
further demonstrated that miR-182 reduced the apoptosis of CRC
cells by performing RNAi screening and functional validation
(17), which functionally suggests
the oncogenic role of miR-182 in CRC. In the present study, we
observed the increased expression of miR-182 in adenomas and
primary CRCs compared to normal colon mucosa. Moreover, increased
miR-182 was correlated with advanced tumor stage, which is similar
to the previous study. Simultaneous suppression of in vitro
cell growth, colony formation, migration, invasion of CRC cells is
shown upon inhibition of miR-182. Collectively with the previous
report, our results highlight the functional characteristics of
miR-182 as an oncomiR in CRC.
Despite that miR-182 has been validated to target
several tumor suppressors including PTEN, MTSS1, Slug and CYLD in
different types of cancers, we sought to reveal the mechanistic
link of miR-182 to target genes contributing to CRC development. We
observed that FoxF2, a member of the forkhead box (FOX) family of
transcription factors, is highly predicted as a miR-182 target
using several bioinformatic algorithms. In previous studies, FoxF2
was described to promote organ development, extracellular matrix
(ECM) synthesis and epithelial-mesenchymal interaction (7,30,31).
It can be activated by hedgehog signaling, and subsequently limits
Bmp and Wnt signaling (7). FoxF2 in
normal prostate stroma could facilitate stomal homeostasis through
upregulation of CXCL12 and regulation of ECM remodeling via
balancing MMPs and TIMPs (5). A
recent study showed that a low level of FoxF2 mRNA is associated
with early-onset metastasis and poor prognosis of patients with
histological grade II and triple-negative breast cancer (6), indicating its clinical value as a
prognostic marker. We demonstrated that FoxF2 is downregulated in
primary CRC tissues and CRC cell lines. Restoration of FoxF2 in CRC
cells significantly suppressed cell growth, colony formation and
cell motility, indicating the tumor-suppressive role of FoxF2.
Since mesenchymal FoxF2 restricted Lgr5-positive stem cells of the
crypt niche and reduced adenoma formation in mouse intestines by
inhibiting Wnt/β-catenin signaling (8), we hypothesized that loss of FoxF2 may
result in increased β-catenin expression and activity. Indeed,
β-catenin expression was experimentally detected to decrease in the
FoxF2-expressing CRC cells as well as in the miR-182-knockdown
cells. Given that miR-182 binds to the 3′UTR of FoxF2 mRNA thus
leading to a decrease in FoxF2 expression, we demonstrated that
aberrantly elevated miR-182 is responsible, at least partly, for
the FoxF2 silencing and increased β-catenin activity in CRC.
More intriguingly, recent studies have shown that
miR-182 stimulates nuclear β-catenin translocation through
repressing SMAD4 in bladder cancer (32), while miR-182 is transactivated by
β-catenin in breast cancer (10),
indicating the important role of the β-catenin/miR-182 feedback
loop in tumor progression. Our results provide evidence for FoxF2
as a mediator involved in this oncogenic loop. In addition, miR-182
is shown to repress RECK, an antagonist of matrix
metalloproteinases (MMPs), while FoxF2 upregulated the expression
of the tissue inhibitor of metalloproteinase 3 (TIMP3), another
strong antagonist of MMPs (5). This
suggests that the miR-182/FoxF2 link may play a role in regulating
ECM remodeling to facilitate tumor cell invasion and
metastasis.
In conclusion, the present study identifies the
regulatory link between miR-182 and FoxF2, and describes a
potential mechanism underlying FoxF2 dysregulation and its
contribution to CRC progression. Additionally, miR-182-induced
downregulation of FoxF2 may partly account for the increased
activity of β-catenin signaling. Inhibition of miR-182 represents a
potential strategy against CRC.
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (81260323), and the
Joint Foundation of Kunming Medical University and Yunnan
Provincial Science and Technology Department (2012FB095).
References
|
1
|
Kaufmann E and Knöchel W: Five years on
the wings of fork head. Mech Dev. 57:3–20. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dong XY, Chen C, Sun X, Guo P, Vessella
RL, Wang RX, Chung LW, Zhou W and Dong JT: FOXO1A is a candidate
for the 13q14 tumor suppressor gene inhibiting androgen receptor
signaling in prostate cancer. Cancer Res. 66:6998–7006. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang H, Meng F, Liu G, Zhang B, Zhu J, Wu
F, Ethier SP, Miller F and Wu G: Forkhead transcription factor
Foxq1 promotes epithelial-mesenchymal transition and breast cancer
metastasis. Cancer Res. 71:1292–1301. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qiao Y, Jiang X, Lee ST, Karuturi RK, Hooi
SC and Yu Q: FOXQ1 regulates epithelial-mesenchymal transition in
human cancers. Cancer Res. 71:3076–3086. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van der Heul-Nieuwenhuijsen L, Dits N, Van
Ijcken W, de Lange D and Jenster G: The FOXF2 pathway in the human
prostate stroma. Prostate. 69:1538–1547. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kong PZ, Yang F, Li L, Li XQ and Feng YM:
Decreased FOXF2 mRNA expression indicates early-onset metastasis
and poor prognosis for breast cancer patients with histological
grade II tumor. PLoS One. 8:e615912013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ormestad M, Astorga J, Landgren H, Wang T,
Johansson BR, Miura N and Carlsson P: Foxf1 and Foxf2 control
murine gut development by limiting mesenchymal Wnt signaling and
promoting extracellular matrix production. Development.
133:833–843. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nik AM, Reyahi A, Pontén F and Carlsson P:
Foxf2 in intestinal fibroblasts reduces numbers of Lgr5+
stem cells and adenoma formation by inhibiting Wnt signaling.
Gastroenterology. 144:1001–1011. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guttilla IK and White BA: Coordinate
regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast
cancer cells. J Biol Chem. 284:23204–23216. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chiang CH, Hou MF and Hung WC:
Up-regulation of miR-182 by β-catenin in breast cancer increases
tumorigenicity and invasiveness by targeting the matrix
metalloproteinase inhibitor RECK. Biochim Biophys Acta.
1830:3067–3076. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu Z, Liu J, Segura MF, Shao C, Lee P,
Gong Y, Hernando E and Wei JJ: MiR-182 overexpression in
tumourigenesis of high-grade serous ovarian carcinoma. J Pathol.
228:204–215. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tsuchiyama K, Ito H, Taga M, Naganuma S,
Oshinoya Y, Nagano K, Yokoyama O and Itoh H: Expression of
microRNAs associated with Gleason grading system in prostate
cancer: miR-182-5p is a useful marker for high grade prostate
cancer. Prostate. 73:827–834. 2013. View Article : Google Scholar
|
|
13
|
Segura MF, Hanniford D, Menendez S, et al:
Aberrant miR-182 expression promotes melanoma metastasis by
repressing FOXO3 and microphthalmia-associated transcription
factor. Proc Natl Acad Sci USA. 106:1814–1819. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lei R, Tang J, Zhuang X, Deng R, Li G, Yu
J, Liang Y, Xiao J, Wang HY, Yang Q and Hu G: Suppression of MIM by
microRNA-182 activates RhoA and promotes breast cancer metastasis.
Oncogene. 33:1287–1296. 2014. View Article : Google Scholar
|
|
15
|
Moskwa P, Buffa FM, Pan Y, et al:
miR-182-mediated downregulation of BRCA1 impacts DNA repair and
sensitivity to PARP inhibitors. Mol Cell. 41:210–220. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang QH, Sun HM, Zheng RZ, Li YC, Zhang
Q, Cheng P, Tang ZH and Huang F: Meta-analysis of microRNA-183
family expression in human cancer studies comparing cancer tissues
with noncancerous tissues. Gene. 527:26–32. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cekaite L, Rantala JK, Bruun J, et al:
MiR-9, -31, and -182 deregulation promote proliferation and tumor
cell survival in colon cancer. Neoplasia. 14:868–879.
2012.PubMed/NCBI
|
|
18
|
Sabates-Bellver J, Van der Flier LG, de
Palo M, et al: Transcriptome profile of human colorectal adenomas.
Mol Cancer Res. 5:1263–1275. 2007. View Article : Google Scholar
|
|
19
|
Jiang X, Tan J, Li J, et al: DACT3 is an
epigenetic regulator of Wnt/β-catenin signaling in colorectal
cancer and is a therapeutic target of histone modifications. Cancer
Cell. 13:529–541. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jorissen RN, Lipton L, Gibbs P, et al: DNA
copy-number alterations underlie gene expression differences
between micro-satellite stable and unstable colorectal cancers.
Clin Cancer Res. 14:8061–8069. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smith JJ, Deane NG, Wu F, et al:
Experimentally derived metastasis gene expression profile predicts
recurrence and death in patients with colon cancer.
Gastroenterology. 138:958–968. 2010. View Article : Google Scholar
|
|
22
|
Watanabe T, Kobunai T, Toda E, et al:
Distal colorectal cancers with microsatellite instability (MSI)
display distinct gene expression profiles that are different from
proximal MSI cancers. Cancer Res. 66:9804–9808. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tsukamoto S, Ishikawa T, Iida S, Ishiguro
M, Mogushi K, Mizushima H, Uetake H, Tanaka H and Sugihara K:
Clinical significance of osteoprotegerin expression in human
colorectal cancer. Clin Cancer Res. 17:2444–2450. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hirata H, Ueno K, Shahryari V, Deng G,
Tanaka Y, Tabatabai ZL, Hinoda Y and Dahiya R: MicroRNA-182-5p
promotes cell invasion and proliferation by down regulating FOXF2,
RECK and MTSS1 genes in human prostate cancer. PLoS One.
8:e555022013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang L, Mao P, Song L, Wu J, Huang J, Lin
C, Yuan J, Qu L, Cheng SY and Li J: miR-182 as a prognostic marker
for glioma progression and patient survival. Am J Pathol.
177:29–38. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun Y, Fang R, Li C, Li L, Li F, Ye X and
Chen H: Hsa-mir-182 suppresses lung tumorigenesis through down
regulation of RGS17 expression in vitro. Biochem Biophys Res
Commun. 396:501–507. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sarver AL, French AJ, Borralho PM, et al:
Human colon cancer profiles show differential microRNA expression
depending on mismatch repair status and are characteristic of
undifferentiated proliferative states. BMC Cancer. 9:4012009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu H, Du L, Wen Z, et al: Up-regulation
of miR-182 expression in colorectal cancer tissues and its
prognostic value. Int J Colorectal Dis. 28:697–703. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Diep CB, Kleivi K, Ribeiro FR, Teixeira
MR, Lindgjaerde OC and Lothe RA: The order of genetic events
associated with colorectal cancer progression inferred from
meta-analysis of copy number changes. Genes Chromosomes Cancer.
45:31–41. 2006. View Article : Google Scholar
|
|
30
|
Wang T, Tamakoshi T, Uezato T, Shu F,
Kanzaki-Kato N, Fu Y, Koseki H, Yoshida N, Sugiyama T and Miura N:
Forkhead transcription factor Foxf2 (LUN)-deficient mice exhibit
abnormal development of secondary palate. Dev Biol. 259:83–94.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Aitola M, Carlsson P, Mahlapuu M, Enerback
S and Pelto-Huikko M: Forkhead transcription factor FoxF2 is
expressed in mesodermal tissues involved in epithelio-mesenchymal
interactions. Dev Dyn. 218:136–149. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hirata H, Ueno K, Shahryari V, Tanaka Y,
Tabatabai ZL, Hinoda Y and Dahiya R: Oncogenic miRNA-182-5p targets
Smad4 and RECK in human bladder cancer. PLoS One. 7:e510562012.
View Article : Google Scholar : PubMed/NCBI
|