1. Introduction
Acute myeloid leukemia (AML) is characterized by
uncontrolled proliferation of myeloid progenitors that exhibit a
severe block in their ability to differentiate into mature
granulocytes or macrophages (1).
The transcription factor CCAAT/enhancer binding protein α (C/EBPα)
is a lineage-specific transcription factor in the hematopoietic
system and is required for the formation of committed myeloid
progenitors from multipotent precursor cells by coupling the direct
transcriptional activation of myeloid-specific genes with the
arrest of cell proliferation (2).
C/EBPα is specifically expressed in granulocytes,
monocytes and eosinophils (3),
although it is also found in hepatocytes, adipocytes and type II
pneumocytes (4). Previous studies
have illustrated the function of C/EBPα in hematopoiesis by
promoting granulocyte and monocyte differentiation (2,5,6).
Studies have eported that C/EBPα expression is detectable at a low
level in the hematopoietic stem cell (HSC) population, and its
expression increases as these cells develop into the common myeloid
progenitor (CMP) and subsequently the granulocyte-monocyte
progenitor (GMP), while conditional C/EBPα deficiency in adult mice
blocked the transition from CMP to GMP, resulting in reduced
formation of both granulocytes and monocytes (7). Non-conditional targeted disruption of
C/EBPα was found to result in a selective block in early
granulocyte maturation, and these mice died at birth due to severe
hypoglycemia (8). Moreover,
knock-in mice with a targeted mutation in the C/EBPα basic region,
which led to its dysfunction, predisposed the mice to a
myeloproliferative disorder (9).
However, when expressed in 32Dcl3 cells, representative of
granulocytic progenitors, exogenous C/EBPα promoted granulopoiesis
(10). These studies suggest that
C/EBPα is a critical regulator of myeloid development. In fact,
growing evidence suggests that the function of C/EBPα is critically
altered in subsets of AML patients based on various factors.
In this review, we summarized the fundamental role
of C/EBPα in myeloid differentiation and the recently identified
mechanisms of its activity.
2. Function of C/EBPα in myeloid
differentiation
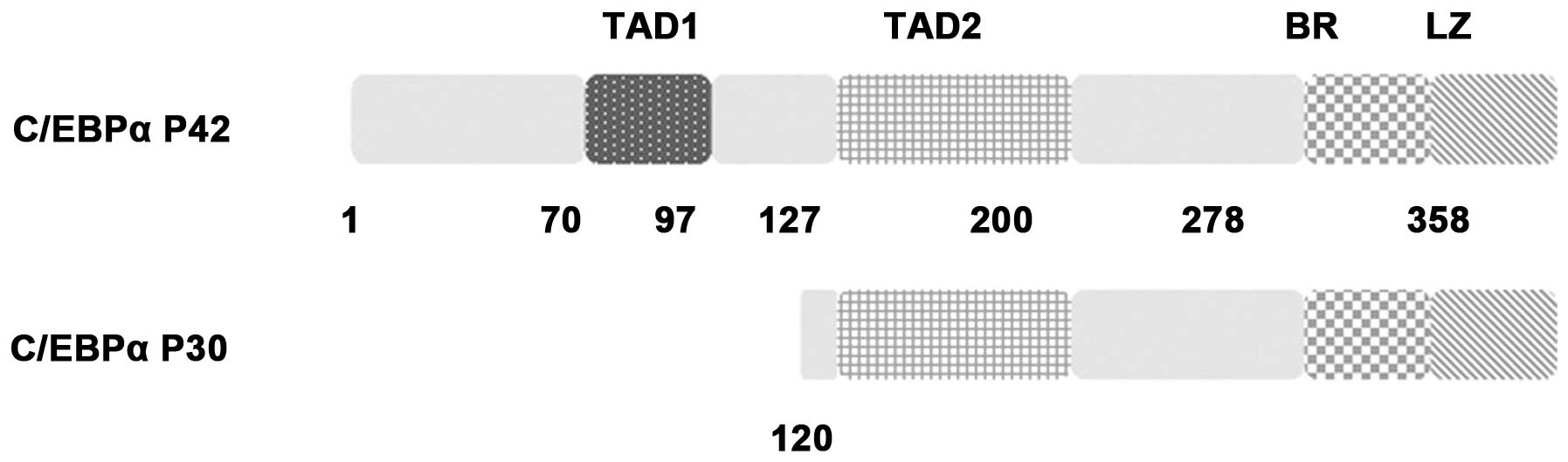
C/EBPα is a member of the basic leucine zipper
(bZIP) transcription factor family, of which several members are
also expressed in the myeloid lineage (e.g., C/EBPβ and C/EBPε)
(2). In mammals, C/EBPα is a
lineage-specific transcription factor that is required for the
formation of committed myeloid progenitors from multipotent
precursor cells. The C/EBPα molecule contains transactivation
domains (TADs) at its N-terminus and a DNA-binding and dimerization
bZIP structure at its C-terminus. Furthermore, C/EBPα is an
intronless gene whose mRNA can be translated from two different AUG
codons giving rise to two distinct isoforms (p42 and p30). p30
lacks two N-terminal transactivation domains that are only present
on p42 (11) (Fig. 1). Unless otherwise indicated, C/EBPα
represents the p42 isoform in the present review.
To date, numerous studies have been reported
regarding the function of C/EBPα in AML. Firstly, genomic mutations
have been detected in the C/EBPα gene in ~5–14% of AML patients
(12,13). Among these mutations, N-terminal
frameshift mutations prematurely truncate the full-length p42 form
while preserving the p30 form, with the latter inhibiting the
remaining wild-type C/EBPα p42 protein in a dominant-negative
manner (12). In addition,
C-terminal in-frame insertions or deletions were found to disrupt
the basic zipper region, thus critically affecting DNA binding
(12,14), which was further illustrated by
reports that the critical basic region residues of the C/EBPα
protein-DNA interaction were identified by an X-ray structure assay
(14). Although there is a
predominant C/EBPα mutation pattern in AML patients, the majority
of AML patients always have more than one C/EBPα mutation (13,15).
In addition, reduced expression could disrupt the function of
C/EBPα for normal hematopoiesis (16). A detailed study showed that deletion
of the C/EBPα gene led to arrest at the CMP to GMP transition,
thereby causing reduced formation of both granulocytes and
monocytes (7). In addition, studies
have reported that C/EBPα heterodimerizes with AP-1 proteins
(C/EBPα:c-Jun or C/EBPα:-c-Fos) for preference in directing
monopoiesis (17), whereas C/EBPα
homodimers cooperate with NF-κB p50 to promote granulopoiesis
(18). Regulation of macrophage and
neutrophil cell fates may also be altered by the relative
PU.1:C/EBPα ratio (19).
3. Regulation of the C/EBPα signaling
pathway
As a critical factor involved in myeloid
differentiation, the function of C/EBPα must be tightly regulated
to maintain the differentiation homeostasis. Detailed study of the
regulation of C/EBPα could also highlight the understanding of the
pathological mechanisms of AML.
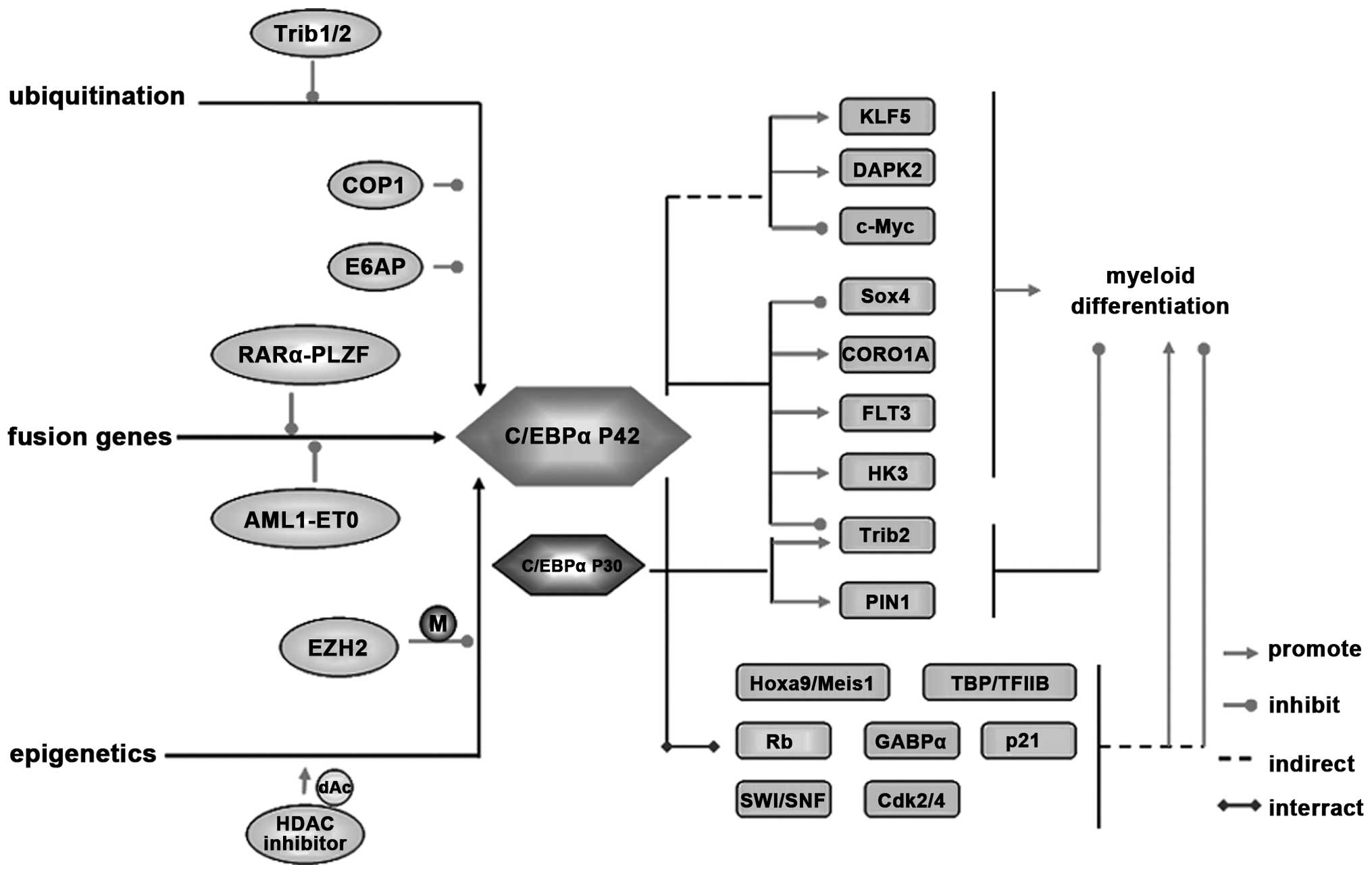
Regulators targeting C/EBPα in myeloid
differentiation and leukemogenesis
Many researches have focused on the factors
regulating C/EBPα expression and its functions in leukemic diseases
from different aspects, such as transcriptional repression by
fusion genes, ubiquitination modification and epigenetic
regulation.
Among the variant translocations in AML, the
t(11:17) translocation is the most frequent, and renders resistance
to all-trans retinoic acid (ATRA) treatment (20). After translocation, RARα is fused to
PLZF to produce two fusion proteins, promyelocytic eukemia zinc
finger-retinoic acid receptor α (PLZF-RARα) and RARα-PLZF, both of
which participate in leukemia development (21). Among them, RARα-PLZF recruits HDAC1
and causes histone H3 deacetylation at C/EBPα target loci, thereby
decreasing the expression of C/EBPα (Fig. 2). In line with this result, HDAC
inhibitors were found to restore C/EBPα expression to a modest
extent (22). In addition to
RARα-PLZF, C/EBPα expression could also be downregulated through
direct transcriptional repression by the fusion oncoprotein ML1-ETO
(23). These findings provide
molecular evidence for a mechanism through which fusion proteins
act as modifier oncogenes that subvert differentiation in the
granulocytic lineage by inhibiting the activity of C/EBPα.
Epigenic modification is envisioned as an important
epigenetic mechanism that regulates the expression of
myeloid-specific genes in the hematopoietic system during
leukemogenesis (24).
Hypermethylation of the C/EBPα promoter was first reported
preferentially in AML-M2 patients (25). The methylation status of the C/EBPα
gene in chronic myeloid leukemia (CML) patients was also
investigated, and the data suggested that aberrant methylation in
the CpG island of the C/EBPα gene promoter could be a common event
in CML (26). The role of EZH2 in
hematopoietic development and leukemia is still controversial. Some
groups suggest that EZH2 acts as a tumor suppressor in the myeloid
lineage (27). However, it was also
reported that ectopic expression of EZH2 causes a block in myeloid
differentiation (28). A recent
study found that EZH2 functions as an oncogene to block
differentiation through suppressing C/EBPα expression. Notably,
C/EBPα was found to be downregulated by EZH2 through methylation
modification of its promoter in MA9-induced leukemia (29). Consistent with the above data, HDAC
inhibitors restore C/EBPα target gene expression (22). A highly significant association was
found between the frequency of C/EBPα gene epigenic modification
and myeloid leukemia, while the role of C/EBPα
methylation/deacetylation in the development, progression and
prognosis in myeloid leukemia warrants further research.
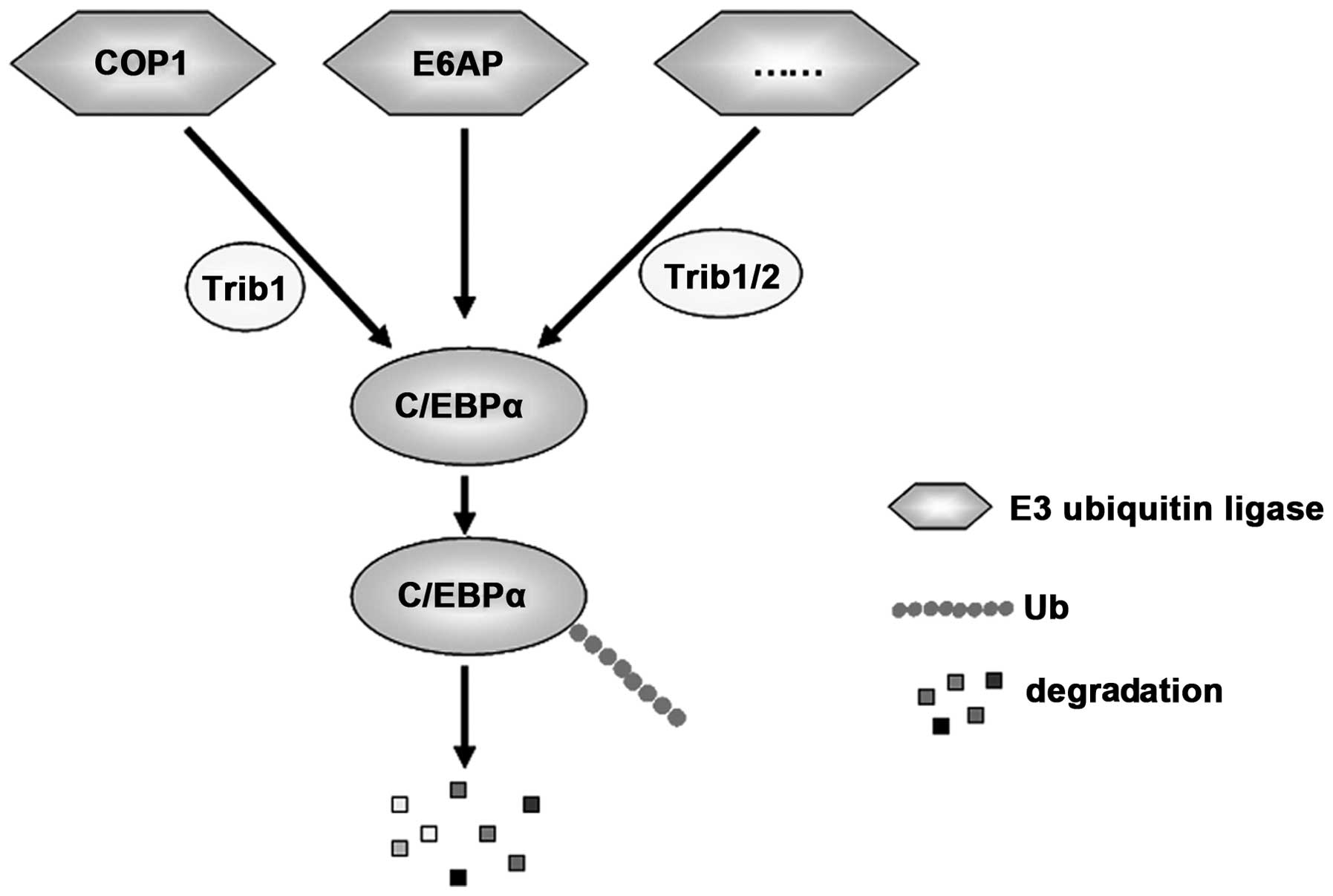
Ubiquitination is an essential posttranslational
modification for the modulation of C/EBPα activity. E3 ligases
specifically targeting lysine 48 (K48)-linked ubiquitination of
C/EBPα could promote the degradation of C/EBPα through the
proteasome and thus terminate the downstream signaling
transduction. Trib1 and Trib2 are two members of the Tribbles
family that function as adapters to recruit E3 ubiquitin ligases
and enhance ubiquitylation of its target protein. Consequently,
Trib1 and Trib2 induce C/EBPα degradation and inhibit its function
(30). E3 ubiquitin ligases,
constitutively photomorphogenic 1 (COP1) (31) and E6-associated protein (E6AP)
(32), were found to promote the
degradation of C/EBPα by promoting its K48-linked
polyubiquitination, thereby blocking myeloid differentiation of
hematopoietic cells for tumorigenesis. Notably, during this
process, COP1, which contains a COP1-binding motif, is recruited by
Trib1 and is essential for downregulation of C/EBPα expression
(31) (Fig. 3). However, whether any
deubiquitinating enzymes exist to specifically remove K48-linked
ubiquitination of C/EBPα and stabilize its expression or whether E3
ligases exist to induce K63-linked polyubiquitination to active
C/EBPα function warrant further investigation. The answer to this
question may help to elucidate the complexities of modulation of
C/EBPα activity through ubiquitination/deubiquitination.
Downstream regulators targeting C/EBPα in
myeloid differentiation and leukemogenesis
In addition to its important functions as a key
target gene, several studies have demonstrated that C/EBPα may
participate in leukemogenesis through regulation of a number of
genes directly or indirectly.
To date, several genes have been identified as being
directly regulated by C/EBPα in myeloid leukemia; for example,
Sox4, which is critical for normal differentiation and expansion of
the lymphoid and myeloid lineages (33,34).
In normal hematopoiesis, C/EBPα expression was found to be
increased over the course of lineage commitment and then suppressed
SOX4 expression through binding to its promoter (35). Importantly, leukemic transformation
by C/EBPα mutation was partially reversed by Sox4 knockdown
(36).
FMS-like tyrosine kinase-3 (FLT3) is a
membrane-bound tyrosine kinase receptor. The interaction between
the receptor FLT3 and its ligand FL led to crucial signaling during
the early stages of the commitment of hematopoietic stem cells
(37). Mutation or overexpression
of the FLT3 gene enhanced the survival and expansion in a variety
of leukemias and was associated with an unfavorable clinical
outcome for AML patients (38).
Kindler et al demonstrated the binding of C/EBPα in human
AML on the FLT3 locus, and defined FLT3 as a direct downstream
effector of C/EBPα. Furthermore, they demonstrated that bi-allelic
C/EBPα mutations may reduce FLT3-mediated leukemogenic signals
(37), which suggests that
regulation of Flt3 expression could depend on strict C/EBPα
activity thresholds in AML.
In addition, glycolytic enzyme hexokinase 3 (HK3)
has been defined as a glycolytic enzyme most frequently expressed
in myeloid cells and represents the dominant hexokinase in
granulocytes accounting for most hexokinase activity (39). By comparing gene profiles in two
cohorts of C/EBPα wild-type and mutant AML patients, C/EBPα was
found to directly regulate HK3 by binding to its promoter (40). Furthermore, activation of HK3
transcription was found to be dependent on C/EBPα during
all-trans retinoic acid (ATRA)-mediated neutrophil
differentiation of APL cells (41).
As a leukocyte-specific gene, CORO1A has been linked
to the inhibition of neutrophil apoptosis, with significantly lower
CORO1A mRNA expression in C/EBPα-mutated AML (42,43).
Recently, C/EBPα was also demonstrated as a direct transcriptional
regulator of CORO1A in APL and C/EBPα-mutated AML patients
(44).
In addition to the above-mentioned downstream
effectors of C/EBPα-p42, there are still several potential genes
regulated by C/EBPα-p30, such as PIN1 and Trib2. PIN1 appears to be
important in tumorigenesis since it was found to be overexpressed
in many types of cancers (45,46).
Evidence for the role of PIN1 in leukemia includes the fact that it
inhibits the ubiquitination of c-Jun, which further blocks
granulocyte differentiation (46,47).
PIN1 was detected as a target of C/EBPα-p30 in AML, as C/EBPα-p30
recruited the transcription factor E2F1 in the PIN1 promoter to
elevate its expression (48). In
addition to the role of Trib2 acting as an upstream effector of
C/EBPα by mediating its proteasomal degradation, recent research
also revealed that Trib2 can also form a feedback regulatory loop
with C/EBPα. In normal myeloid progenitor cells, C/EBPα-p42 was
found to bind to the Trib2 promoter and inhibit Trib2 activation.
Conversely, C/EBPα-p30 activated the Trib2 promoter in preleukemic
cells resulting in elevated Trib2 expression, ultimately
contributing to the degradation of C/EBPα-p42 and uncontrolled
proliferation in AML (49). That
is, the exact role of Trib2 depends on the activity of C/EBPα-p30
or C/EBPα-p42 in its specific context. However, the mechanism of
the switch between these two isoforms (C/EBPα-p30 or C/EBPα-p42) is
still unknown.
In addition to binding to the promoter of several
genes to participate directly in leukemogenesis, C/EBPα also
indirectly regulates certain genes over the course. For example,
transcription factor krüppel-like factor 5 (KLF5), an essential
factor for granulocytic differentiation, was found to have a low
level in AML (41,50). A study reported that KLF5 is
indirectly regulated by C/EBPα, with its activation dependent on
C/EBPα during ATRA-mediated neutrophil differentiation in APL cells
(41).
DAPK2 is a proapoptotic protein that is mainly
expressed in hematopoietic tissue. In addition to participating in
different cell death pathways (51,52),
Rizzi et al (53) and Fang
et al (54) found a specific
function for DAPK2 as an enhancer of neutrophil and erythroid
differentiation. Other studies further confirmed that DAPK2 in
myeloid cells is dependent on C/EBPα during granulocytic
differentiation and this process seems to be indirect (55). Moreover, C/EBPα interferes with E2F1
transactivation of the c-Myc promoter in AML (56), which may influence proliferation and
differentiation in HL-60 cells through VEGF (57) (Fig.
4).
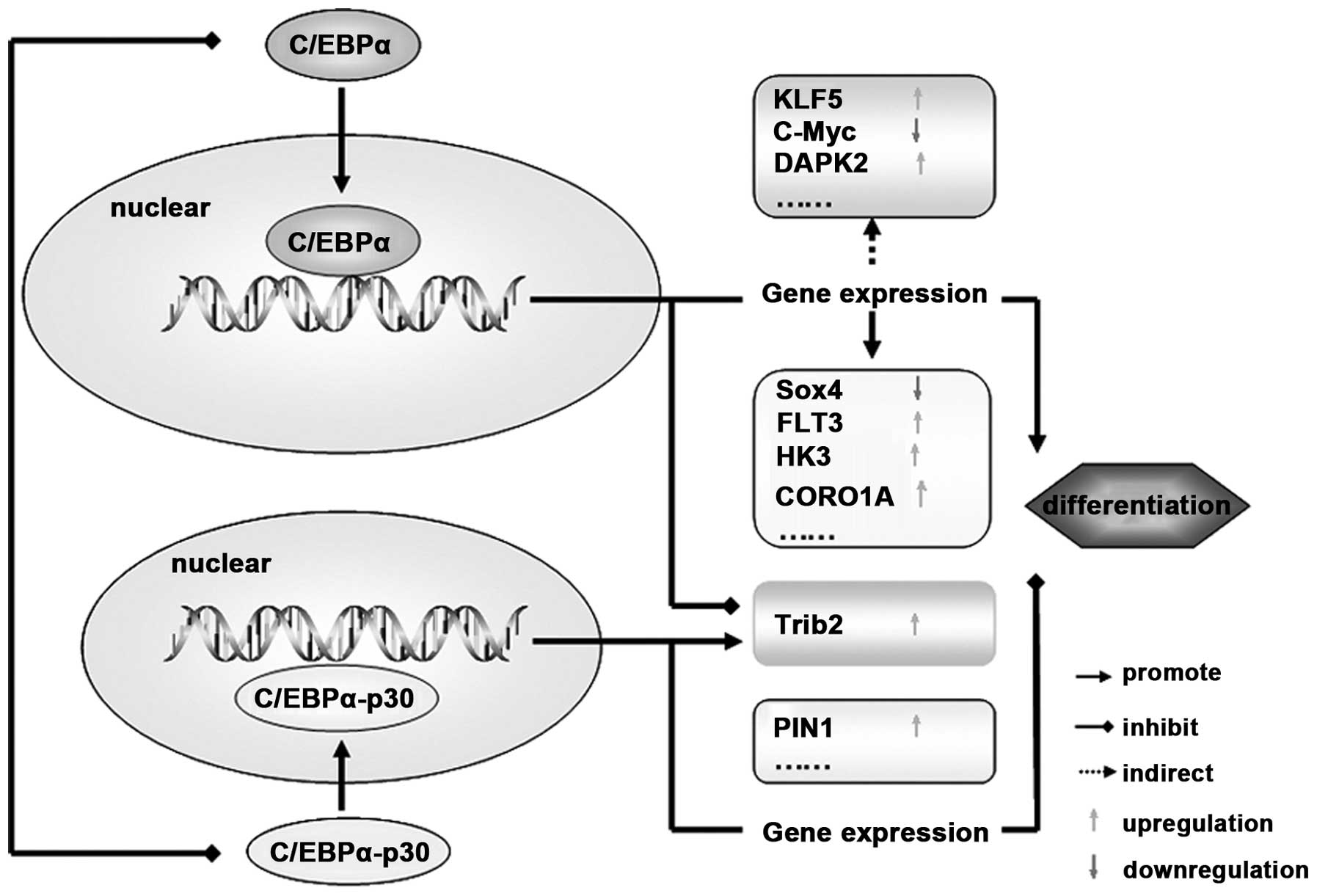 | Figure 4C/EBPα functions though regulation of
its target genes. After its translocation into the nucleus,
C/EBPα-p42 promotes cell differentiation by regulating the gene
expression directly (such as Sox4, FLT3, HK3 and CORO1A) or
indirectly (such as KLF5, DAPK2 and C-myc). In addition, C/EBPα-p30
isoform blocks cell differentiation by targeting PIN1 and Trib2.
Notably, Trib2 can be regulated by both C/EBPα-p42 and C/EBPα-p30
isoforms, and the switch mechanisms between them need further
studies. C/EBPα, CCAAT/enhancer binding protein α; FLT3, FMS-like
tyrosine kinase-3; HK3, hexokinase 3; KLF5, krüppel-like factor
5. |
A number of new molecular genetic abnormalities have
been identified in AML in the last few decade. Further studies are
needed to analyze the interrelation between them involving C/EBPα,
and a pivotal co-target gene would bear clinical significance.
Collaborating factors with C/EBPα
As mentioned earlier, C/EBPα-p30 directly activates
Trib2 (49) and PIN1 (48) expression, by cooperating with E2F1
in AML. In addition, C/EBPα cooperates with several other proteins
in the myeloid lineage, such as Hoxa9/Meis1. Homeobox A9 (HOXA9) is
a homeodomain-containing transcription factor that plays a key role
in HSC expansion and is commonly deregulated in human acute
leukemias (58). Overexpression of
HOXA9 always exists along with its cofactor meis homeobox 1 (MEIS1)
in the pattern of Hoxa9/Meis1 in AML. Recent studies suggest that
C/EBPα acts as a pioneer transcription factor in
Hoxa9/Meis1-mediated leukemogenesis through regulating its target
genes, Cdkn2a/b (59) and
Sox4 (60).
Previous studies have also demonstrated that C/EBPα
interacts with its different binding partners, including TBP and
TFIIB (basal transcription initiation factors) (61), the SWI/SNF complexes (chromatin
remodeling complexes) (62), Rb
(tumor-suppressor protein) (63),
Cdk2 and Cdk4 (cyclin-dependent kinases), p21 (cyclin-dependent
kinase inhibitor) (64), GABPα
(cell cycle regulator and transcription factor) (65). All of these may play a crucial role
during the process of leukemogenesis.
Small molecules targeting C/EBPα
In search for small molecules that are able to
reverse the low expression of the C/EBPα signature, a connectivity
map was applied. This analysis predicted positive connectivity
between the C/EBPα activation signature and histone deacetylase
inhibitors. The results showed that histone deacetylase inhibitors
reactivated the expression of the C/EBPα signature and promoted
granulocytic differentiation of primary samples from the C/EBPα
dysfunctional subset harboring biallelic C/EBPα mutations (66), which indicated that HDAC inhibitors
could represent a promising therapeutic approach in this particular
subtype of AML (67,68). Cytarabine (or Ara-C) is a pyrimidine
antagonist, which interferes with DNA synthesis and is used in
upfront and salvage regimens for AML (69). To improve the cytotoxic activity of
Ara-C treatment, various novel drug combinations have been explored
(70,71). Recently, it was demonstrated that
miR-181a could sensitize a chemotherapy-resistant HL60 cell line to
Ara-C treatment (72). Zhao et
al found that the C/EBPα-p30 isoform could bind to the
miR-181a-1 promoter to upregulate its expression. Furthermore,
lenalidomide, a drug approved for myelodysplastic syndromes and
multiple myeloma, sensitized leukemic cells to cytarabine (Ara-C)
chemotherapy by enhancing translation of the C/EBPα-p30 isoform as
well as miR-181a levels (73).
Additionally, Ko et al found that the methylation status of
let-7a-3 was inversely correlated with the methylation status in
the distal promoter region of C/EBPα in AML (74).
Therefore, it may be promising to design various
small molecules targeting the C/EBPα signaling pathway for the
treatment of AML.
4. Conclusion
In this review, we summarized the regulatory
mechanisms and the functional targets of C/EBPα in AML (Fig. 5). A more detailed molecular analysis
of C/EBPα will ultimately highlight a number of new oncogenes that
may supplement the prognostic information obtained by conventional
karyotyping. Furthermore, targeted therapies should interfere with
C/EBPα initiating or cooperating proteins, to improve the treatment
of AML. Moreover, as growing evidence implicates aberrant C/EBPα
activity in a variety of diseases, including solid tumors and
rheumatoid arthritis, small molecular compounds specific for C/EBPα
may provide potential strategies for the therapeutic intervention
of a variety of diseases.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of China (nos. 81300426, 30771103 and
81172792), the Project for Shandong Medical and Health science and
Technology Plan Project (2013Ws0365), and the ‘Twelfth Five-Year’
National Science and Technology Support Program (2013BaI07B02).
References
|
1
|
Roe JS and Vakoc CR: C/EBPα: Critical at
the origin of leukemic transformation. J Exp Med. 211:1–4. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rosenbauer F and Tenen DG: Transcription
factors in myeloid development: Balancing differentiation with
transformation. Nat Rev Immunol. 7:105–117. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Scott LM, Civin CI, Rorth P and Friedman
AD: A novel temporal expression pattern of three C/EBP family
members in differentiating myelomonocytic cells. Blood.
80:1725–1735. 1992.PubMed/NCBI
|
|
4
|
Birkenmeier EH, Gwynn B, Howard S, Jerry
J, Gordon JI, Landschulz WH and McKnight SL: Tissue-specific
expression, developmental regulation, and genetic mapping of the
gene encoding CCAAT/enhancer binding protein. Genes Dev.
3:1146–1156. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Koschmieder S, Halmos B, Levantini E and
Tenen DG: Dysregulation of the C/EBPalpha differentiation pathway
in human cancer. J Clin Oncol. 27:619–628. 2009. View Article : Google Scholar :
|
|
6
|
Mueller BU and Pabst T: C/EBPalpha and the
pathophysiology of acute myeloid leukemia. Curr Opin Hematol.
13:7–14. 2006. View Article : Google Scholar
|
|
7
|
Zhang P, Iwasaki-Arai J, Iwasaki H, et al:
Enhancement of hematopoietic stem cell repopulating capacity and
self-renewal in the absence of the transcription factor C/EBP
alpha. Immunity. 21:853–863. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang DE, Zhang P, Wang ND, Hetherington
CJ, Darlington GJ and Tenen DG: Absence of granulocyte
colony-stimulating factor signaling and neutrophil development in
CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad
Sci USA. 94:569–574. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Porse BT, Bryder D, Theilgaard-Mönch K,
Hasemann MS, Anderson K, Damgaard I, Jacobsen SE and Nerlov C: Loss
of C/EBP alpha cell cycle control increases myeloid progenitor
proliferation and transforms the neutrophil granulocyte lineage. J
Exp Med. 202:85–96. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Scott E, Sawyers CL and Friedman
AD: C/EBPalpha bypasses granulocyte colony-stimulating factor
signals to rapidly induce PU.1 gene expression, stimulate
granulocytic differentiation, and limit proliferation in 32D cl3
myeloblasts. Blood. 94:560–571. 1999.PubMed/NCBI
|
|
11
|
Bereshchenko O, Mancini E, Moore S, Bilbao
D, Månsson R, Luc S, Grover A, Jacobsen SE, Bryder D and Nerlov C:
Hematopoietic stem cell expansion precedes the generation of
committed myeloid leukemia-initiating cells in C/EBPalpha mutant
AML. Cancer Cell. 16:390–400. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pabst T, Mueller BU, Zhang P, Radomska HS,
Narravula S, Schnittger S, Behre G, Hiddemann W and Tenen DG:
Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer
binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat
Genet. 27:263–270. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Preudhomme C, Sagot C, Boissel N, et al:
ALFA Group: Favorable prognostic significance of CEBPA mutations in
patients with de novo acute myeloid leukemia: A study from the
Acute Leukemia French Association (ALFA). Blood. 100:2717–2723.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Miller M, Shuman JD, Sebastian T, Dauter Z
and Johnson PF: Structural basis for DNA recognition by the basic
region leucine zipper transcription factor CCAAT/enhancer-binding
protein alpha. J Biol Chem. 278:15178–15184. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pabst T and Mueller BU: Transcriptional
dysregulation during myeloid transformation in AML. Oncogene.
26:6829–6837. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pabst T and Mueller BU: Complexity of
CEBPA dysregulation in human acute myeloid leukemia. Clin Cancer
Res. 15:5303–5307. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cai DH, Wang D, Keefer J, Yeamans C,
Hensley K and Friedman AD: C/EBP alpha:AP-1 leucine zipper
heterodimers bind novel DNA elements, activate the PU.1 promoter
and direct monocyte lineage commitment more potently than C/EBP
alpha homodimers or AP-1. Oncogene. 27:2772–2779. 2008. View Article : Google Scholar
|
|
18
|
Wang D, Paz-Priel I and Friedman AD:
NF-kappa B p50 regulates C/EBP alpha expression and inflammatory
cytokine-induced neutrophil production. J Immunol. 182:5757–5762.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dahl R, Walsh JC, Lancki D, Laslo P, Iyer
SR, Singh H and Simon MC: Regulation of macrophage and neutrophil
cell fates by the PU.1:C/EBPalpha ratio and granulocyte
colony-stimulating factor. Nat Immunol. 4:1029–1036. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Licht JD, Chomienne C, Goy A, et al:
Clinical and molecular characterization of a rare syndrome of acute
promyelocytic leukemia associated with translocation (11;17).
Blood. 85:1083–1094. 1995.PubMed/NCBI
|
|
21
|
Chen Z, Brand NJ, Chen A, Chen SJ, Tong
JH, Wang ZY, Waxman S and Zelent A: Fusion between a novel
Krüppel-like zinc finger gene and the retinoic acid receptor-alpha
locus due to a variant t(11;17) translocation associated with acute
promyelocytic leukaemia. EMBO J. 12:1161–1167. 1993.PubMed/NCBI
|
|
22
|
Girard N, Tremblay M, Humbert M, Grondin
B, Haman A, Labrecque J, Chen B, Chen Z, Chen SJ and Hoang T:
RARα-PLZF oncogene inhibits C/EBPα function in myeloid cells. Proc
Natl Acad Sci USA. 110:13522–13527. 2013. View Article : Google Scholar
|
|
23
|
Pabst T, Mueller BU, Harakawa N, Schoch C,
Haferlach T, Behre G, Hiddemann W, Zhang DE and Tenen DG: AML1-ETO
downregulates the granulocytic differentiation factor C/EBP alpha
in t(8;21) myeloid leukemia. Nat Med. 7:444–451. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Baer C, Claus R, Frenzel LP, et al:
Extensive promoter DNA hypermethylation and hypomethylation is
associated with aberrant microRNA expression in chronic lymphocytic
leukemia. Cancer Res. 72:3775–3785. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chim CS, Wong AS and Kwong YL: Infrequent
hypermethylation of CEBPA promotor in acute myeloid leukaemia. Br J
Haematol. 119:988–990. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Annamaneni S, Kagita S, Gorre M, Digumarti
RR, Satti V and Battini MR: Methylation status of CEBPA gene
promoter in chronic myeloid leukemia. Hematology. 19:42–44. 2014.
View Article : Google Scholar
|
|
27
|
Nikoloski G, Langemeijer SM, Kuiper RP, et
al: Somatic mutations of the histone methyltransferase gene EZH2 in
myelodysplastic syndromes. Nat Genet. 42:665–667. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Herrera-Merchan A, Arranz L, Ligos JM, de
Molina A, Dominguez O and Gonzalez S: Ectopic expression of the
histone methyltransferase Ezh2 in haematopoietic stem cells causes
myeloproliferative disease. Nat Commun. 3:6232012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thiel AT, Feng Z, Pant DK, Chodosh LA and
Hua X: The trithorax protein partner menin acts in tandem with EZH2
to suppress C/EBPα and differentiation in MLL-AF9 leukemia.
Haematologica. 98:918–927. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dedhia PH, Keeshan K, Uljon S, Xu L, Vega
ME, Shestova O, Zaks-Zilberman M, Romany C, Blacklow SC and Pear
WS: Differential ability of Tribbles family members to promote
degradation of C/EBPalpha and induce acute myelogenous leukemia.
Blood. 116:1321–1328. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yoshida A, Kato JY, Nakamae I and
Yoneda-Kato N: COP1 targets C/EBPα for degradation and induces
acute myeloid leukemia via Trib1. Blood. 122:1750–1760. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pal P, Lochab S, Kanaujiya JK, Kapoor I,
Sanyal S, Behre G and Trivedi AK: E6AP, an E3 ubiquitin ligase
negatively regulates granulopoiesis by targeting transcription
factor C/EBPα for ubiquitin-mediated proteasome degradation. Cell
Death Dis. 4:e5902013. View Article : Google Scholar
|
|
33
|
Sandoval S, Kraus C, Cho EC, et al: Sox4
cooperates with CREB in myeloid transformation. Blood. 120:155–165.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Aue G, Du Y, Cleveland SM, et al: Sox4
cooperates with PU.1 haploinsufficiency in murine myeloid leukemia.
Blood. 118:4674–4681. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fung TK, Leung AY and So CW: Sox4you: A
new player in C/EBPα leukemia. Cancer Cell. 24:557–559. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang H, Alberich-Jorda M, Amabile G, et
al: Sox4 is a key oncogenic target in C/EBPα mutant acute myeloid
leukemia. Cancer Cell. 24:575–588. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kindler T, Lipka DB and Fischer T: FLT3 as
a therapeutic target in AML: Still challenging after all these
years. Blood. 116:5089–5102. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kelly LM, Liu Q, Kutok JL, Williams IR,
Boulton CL and Gilliland DG: FLT3 internal tandem duplication
mutations associated with human acute myeloid leukemias induce
myeloproliferative disease in a murine bone marrow transplant
model. Blood. 99:310–318. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Federzoni EA, Valk PJ, Torbett BE,
Haferlach T, Löwenberg B, Fey MF and Tschan MP: PU.1 is linking the
glycolytic enzyme HK3 in neutrophil differentiation and survival of
APL cells. Blood. 119:4963–4970. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Federzoni EA, Humbert M, Torbett BE, Behre
G, Fey MF and Tschan MP: CEBPA-dependent HK3 and KLF5 expression in
primary AML and during AML differentiation. Sci Rep. 4:42612014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Diakiw SM, Kok CH, To LB, Lewis ID, Brown
AL and D’Andrea RJ: The granulocyte-associated transcription factor
Krüppel-like factor 5 is silenced by hypermethylation in acute
myeloid leukemia. Leuk Res. 36:110–116. 2012. View Article : Google Scholar
|
|
42
|
Chan KT, Creed SJ and Bear JE: Unraveling
the enigma: Progress towards understanding the coronin family of
actin regulators. Trends Cell Biol. 21:481–488. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Moriceau S, Kantari C, Mocek J, et al:
Coronin-1 is associated with neutrophil survival and is cleaved
during apoptosis: Potential implication in neutrophils from cystic
fibrosis patients. J Immunol. 182:7254–7263. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Federzoni EA, Humbert M, Valk PJ, Behre G,
Leibundgut EO, Torbett BE, Fey MF and Tschan MP: The actin-binding
protein CORO1A is a novel PU.1 (SPI1)- and CEBPA-regulated gene
with significantly lower expression in APL and CEBPA-mutated AML
patients. Br J Haematol. 160:855–859. 2013. View Article : Google Scholar
|
|
45
|
Bao L, Kimzey A, Sauter G, Sowadski JM, Lu
KP and Wang DG: Prevalent overexpression of prolyl isomerase Pin1
in human cancers. Am J Pathol. 164:1727–1737. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T,
Petkova V and Lu KP: Pin1 is overexpressed in breast cancer and
cooperates with Ras signaling in increasing the transcriptional
activity of c-Jun towards cyclin D1. EMBO J. 20:3459–3472. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rinehart-Kim J, Johnston M, Birrer M and
Bos T: Alterations in the gene expression profile of MCF-7 breast
tumor cells in response to c-Jun. Int J Cancer. 88:180–190. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pulikkan JA, Dengler V, Peer Zada AA,
Kawasaki A, Geletu M, Pasalic Z, Bohlander SK, Ryo A, Tenen DG and
Behre G: Elevated PIN1 expression by C/EBPalpha-p30 blocks
C/EBPalpha-induced granulocytic differentiation through c-Jun in
AML. Leukemia. 24:914–923. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Rishi L, Hannon M, Salomè M, et al:
Regulation of Trib2 by an E2F1-C/EBPα feedback loop in AML cell
proliferation. Blood. 123:2389–2400. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Humbert M, Halter V, Shan D, Laedrach J,
Leibundgut EO, Baerlocher GM, Tobler A, Fey MF and Tschan MP:
Deregulated expression of Kruppel-like factors in acute myeloid
leukemia. Leuk Res. 35:909–913. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Britschgi A, Trinh E, Rizzi M, Jenal M,
Ress A, Tobler A, Fey MF, Helin K and Tschan MP: DAPK2 is a novel
E2F1/KLF6 target gene involved in their proapoptotic function.
Oncogene. 27:5706–5716. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Britschgi A, Simon HU, Tobler A, Fey MF
and Tschan MP: Epigallocatechin-3-gallate induces cell death in
acute myeloid leukaemia cells and supports all-trans retinoic
acid-induced neutrophil differentiation via death-associated
protein kinase 2. Br J Haematol. 149:55–64. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rizzi M, Tschan MP, Britschgi C, et al:
The death-associated protein kinase 2 is up-regulated during normal
myeloid differentiation and enhances neutrophil maturation in
myeloid leukemic cells. J Leukoc Biol. 81:1599–1608. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fang J, Menon M, Zhang D, Torbett B,
Oxburgh L, Tschan M, Houde E and Wojchowski DM: Attenuation of
EPO-dependent erythroblast formation by death-associated protein
kinase-2. Blood. 112:886–890. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Humbert M, Federzoni EA, Britschgi A, et
al: The tumor suppressor gene DAPK2 is induced by the myeloid
transcription factors PU.1 and C/EBPα during granulocytic
differentiation but repressed by PML-RARα in APL. J Leukoc Biol.
95:83–93. 2014. View Article : Google Scholar :
|
|
56
|
Porse BT, Pedersen TA, Xu X, Lindberg B,
Wewer UM, Friis-Hansen L and Nerlov C: E2F repression by C/EBPalpha
is required for adipogenesis and granulopoiesis in vivo. Cell.
107:247–258. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Song G, Li Y, Zhang Z, et al: c-myc but
not Hif-1α-dependent downregulation of VEGF influences the
proliferation and differentiation of HL-60 cells induced by ATRA.
Oncol Rep. 29:2378–2384. 2013.PubMed/NCBI
|
|
58
|
Eklund E: The role of Hox proteins in
leukemogenesis: Insights into key regulatory events in
hematopoiesis. Crit Rev Oncog. 16:65–76. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kroon E, Krosl J, Thorsteinsdottir U,
Baban S, Buchberg AM and Sauvageau G: Hoxa9 transforms primary bone
marrow cells through specific collaboration with Meis1a but not
Pbx1b. EMBO J. 17:3714–3725. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Collins C, Wang J, Miao H, Bronstein J,
Nawer H, Xu T, Figueroa M, Muntean AG and Hess JL: C/EBPα is an
essential collaborator in Hoxa9/Meis1-mediated leukemogenesis. Proc
Natl Acad Sci USA. 111:9899–9904. 2014. View Article : Google Scholar
|
|
61
|
Nerlov C and Ziff EB: CCAAT/enhancer
binding protein-alpha amino acid motifs with dual TBP and TFIIB
binding ability co-operate to activate transcription in both yeast
and mammalian cells. EMBO J. 14:4318–4328. 1995.PubMed/NCBI
|
|
62
|
Müller C, Calkhoven CF, Sha X and Leutz A:
The CCAAT enhancer-binding protein alpha (C/EBPalpha) requires a
SWI/SNF complex for proliferation arrest. J Biol Chem.
279:7353–7358. 2004. View Article : Google Scholar
|
|
63
|
Chen PL, Riley DJ, Chen Y and Lee WH:
Retinoblastoma protein positively regulates terminal adipocyte
differentiation through direct interaction with C/EBPs. Genes Dev.
10:2794–2804. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang H, Iakova P, Wilde M, Welm A, Goode
T, Roesler WJ and Timchenko NA: C/EBPalpha arrests cell
proliferation through direct inhibition of Cdk2 and Cdk4. Mol Cell.
8:817–828. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Shimokawa T, Nunomura S, Enomoto Y and Ra
C: Amino acid residues in the beta3 strand and subsequent loop of
the conserved ETS domain that mediate basic leucine zipper (bZIP)
recruitment and potentially distinguish functional attributes of
ETS proteins. Biochem J. 430:129–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shimokawa T, Nunomura S, Fujisawa D and Ra
C: Identification of the C/EBPα C-terminal tail residues involved
in the protein interaction with GABP and their potency in myeloid
differentiation of K562 cells. Biochim Biophys Acta.
1829:1207–1217. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lin RJ, Nagy L, Inoue S, Shao W, Miller WH
Jr and Evans RM: Role of the histone deacetylase complex in acute
promyelocytic leukaemia. Nature. 391:811–814. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zapotocky M, Mejstrikova E, Smetana K,
Stary J, Trka J and Starkova J: Valproic acid triggers
differentiation and apoptosis in AML1/ETO-positive leukemic cells
specifically. Cancer Lett. 319:144–153. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tallman MS, Gilliland DG and Rowe JM: Drug
therapy for acute myeloid leukemia. Blood. 106:1154–1163. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chen P, Aimiuwu J, Xie Z, Wei X, Liu S,
Klisovic R, Marcucci G and Chan KK: Biochemical modulation of
aracytidine (Ara-C) effects by GTI-2040, a ribonucleotide reductase
inhibitor, in K562 human leukemia cells. AAPS J. 13:131–140. 2011.
View Article : Google Scholar :
|
|
71
|
Uy GL, Rettig MP, Motabi IH, et al: A
phase 1/2 study of chemosensitization with the CXCR4 antagonist
plerixafor in relapsed or refractory acute myeloid leukemia. Blood.
119:3917–3924. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bai H, Cao Z, Deng C, Zhou L and Wang C:
miR-181a sensitizes resistant leukaemia HL-60/Ara-C cells to Ara-C
by inducing apoptosis. J Cancer Res Clin Oncol. 138:595–602. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Hickey CJ, Schwind S, Radomska HS, et al:
Lenalidomide-mediated enhanced translation of C/EBPα-p30 protein
up-regulates expression of the antileukemic microRNA-181a in acute
myeloid leukemia. Blood. 121:159–169. 2013. View Article : Google Scholar :
|
|
74
|
Ko YC, Fang WH, Lin TC, Hou HA, Chen CY,
Tien HF and Lin LI: MicroRNA let-7a-3 gene methylation is
associated with karyotyping, CEBPA promoter methylation, and
survival in acute myeloid leukemia. Leuk Res. 38:625–631. 2014.
View Article : Google Scholar : PubMed/NCBI
|