Introduction
Monopolar spindle-one-binder (MOB) family proteins
are conserved from yeasts to human and function as the regulators
of signaling pathways involved in cell mitosis, apoptosis and
morphogenesis (1–9). Human MOB2 (hMOB2) is a member of the
hMOB family of proteins, which contains at least 6 distinct hMOBs
(hMOB1A, hMOB1B, hMOB2, hMOB3A, hMOB3B and hMOB3C) (2,10).
Among them, hMOB1A and hMOB1B have been characterized as the
putative tumor suppressors in tumor cell proliferation, apoptosis
and centrosome duplication through regulating the activation of the
NDR kinase/large tumor suppressor kinase (2,4–7,11),
while the biological roles of other hMOBs are less well defined. To
date, hMOB2 has also been widely termed hepatocellular
carcinoma-associated gene 2 (HCCA2), and it is involved in the
hepatocellular carcinoma development and progression (12). Nevertheless, data from the locus on
the chromosome and the open reading frame (ORF) of mRNA of the
hMOB2 gene (GenBank Accession: NM_053005, at position 11p15.5) and
HCCA2 gene (GenBank Accession: AF206328, at position 1q22) revealed
clear differences between them, suggesting that the biological
behavior of hMOB2 may be different from the HCCA2. Therefore, it is
currently unclear whether hMOB2 is involved in tumor development
and progression.
In Drosophila, MOB2 plays a role in wing hair
morphogenesis by interacting with the related tricornered and warts
kinases (13,14) and is involved in the photoreceptor
cell development and rhabdomere formation by regulating the actin
cytoskeleton rearrangement (15).
The study also showed that mouse MOB2 promotes the neurite
formation through regulating the actin cytoskeleton rearrangement
(16). Currently, hMOB2 has been
characterized to be involved in the regulation of hMOB1 by
competing for the NDR1/2 kinases (8), which have already been linked to the
actin cytoskeleton (17), although
the mechanism involved is only partly understood. Since the dynamic
remodeling of the actin cytoskeleton is also fundamental for many
physiological processes like cell adhesion and cell motility
(18), which contribute to tumor
cell migration, invasion and metastasis (19), a better understanding of
hMOB2-induced changes in the cytoskeletal regulation, cell adhesion
and motility may result in better insight into tumor
metastasis.
In the present study, we evaluated the effect of
hMOB2 on tumor cell migration, invasion and cell-matrix adhesion.
We found that hMOB2 overexpression appeared to significantly
inhibit cell motility and promoted cell-matrix adhesion, while
hMOB2 knockdown decreased not only the cell motility, but also the
cell-matrix adhesion. Furthermore, we also demonstrated that both
hMOB2 overexpression and knockdown altered assembly of the focal
adhesions and the actin cyto-skeleton rearrangement presumably
through regulating the focal adhesion kinase (FAK)-Src-paxillin
signaling pathway, unveiling a novel mechanism of cell motility and
cell-matrix adhesion regulation induced by hMOB2.
Materials and methods
Cell lines and culture conditions
The human hepatocellular carcinoma cell lines HepG2
and SMMC-7721, and the human embryonic kidney (HEK) 293T cells were
obtained from the Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China), then cultured in Dulbecco’s modified
Eagle’s medium (DMEM; Gibco) supplemented with 10% heat-inactivated
fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml
streptomycin and maintained in a humidified incubator at 37°C with
5% CO2.
Production of recombinant
lentiviruses
To construct the lentiviral vector expressing hMOB2,
the transfer vector pGC-hMOB2 was constructed by cloning the
full-length of hMOB2 cDNA, amplified by polymerase chain reaction
(PCR) from the hMOB2 cDNA clone which was obtained from the
GeneChem Corporation (Shanghai, China) with the primers
5′-GAGGATCCCCGGGTACCGGTCGCCACCATGGACT GGCTCATGGGGAAG-3′ (sense),
and 5′-TCCTTGTAGTCC ATACCTCTCTCCTTCACGTGGTTCTG-3′ (antisense), into
the AgeI sites of pGV287 purchased from the GeneChem
Corporation.
To construct the lentiviral vectors expressing siRNA
targeting hMOB2, four different siRNA targeting sequences directed
against hMOB2 were selected under the guide of siRNA designing
software provided by GenScript. The sequences containing the hMOB2
siRNA targets (underlined sequence) were as follows:
5′-CCGGCATCACCGACTTCCAGTTCAACTCGAGTTGAACTGGAAGTCGGTGATGTTTTTG-3′
(sense) and 5′-AATTCAAAAACATCACCGACTTCCAGTTCAACTCGAGTTGAACTGGAAGTCGGTGA-3′
(antisense) for sihMOB2-1; 5′-CCGGCAGAGATTGACCTTAACGAGTCTCGAGACTCGTTAAGGTCAATCTCTGTTTTTG-3′
(sense), and 5′-AATTCAAAAACAGAGATTGACCTTAACGAGTCTCGAGACTCGTTAAGGTCAATCTC-3′
(anti-sense), for sihMOB2-2; 5′-CCGGCATGTGCAACACACAGTACTACTCGAGTAGTACTGTGTGTTGCACATGTTTTTG-3′
(sense), and 5′-AATTCAAAAACATGTGCAACACACAGTACTACTCGAGTAGTACTGTGTGTTGCACA-3′
(antisense), for sihMOB2-3; 5′-CCGGCAGCTGGTGACGGATGAGGACCTCGAGGTCCTCATCCGTCACCAGCTGTTTTTG-3′
(sense), and 5′-AATTCAAAAACAGCTGGTGACGGATGAGGACCTCGAGGTCCTCATCCGTCACCAGC-3′
(antisense), for sihMOB2-4. Then, the primers were synthesized and
cloned into the AgeI and EcoRI sites of pGV115,
purchased from the GeneChem Corporation.
For the lentiviral production, the lentiviruses
encoding hMOB2 (LV-hMOB2) and siRNA against hMOB2 (LV-sihMOB2-1,
LV-sihMOB2-2, LV-sihMOB2-3 and LV-sihMOB2-4) were produced by
co-transfection of the lentiviral transfer vectors together with
the packaging system pHelper 1.0 and 2.0 vectors purchased from the
GeneChem Corporation into the HEK293T cells using Lipofectamine
2000 (Invitrogen, USA). The lentivirus particles were harvested and
purified, and the infectious titers were determined by GeneChem
Corporation. Control lentivirus (LV-CTL) and the non-silencing
siRNA control lentivirus (LV-siCTL) were also generated by GeneChem
Corporation and performed as the empty vectors for overexpression
and knockdown, respectively.
For lentiviral infection, the HepG2 and SMMC-7721
cells were plated at a concentration of 1×105 cells in
the 6-well culture plates, and were then infected with indicated
lentiviruses at a MOI of 20 and 30 in the presence of Polybrene (8
μg/ml), respectively. The infected cells continued to be
cultured for over 96 h in DMEM supplemented with 10% FBS. The green
fluorescence protein (GFP), which was co-expressed in all the
lentivirus-infected cells, served as a selection marker to indicate
the successfully infected HepG2 or SMMC-7721 cells. To enrich the
GFP-positive cells, the cells were sorted in the FACSCalibur
(Becton-Dickinson), and the GFP-positive cells were returned to
culture immediately, and were considered for the successful
lentivirus-transduction in following experiments.
Cell migration and invasion assays
For the Transwell migration assay, the 24-well
Transwell chambers with 8.0-μm pore size of the porous
membrane (Costar, USA) were performed. Briefly, the
lentivirus-transduced cells were serum-starved for 24 h and then
3×104 cells in a volume of 200 μl suspended in
serum-free DMEM were seeded onto the upper chamber, which was
placed into the lower chamber containing 600 μl of DMEM
supplemented with 10% FBS. After incubation for 24 h at 37°C, the
cells on the upper surface of the chambers were removed, and the
cells that migrated to the lower surface of the filter were washed
with phosphate-buffered saline (PBS), fixed with methanol for 15
min and stained with 0.1% crystal violet for 20 min. The migrated
cells in at least five randomly selected fields at ×200
magnification were quantified, and images were captured using a
phase contrast microscope equipped with a digital image capturing
system. A cell invasion assay was also performed in the Transwell
chambers as previously described with the minor modification that
the upper chambers were precoated with 100 μg/ml Matrigel
(BD Biosciences), and 200 μl of 5×104 cells
suspended in a serum-free DMEM were added to the upper chamber. The
assays were performed in triplicate for each experiment and
repeated three times.
Cell-matrix adhesion assay
For the cell-matrix adhesion assay, wells of 96-well
culture plates were coated with 10 μg/ml collagen type I (BD
Biosciences) overnight at 4°C, blocked with 1% heat-denatured
bovine serum albumin (BSA) in PBS at 37°C for 1 h, and were then
washed with PBS. The lentivirus-transduced HepG2 and SMMC-7721
cells were harvested, suspended at 5×105 cells/ml in
DMEM supplemented with 0.1% BSA and 100 μl of the cell
suspension were added to the coated well. After incubation at 37°C
for 45 min, non-adherent cells were gently removed by being washed
four times with PBS. The numbers of the adherent cells were
quantified by the Cell Counting Kit-8 (CCK-8; Obio Technology,
Shanghai, China) assay according to the manufacturer’s instruction
followed by absorbance measurement at 450 nm using a
multifunctional microplate reader (Bio-Tek, USA). The assays were
performed in 6-wells and repeated five times.
RNA preparation and real-time reverse
transcription quantitative PCR (RT-qPCR)
Total RNA was isolated from the
lentivirus-transduced cells using the TRIzol reagent (Invitrogen),
and reverse transcribed using the HiScript First Strand cDNA
Synthesis kit (Vazyme, Nanjing, China) according to the
manufacturer’s instruction. RT-qPCR was used to determine the hMOB2
mRNA levels on an ABI 7500 Real-Time PCR System (Applied
Biosystems, Carlsbad, CA, USA) using the AceQ® qPCR
SYBR®-Green Master Mix kit (Vazyme) according to the
manufacturer’s instruction. The primers for hMOB2 were:
5′-TTCCACCACATCAACCTGCA GTA-3′ (sense), and
5′-GGAGCTCATGACGAAGTCAACG TA-3′ (antisense); for GAPDH,
5′-GCACCGTCAAGGCTGA GAAC-3′ (sense), and 5′-TGGTGAAGACGCCAGTGGA-3′
(antisense), were used to run the RT-qPCR. The relative levels of
the hMOB2 mRNA normalized to GAPDH mRNA were evaluated by
2−ΔΔCt.
Western blot assay
The lentivirus-transduced cells were lysed in RIPA
buffer (Beyotime, China) containing 1 μmol
phenylmethylsulfonyl fluoride and a protease and phosphatase
inhibitor cocktail (Roche). After determination of the protein
concentration with the Bradford method, the protein samples were
electrophoresed by SDS-polyacrylamide gel electrophoresis
(SDS-PAGE), and were then transferred onto the polyvinylidene
fluoride membrane (PVDF, Millipore), which was subsequently blocked
with 5% non-fat dried milk or 3% BSA for tyrosine phosphorylation
blots in TBS for 1 h at room temperature, and incubated with the
indicated primary antibodies at 4°C overnight. The primary
antibodies used in the present study were: poly-clonal rabbit
anti-hMOB2 (1:800; Abcam); monoclonal rabbit anti-FAK, polyclonal
rabbit anti-pY397FAK, polyclonal rabbit anti-PY925FAK, monoclonal
rabbit anti-Src, polyclonal rabbit anti-pY527Src (1:1,000; Cell
Signaling Technology); monoclonal mouse anti-paxillin, polyclonal
goat anti-pY118paxillin, monoclonal mouse anti-GAPDH (1:200; Santa
Cruz Biotechnology). The PVDF membrane was then washed three times
with TBS with 0.1% Tween-20 (TBST) followed by incubation for 2 h
at room temperature with horseradish peroxidase-conjugated goat
anti-rabbit IgG, goat anti-mouse IgG or donkey anti-goat IgG
antibodies (1:2,000; KangChen Bio-tech, Shanghai, China). The
protein bands were detected by enhanced chemiluminescence using the
Pierce ECL Plus Western Blotting Substrate kit (Thermo
Scientific).
Immunofluorescence analysis
The lentivirus-transduced cells were plated onto
glass coverslips in 24-well culture plates and maintained in DMEM
supplemented with 10% FBS for 24 h. The cells were washed with PBS,
fixed with 4% para-formaldehyde diluted in PBS for 30 min,
permeabilized with 0.5% Triton X-100 in PBS for 10 min, and then
blocked with 3% BSA in PBS for 1 h at room temperature. After
incubation overnight at 4°C with monoclonal mouse anti-paxillin
antibody diluted 1:100 in blocking solution, the cells were washed
three times with PBS and subsequently incubated with the
Rhodamine-conjugated goat anti-mouse IgG (1:1,000; Biosource
International) for 2 h at room temperature in the dark. Actin was
visualized using Rhodamine-conjugated phalloidin (Sigma) at 37°C
for 1 h at room temperature and then gently washed in PBS. All the
cells were counterstained with DAPI (Sigma) for nucleus. After
being washed thoroughly with PBS, the images were captured using a
fluorescent microscope equipped with a digital image capturing
system.
Statistical analysis
Data are expressed as the means ± standard deviation
(SD). Statistical significance was determined by the Student’s
t-test, and the p-values <0.05 were considered to indicate a
statistically significant result.
Results
Expression of hMOB2 after lentiviral
transduction
The overexpression and knockdown of hMOB2 in the
lentivirus-transduced cells were initially examined. The levels of
hMOB2 mRNA from the total RNA extracts were quantified using
RT-qPCR, normalized to GAPDH mRNA and the hMOB2 protein levels from
the whole cell extracts were determined by western blot assay. As
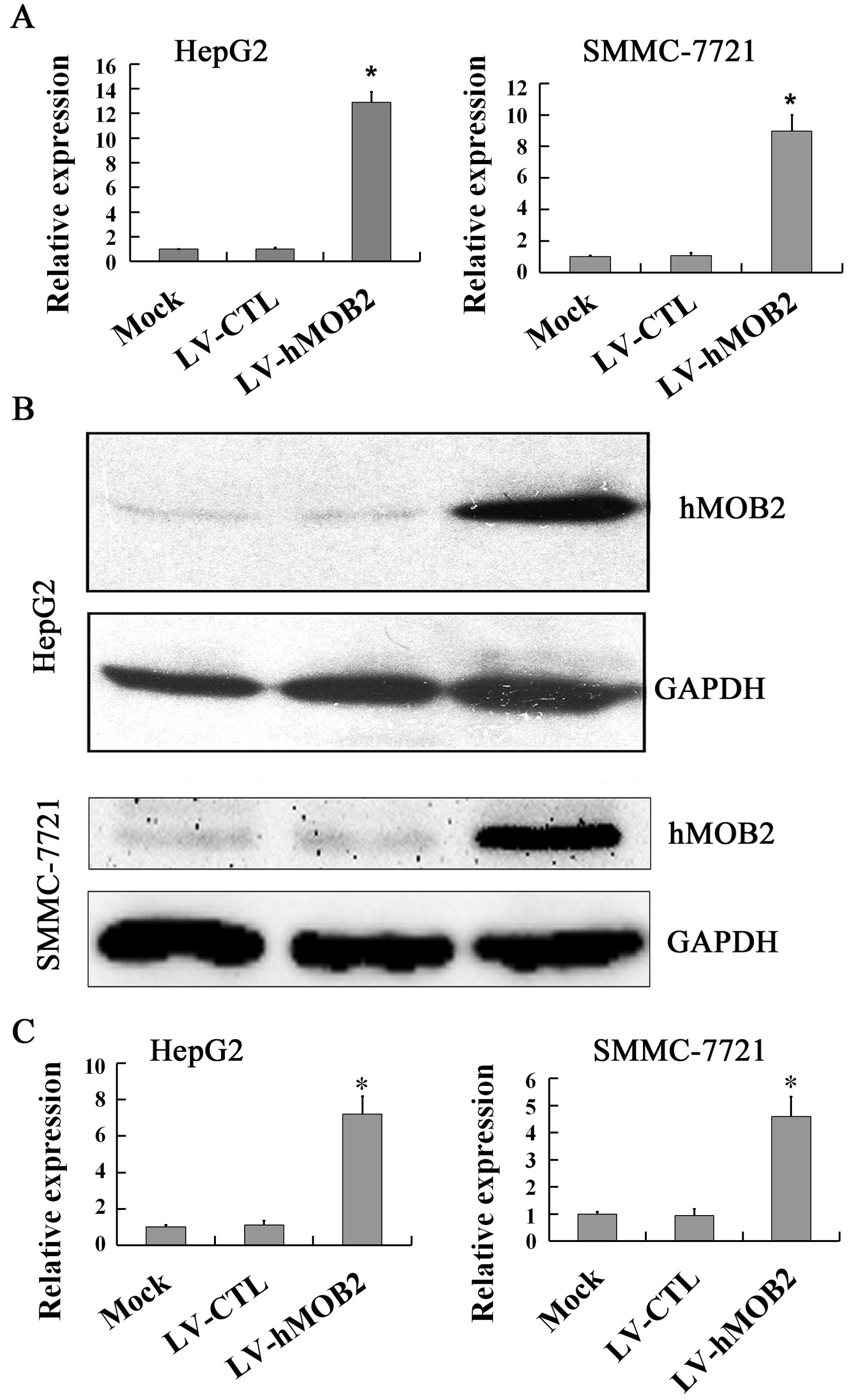
shown in Figs. 1 and 2, overexpression of hMOB2 was observed in
LV-hMOB2 trans-duced HepG2 and SMMC-772 cells both in mRNA
(Fig. 1A) and protein levels
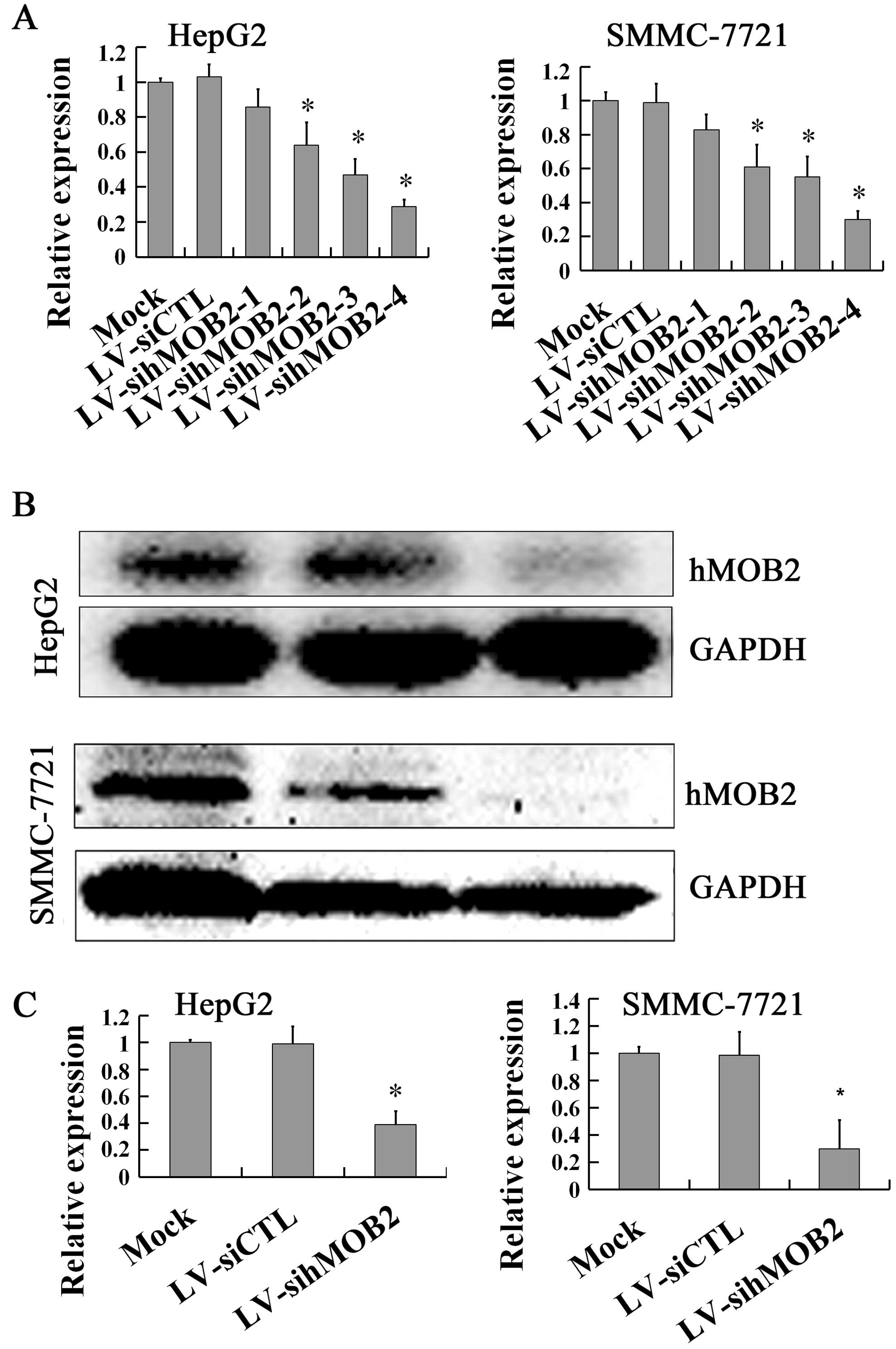
(Fig. 1B and C), while knockdown of
hMOB2 expression by LV-sihMOB2-4 was achieved with the most
efficient inhibition at the mRNA (Fig.
2A) and the protein levels (Fig.
2B and C) both in HepG2 and SMMC-772 cells. These results
demonstrated that the indicated lentiviruses were successfully
transduced into the HepG2 and SMMC-772 cells, and hMOB2 was
overexpressed in LV-hMOB2-transduced, and the most efficient
knockdown of hMOB2 was shown in the LV-sihMOB2-4-transduced cells.
Therefore, LV-hMOB2 and LV-sihMOB2-4 (following named LV-sihMOB2)
could be used in the following experiments.
Effect of hMOB2 on cell migration and
invasion
To evaluate the role of hMOB2 in cell migration and
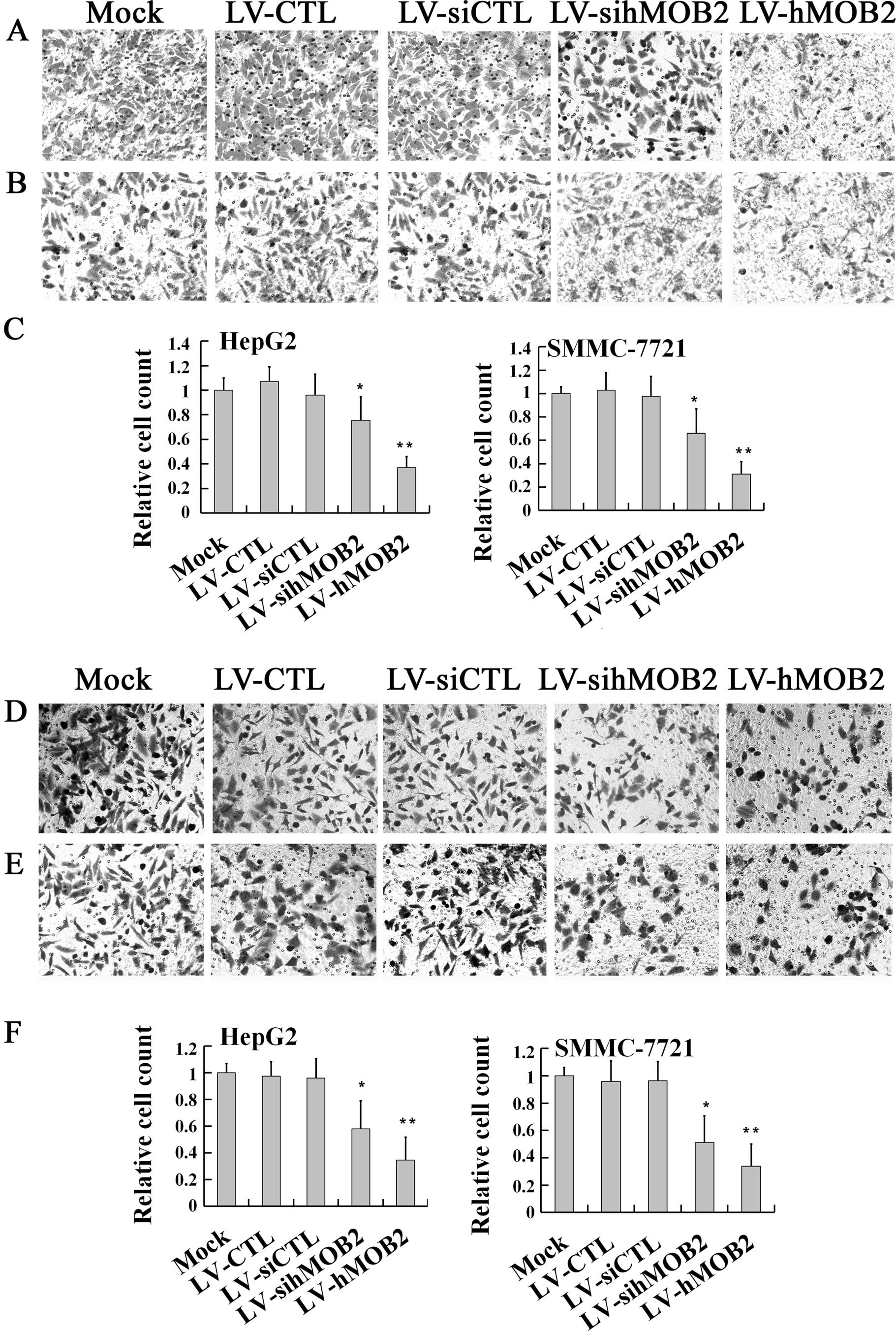
invasion, we first performed a Transwell migration assay. As shown
in Fig. 3, both HepG2 and SMMC-772
cells infected with LV-hMOB2 showed dramatic inhibition of the cell
migratory ability, when compared with those transduced with the
empty vectors (LV-CTL and LV-siCTL) and the mock. Notably,
knockdown of the hMOB2 expression by LV-sihMOB2 also decreased the
motile cells either in the HepG2 or the SMMC-772 cells (Fig. 3A–C). No significant differences in
the cell migration were found among the mock and the empty
vector-transduced cells. Similar effects of the hMOB2
over-expression and knockdown on cell invasion were also found in
the Transwell cell invasion assay (Fig.
3D–F). Together, the results demonstrated that both
overexpression and knockdown of hMOB2 attenuated the general
capacity of cell migration and invasion.
Effect of hMOB2 on the cell-matrix
adhesion
Since cell migration and invasion require
cell-extracellular matrix (ECM) adhesion and then detachment from
ECM (19,20), we next investigated the effect of
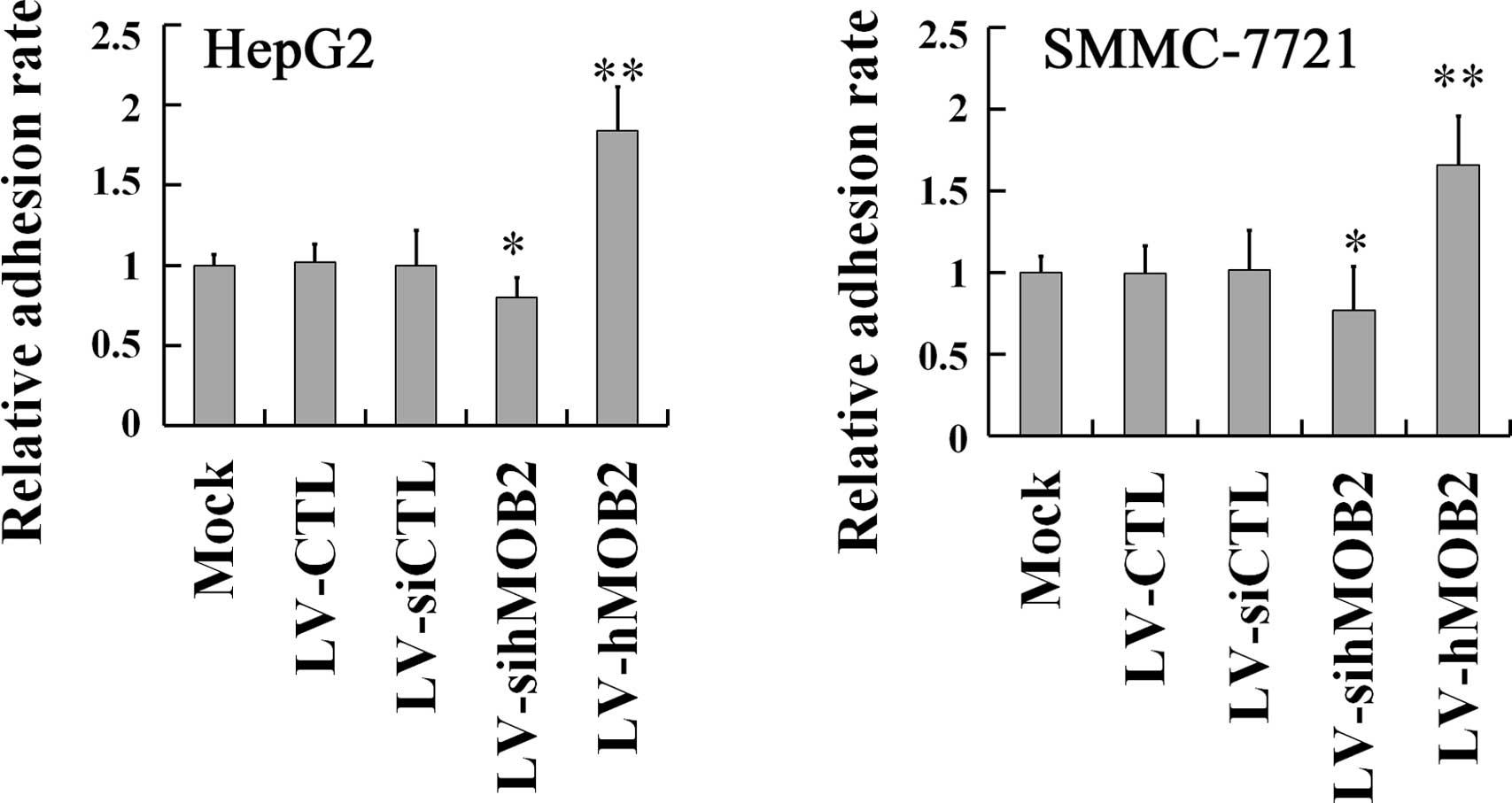
hMOB2 on the cell-matrix adhesion. As shown in Fig. 4, hMOB2 overexpression obviously
promoted both the HepG2 and SMMC-772 cell adhesion onto the
collagen type I matrix, while hMOB2 knockdown decreased the
adhesive behavior either in HepG2 or SMMC-772 cells, when compared
with those in the empty vector-transduced cells and the mock. No
significant difference in the cell-matrix adhesion was observed
among the mock and the empty vector-transduced cells. Given the
results of the cell motility and the cell-matrix adhesion assays,
reduction of the cell motility by hMOB2 overexpression may be the
consequence of the strong or excessive cell-matrix adhesion, while
the decrease in the cell motility caused by hMOB2 knockdown is most
likely attributed to the decrease of the cell-matrix adhesion.
Effect of hMOB2 on the focal adhesion and
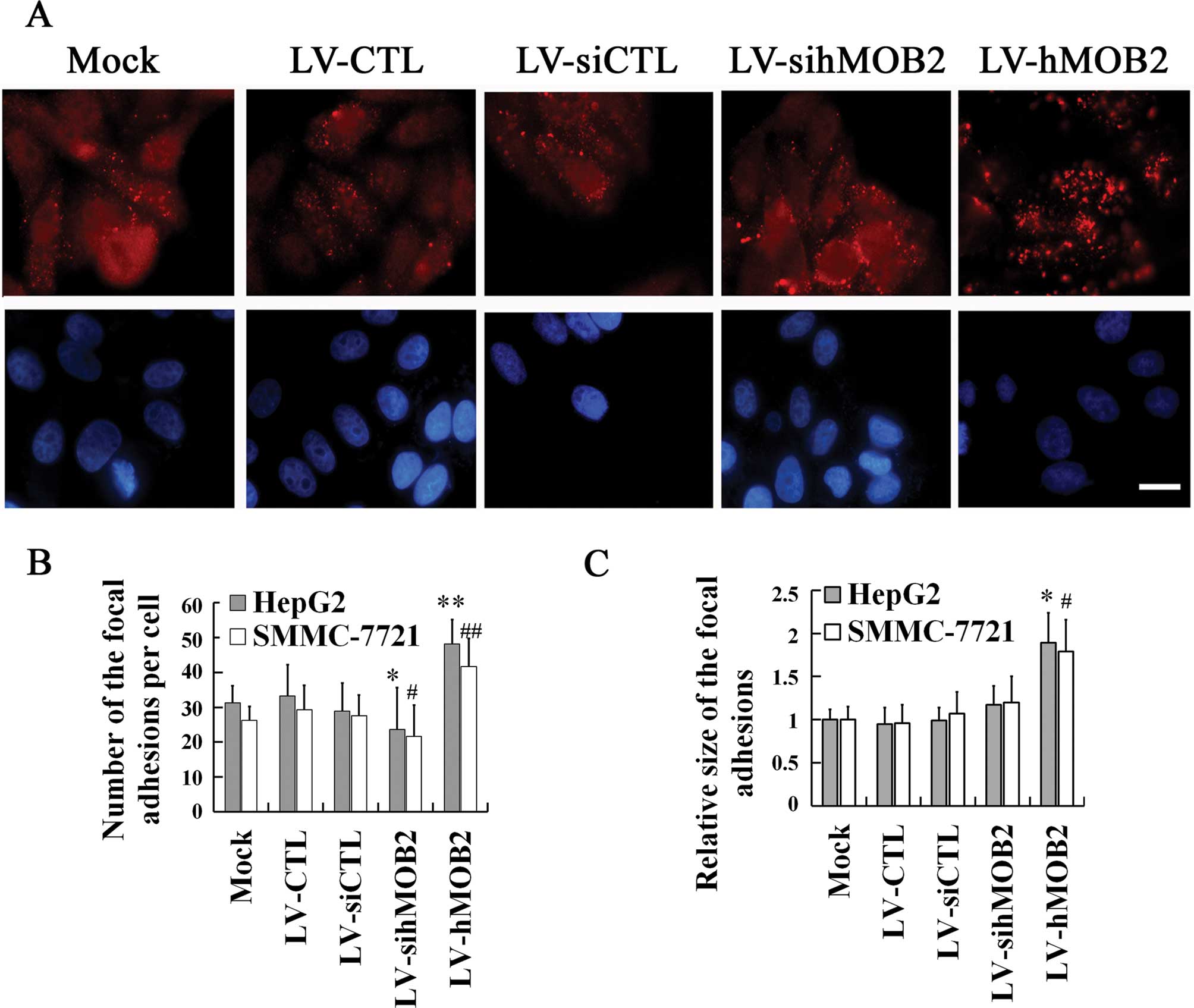
the actin cytoskeleton rearrangement
The focal adhesions are cellular loci through which
the actin filaments establish adhesive links between cell and ECM,
and the dynamic assembly and disassembly of focal adhesion is
essential for cell-matrix adhesion and cell motility (21–23).
To further verify the implication of the hMOB2 expression in the
cell-matrix adhesion and cell motility, we examined the focal
adhesion formation by staining with paxillin (a marker of mature
and nascent focal adhesion) and the remodeling of the actin
cytoskeleton by staining with phalloidin in the
lentivirus-transduced cells. In Fig.
5, hMOB2 overexpression significantly increased both the number
and the size of the focal adhesions and exhibited diffuse
distribution of the focal adhesion throughout the HepG2 cells,
while the hMOB2 knockdown reduced the number of the focal adhesions
producing a prominent peripheral distribution of the HepG2 cells,
when compared with those in the empty vector-transduced cells and
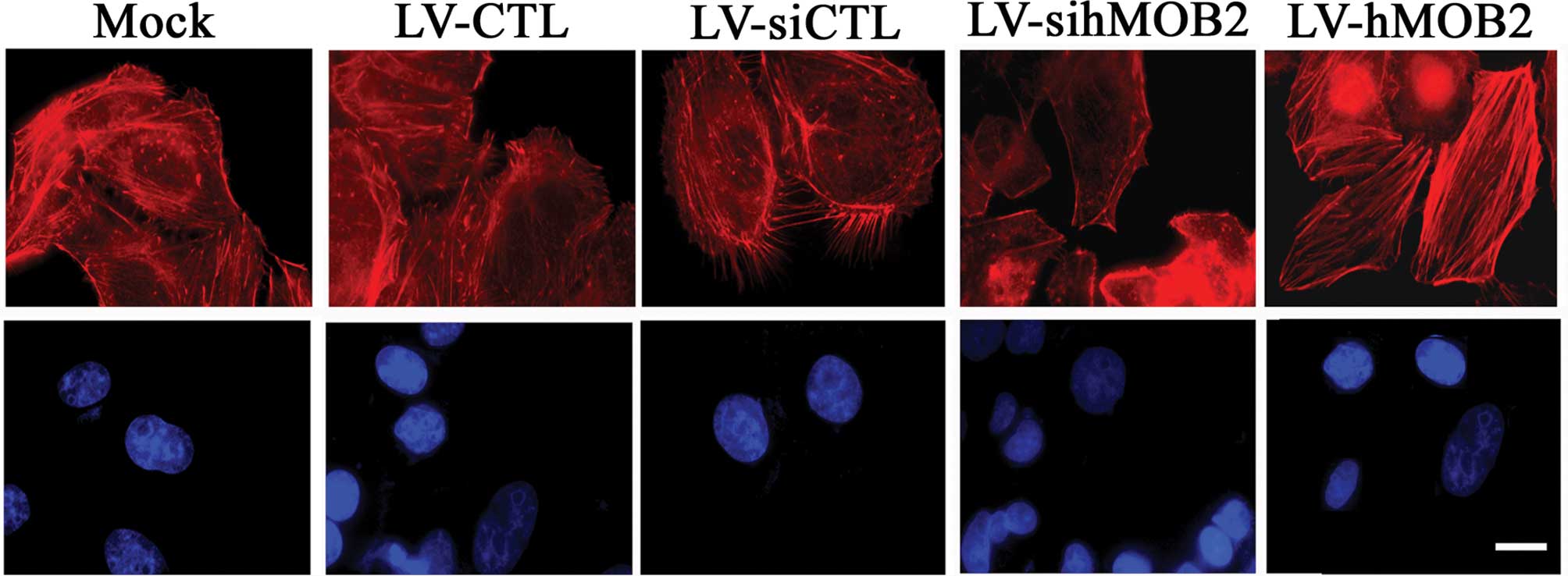
the mock. In parallel, more stress fibers and less filopodia and
lamellipodia at the leading edge were observed in the
hMOB2-overexpressing HepG2 cells than those in the empty
vector-transduced cells and the mock, whereas the hMOB2 knockdown
resulted in the dissociation of the centrally located stress fibers
and exhibited a peripheral distribution of the actin filaments
(Fig. 6). No significant
differences in the focal adhesion formation and the actin
cytoskeleton rearrangement were observed among the mock and the
empty vector-transduced cells. Similar results were also found in
the SMMC-772 cells (data not shown). These data suggested that the
altered assembly of the focal adhesions and the actin cytoskeleton
rearrangement caused by hMOB2 overexpression and knockdown are
possibly involved in the regulation of cell-matrix adhesion and
cell motility.
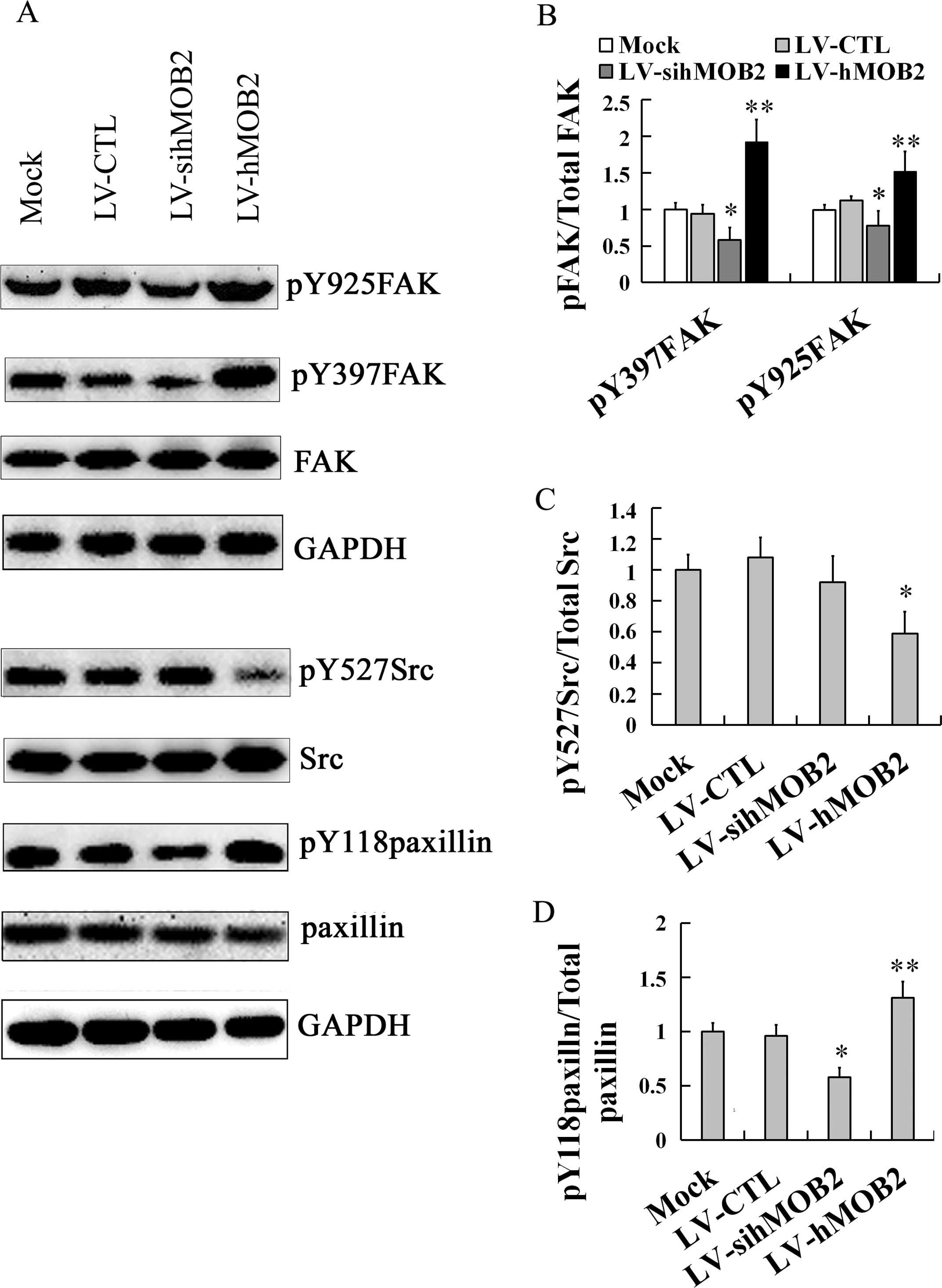
Effect of hMOB2 on the FAK-Src-paxillin
signaling pathway
Since cell-matrix adhesion and cell motility are
dependent on the dynamic assembly and disassembly of focal
adhesions and the remodeling of the actin cytoskeleton, which are
regulated by a variety of signaling molecules, including FAK, Src
and paxillin (24–26), we further performed western blot
assay to evaluate the effects of hMOB2 on the expression of these
signaling molecules. As shown in Fig.
7, hMOB2 overexpression upregulated the expression of
phosporylation of FAK at Y397 (pY397FAK) and Y925 (pY925FAK), and
phosporylation of paxillin at Y118 (pY118paxillin), and
downregulated the phosphorylation of Src at Y527 (pY527Src) while
hMOB2 knockdown obviously downregulated the expression of pY397FAK,
pY925FAK and pY118paxillin, and resulted in only a slight but a
non-significant reduction of the expression of the pY527Src, and no
significant difference was found either in the LV-CTL-transduced or
the LV-siCTL-transduced cells (data not shown). Neither hMOB2
overexpression nor hMOB2 knockdown had significant effects on the
expression of the total FAK, Src and paxillin. Similar results were
also found in the SMMC-772 cells (data not shown). Thus, these
results suggest that FAK-Src-paxillin signaling pathway regulated
by hMOB2 expression is likely involved in the regulation to the
cell-matrix adhesion and cell motility.
Discussion
The findings presented here revealed a currently
unknown role for hMOB2 in regulation of the cell-matrix adhesion
and cell motility. The present study demonstrated that
overexpression of hMOB2 significantly decreased the cell migration
and invasion, but promoted the cell-matrix adhesion, while hMOB2
knockdown decreased not only the cell motility, but also the
cell-matrix adhesion, revealing a novel regulatory mechanism of
cell motility and cell-matrix adhesion induced by hMOB2 expression.
Functional studies further disclosed an important role of hMOB2 in
the modulation of the focal adhesion formation, the remodeling of
the actin cytoskeleton and the FAK-Src-paxillin signaling. We
demonstrated that the altered cell motility and cell-matrix
adhesion induced by hMOB2 expression was likely caused by its
regulation to the assembly of focal adhesions and the actin
cytoskeleton rearrangement through the activation of the
FAK-Src-paxillin signal pathway.
Although hMOB2 has been reported to function as an
inhibitor of NDR1/2 kinases in a binding-dependent manner (8), which has already been linked to the
actin cytoskeleton (17), the
function of hMOB2 expression in tumor cell migration and adhesion
has not been addressed. Previous studies have demonstrated that
mouse Mob2 and NDR activation in neuronal function have already
been linked to the actin cytoskeleton rearrangement (16,17),
which is intimately associated with cell motility and adhesion.
Therefore, our initial experiments were performed to investigate
whether hMOB2 affects tumor cell migration and invasion. Our
findings that hMOB2 overexpression obviously suppressed the cell
migration and invasion and that hMOB2 knockdown also decreased the
cell motility either in HepG2 or SMMC-772 cells suggests a
mechanism by which overexpression and knockdown of hMOB2 attenuated
the general capacity of cell motility. Since cell migration and
invasion require cell-ECM adhesion and then release from ECM,
decreased and strong or excessive cell-ECM adhesion will inhibit
cell motility (20,27). Next, it was evaluated whether hMOB2
overexpression and knockdown inhibited the cell motility through
its regulation for the cell-matrix adhesion. Our findings that
hMOB2 overexpression significantly promoted cell-matrix adhesion
and that hMOB2 knockdown decreased cell-matrix adhesion raised the
exciting possibility that the decrease in the cell motility by
hMOB2 overexpression may be attributed to the strong or excessive
cell-matrix adhesion, while the reduction in the cell motility
caused by hMOB2 knockdown is most likely the consequence of the
decrease of the cell-matrix adhesion.
Cell motility and cell-ECM adhesion depend on the
dynamic assembly and disassembly of the focal adhesions (20,22,23,28–30),
which connect the bundles of actin filaments from cell to ECM and
are often influenced by the formation of stress fibers, and the
dynamic assembly of the actin cytoskeleton plays an essential role
in cell migration and adhesion (18,31).
It has also been well characterized that the strong stress fibers
provide large focal adhesions, while weak stress fibers show small
focal adhesions (31). Thus, we
further performed in immunofluorescence assay to examine the
effects of hMOB2 on the focal adhesions and the actin cytoskeleton
rearrangement. We found that the increased number and size of focal
adhesions which are diffusively distributed throughout the cell,
and the robust stress fibers assembly as well as the decreased
formation of filopodia and lamellipodia at the leading edge were
exhibited in the hMOB2 overexpressing cells, suggesting that hMOB2
overexpression led to the induction of the number and the
enlargement of the focal adhesions. This is consistent with the
enhanced stress fiber formation and the reduction of the formation
of filopodia and lamellipodia and exhibits the characteristics of
the adhesive cells and makes sense with respect to the promoted the
cell-matrix adhesion and subsequently suppressed the cell motility
as mentioned above. The decreased number of the focal adhesions and
the reduction of centrally located stress fiber assembly were
observed in the hMOB2 knockdown cells in which a prominent
peripheral distribution of the focal adhesions and the actin
filaments was shown, which is consistent with the decrease of the
cell-matrix adhesion and subsequently suppressed the cell motility.
Therefore, our observations clearly supported the notion that the
stress fiber formation is directly associated with the cell-matrix
adhesion via the focal adhesions, while the formation of the
filopodia and lamellipodia at the leading edge is associated with
cell motility (32–34). Collectively, these results suggest
that the modulation of cell motility and cell-matrix adhesion by
hMOB2 overexpression and knockdown is possibly mediated through
regulating the assembly of focal adhesions and the actin
cytoskeleton reorganization, providing new insight into the
function of hMOB2 in cell-matrix adhesion and cell motility.
It is generally understood for most cell types that
the dynamic assembly and disassembly of the focal adhesion and the
actin cytoskeleton are regulated by the FAK-Src signaling molecules
(21–23), and the FAK-mediated focal adhesions
linkage to the actin cytoskeleton were partially via binding to
paxillin (22,23,28,29).
Upon activation of the FAK-Src signaling, autophosphorylation of
FAK at Y397 creates a high-affinity binding site for Src and causes
Src recruitment and activation, which thereafter phosphorylates
other sites of FAK including Y925 resulting in the further
activation of FAK, and other cellular scaffolding molecules
including paxillin, thereby acting to modulate the focal adhesion
dynamics and the actin cytoskeleton rearrangement during
cell-matrix adhesion and motility (23,24,31,35–40).
Based on our findings that hMOB2 overexpression and knockdown
regulated the cell motility and cell-matrix adhesion, we further
evaluated the expression of some molecules involved in the FAK-Src
signaling. We found that hMOB2 overexpression promoted
autophosporylation of the FAK at Y397, downregulated the expression
of pY527Src [the phosphorylation of Y527 inhibits Src activities
(41)], and subsequently
upregulated the expression of pY925FAK and pY118paxillin, whereas
hMOB2 knockdown resulted in the downregulation of the expression of
pY397FAK, pY925FAK and pY118paxillin, and a slight but
non-significant reduction of the pY527Src. These data suggest that
the altered cell-matrix adhesion and cell motility induced by hMOB2
overexpression and knockdown probably requires functional
FAK-Src-paxillin signaling molecules to regulate the focal adhesion
formation and the actin cytoskeleton rearrangement, although
details need further study.
In summary, the data presented here demonstrated
that hMOB2 modulated the cell motility and the cell-matrix adhesion
as the result of the regulation of the focal adhesion formation as
well as the actin cytoskeleton rearrangement through the activation
of the FAK-Src-paxillin signal pathway. Although further study is
required to address the underlying mechanisms as to how hMOB2
expression coordinates the FAK-Src-paxillin signal pathway, it is
likely that hMOB2 modulates cell-matrix adhesion and motility,
which will likely contribute to our insight into tumor metastasis
and progression. These findings may reinforce a role for hMOB2 as a
new molecular target for metastatic carcinoma.
Acknowledgments
The present study was supported in part by the
Yangzhou Science and Technology Project (no. YZ2008095), the
Science and Technology Innovation Fund Projects of Yangzhou
University Student, the Yangzhou Vocational University Research
Project (no. 07z10) and the National Nature Science Foundation of
China (no. 81172278).
References
|
1
|
Hergovich A, Kohler RS, Schmitz D,
Vichalkovski A, Cornils H and Hemmings BA: The MST1 and hMOB1 tumor
suppressors control human centrosome duplication by regulating NDR
kinase phosphorylation. Curr Biol. 19:1692–1702. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hergovich A: MOB control: Reviewing a
conserved family of kinase regulators. Cell Signal. 23:1433–1440.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stavridi ES, Harris KG, Huyen Y, Bothos J,
Verwoerd PM, Stayrook SE, Pavletich NP, Jeffrey PD and Luca FC:
Crystal structure of a human Mob1 protein: Toward understanding
Mob-regulated cell cycle pathways. Structure. 11:1163–1170. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ponchon L, Dumas C, Kajava AV, Fesquet D
and Padilla A: NMR solution structure of Mob1, a mitotic exit
network protein and its interaction with an NDR kinase peptide. J
Mol Biol. 337:167–182. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lai ZC, Wei X, Shimizu T, Ramos E,
Rohrbaugh M, Nikolaidis N, Ho LL and Li Y: Control of cell
proliferation and apoptosis by mob as tumor suppressor, mats. Cell.
120:675–685. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mrkobrada S, Boucher L, Ceccarelli DF,
Tyers M and Sicheri F: Structural and functional analysis of
Saccharomyces cerevisiae Mob1. J Mol Biol. 362:430–440. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ho LL, Wei X, Shimizu T and Lai ZC: Mob as
tumor suppressor is activated at the cell membrane to control
tissue growth and organ size in Drosophila. Dev Biol. 337:274–283.
2010. View Article : Google Scholar
|
|
8
|
Kohler RS, Schmitz D, Cornils H, Hemmings
BA and Hergovich A: Differential NDR/LATS interactions with the
human MOB family reveal a negative role for human MOB2 in the
regulation of human NDR kinases. Mol Cell Biol. 30:4507–4520. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tavares A, Gonçalves J, Florindo C,
Tavares AA and Soares H: Mob1: Defining cell polarity for proper
cell division. J Cell Sci. 125:516–527. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chow A, Hao Y and Yang X: Molecular
characterization of human homologs of yeast MOB1. Int J Cancer.
126:2079–2089. 2010.
|
|
11
|
Florindo C, Perdigão J, Fesquet D,
Schiebel E, Pines J and Tavares AA: Human Mob1 proteins are
required for cytokinesis by controlling microtubule stability. J
Cell Sci. 125:3085–3090. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang ZX, Wang HY and Wu MC: Identification
and characterization of a novel human hepatocellular
carcinoma-associated gene. Br J Cancer. 85:1162–1167. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He Y, Emoto K, Fang X, Ren N, Tian X, Jan
YN and Adler PN: Drosophila Mob family proteins interact with the
related tricor-nered (Trc) and warts (Wts) kinases. Mol Biol Cell.
16:4139–4152. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Emoto K, He Y, Ye B, Grueber WB, Adler PN,
Jan LY and Jan YN: Control of dendritic branching and tiling by the
Tricornered-kinase/Furry signaling pathway in Drosophila sensory
neurons. Cell. 119:245–256. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu LY, Lin CH and Fan SS: Function of
Drosophila mob2 in photoreceptor morphogenesis. Cell Tissue Res.
338:377–389. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin CH, Hsieh M and Fan SS: The promotion
of neurite formation in Neuro2A cells by mouse Mob2 protein. FEBS
Lett. 585:523–530. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stork O, Zhdanov A, Kudersky A, Yoshikawa
T, Obata K and Pape HC: Neuronal functions of the novel
serine/threonine kinase Ndr2. J Biol Chem. 279:45773–45781. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Akakura S and Gelman IH: Pivotal role of
AKAP12 in the regulation of cellular adhesion dynamics: Control of
cytoskeletal architecture, cell migration, and mitogenic signaling.
J Signal Transduct. 2012:5291792012.PubMed/NCBI
|
|
19
|
Lock JG, Wehrle-Haller B and Strömblad S:
Cell-matrix adhesion complexes: Master control machinery of cell
migration. Semin Cancer Biol. 18:65–76. 2008. View Article : Google Scholar
|
|
20
|
Zong F, Fthenou E, Mundt F, Szatmári T,
Kovalszky I, Szilák L, Brodin D, Tzanakakis G, Hjerpe A and Dobra
K: Specific syndecan-1 domains regulate mesenchymal tumor cell
adhesion, motility and migration. PLoS One. 6:e148162011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Geiger B, Spatz JP and Bershadsky AD:
Environmental sensing through focal adhesions. Nat Rev Mol Cell
Biol. 10:21–33. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lanning NJ, Su HW, Argetsinger LS and
Carter-Su C: Identification of SH2B1β as a focal adhesion protein
that regulates focal adhesion size and number. J Cell Sci.
124:3095–3105. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Webb DJ, Donais K, Whitmore LA, Thomas SM,
Turner CE, Parsons JT and Horwitz AF: FAK-Src signalling through
paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell
Biol. 6:154–161. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fraley SI, Feng Y, Krishnamurthy R, Kim
DH, Celedon A, Longmore GD and Wirtz D: A distinctive role for
focal adhesion proteins in three-dimensional cell motility. Nat
Cell Biol. 12:598–604. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
van Nimwegen MJ and van de Water B: Focal
adhesion kinase: A potential target in cancer therapy. Biochem
Pharmacol. 73:597–609. 2007. View Article : Google Scholar
|
|
26
|
Mitra SK and Schlaepfer DD:
Integrin-regulated FAK-Src signaling in normal and cancer cells.
Curr Opin Cell Biol. 18:516–523. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ishikawa T and Kramer RH: Sdc1 negatively
modulates carcinoma cell motility and invasion. Exp Cell Res.
316:951–965. 2010. View Article : Google Scholar :
|
|
28
|
Geiger B, Bershadsky A, Pankov R and
Yamada KM: Transmembrane crosstalk between the extracellular matrix
- cytoskeleton crosstalk. Nat Rev Mol Cell Biol. 2:793–805. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hynes RO: Integrins: Bidirectional,
allosteric signaling machines. Cell. 110:673–687. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gupton SL and Waterman-Storer CM:
Spatiotemporal feedback between actomyosin and focal-adhesion
systems optimizes rapid cell migration. Cell. 125:1361–1374. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hotulainen P and Lappalainen P: Stress
fibers are generated by two distinct actin assembly mechanisms in
motile cells. J Cell Biol. 173:383–394. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hall A: Rho GTPases and the actin
cytoskeleton. Science. 279:509–514. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ridley AJ: Rho GTPases and cell migration.
J Cell Sci. 114:2713–2722. 2001.PubMed/NCBI
|
|
34
|
Machacek M, Hodgson L, Welch C, Elliott H,
Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM and Danuser G:
Coordination of Rho GTPase activities during cell protrusion.
Nature. 461:99–103. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu J, Di G, Wu CT, Hu X and Duan H:
CEACAM1 inhibits cell-matrix adhesion and promotes cell migration
through regulating the expression of N-cadherin. Biochem Biophys
Res Commun. 430:598–603. 2013. View Article : Google Scholar
|
|
36
|
Luo M and Guan JL: Focal adhesion kinase:
A prominent determinant in breast cancer initiation, progression
and metastasis. Cancer Lett. 289:127–139. 2010. View Article : Google Scholar :
|
|
37
|
Zhao J and Guan JL: Signal transduction by
focal adhesion kinase in cancer. Cancer Metastasis Rev. 28:35–49.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Van Slambrouck S, Jenkins AR, Romero AE
and Steelant WF: Reorganization of the integrin α2 subunit controls
cell adhesion and cancer cell invasion in prostate cancer. Int J
Oncol. 34:1717–1726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Van Slambrouck S, Grijelmo C, De Wever O,
Bruyneel E, Emami S, Gespach C and Steelant WF: Activation of the
FAK-src molecular scaffolds and p130Cas-JNK signaling cascades by
α1-integrins during colon cancer cell invasion. Int J Oncol.
31:1501–1508. 2007.PubMed/NCBI
|
|
40
|
Crosara-Alberto DP, Inoue RY and Costa CR:
FAK signalling mediates NF-kappaB activation by mechanical stress
in cardiac myocytes. Clin Chim Acta. 403:81–86. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bjorge JD, Jakymiw A and Fujita DJ:
Selected glimpses into the activation and function of Src kinase.
Oncogene. 19:5620–5635. 2000. View Article : Google Scholar : PubMed/NCBI
|