Introduction
Breast cancer continues to be the most common type
of cancer diagnosed among women in China, and it was estimated that
there were ~232,340 new cases of invasive breast cancer in 2013
(1). The American Cancer Society
projected ~39,620 breast cancer-associated mortalities in 2013
(2). In the past three decades,
breast cancer has been the second leading cause of cancer death
after lung cancer (3). However,
breast cancer is a complex disease entity with different biological
characteristics and clinical behavior. Many studies have shown that
there are no treatment guidelines for triple-negative breast
cancer.
Adjuvant endocrine therapy is an important method
used for the comprehensive treatment of breast cancer (4). Chemotherapy for breast cancer is
closely associated with hormone levels. Tamoxifen (TAM) is
non-synthetic cholesterol antiestrogen, which is the basic drug
utilized in adjuvant endocrine therapy for breast cancer. TAM is
widely used to treat breast cancer patients (5). However, the differences in the gene
polymorphism of patients who are administered the drug may lead to
individual differences regarding efficacy and toxicity (6). Genetic factors play an important role
in these differences.
Organic anion transporting polypeptide 1B1 (OATP1B1)
is a type of intake body, distributed in multiple human organs and
particularly distributed in specific organs (7). It belongs to the superfamily of solute
uptake, of which the encoding genes are known as the SLCO1B1
gene collectively. In the Chinese population, mutation frequencies
of OATP1B1 388A>G and OATP1B1 521T>C are 73.4 and 14.0%,
respectively, which are similar to those of the Japanese population
(8,9). The incidence rate of OATP1B1 521T>C
is 16% in Asians and 18% in Caucasians, thus this incidence rate is
varied based on ethnicity (10).
Studies have shown that in African-Americans, the mutation rate of
OATP1B1 388G is 2.5-fold that of Europeans and Americans, with the
mutation rate of OATP1B1 521C being higher among the Finnish
population. The mutations of OATP1B1*15 (388G and 521C) is 43.4% in
the Japanese population simultaneously. However, the incidence rate
of OATP1B1*15 is 14% in Caucasians (11).
The majority of current studies concerning the
genetic polymorphism of OATPlB1 have focused on statin drugs, which
enter the liver cells to exert an inhibitory effect on HMG-CoA
reductase. Therefore, OATP1B1 is important in most internalization
processes of statin drugs into hepatocytes (12). Recent findings have shown that the
genetic polymorphism of OATPlB1 affects the pharmacokinetic process
of irinotecan in cancer patients significantly, including
OATP1B1*la, OATP1B1*lb, OATP1B1*5 and OATP1B1*15 (13,14).
There were no significant effects on simvastatin
pharmacokinetics by SLCO1B1 388A>G, SLCO1B1 521T>C, SLCO1B1
11187G>A, SLCO1B1 571T>C and SLCO1B1 597C>T. Ritonavir
intracellular concentrations were associated with OATP1B1 521T>C
polymorphism (15). Previous
results suggest that the gene mutation of OATP1B1 521T>C
influences the transport of rosuvastatin (16). However, no association between the
OATP1B1 521T>C polymorphism and the cholesterol synthesis
response and rosuvastatin plasma concentration has been identified
(17). OATP1B1 388A>G and
521T>C have been shown to lead to altered pharmaco-kinetics of
pravastatin. The pharmacokinetics of irinotecan, estrone-3-sulfate
and estradiol-17β-glucuronide also affect OATP1B1 388A>G and
521T>C, although without statistical significance (18).
In the present study, we found that the specific
mutation of OATP1Bl significantly reduced the uptake activity of
OATP1Bl protein, which may affect the internalization of many
endogenous substances and drug uptake into hepatocytes and
significantly influence the metabolism and efficacy of many drugs.
OATP1Bl also influences the adverse reactions of some drugs and the
interaction between drugs. A gene polymorphism expression platform
of OATP1Bl 388GG and 521CC was also established and utilized to
determine whether OATP1Bl 388GG and OATP1Bl 521CC gene
polymorphisms can affect TAM treatment of breast cancer in
vitro.
Materials and methods
Total RNA extraction
Approximately 100 mg tissue was ground into a powder
in liquid nitrogen, and moderate RNAiso Plus (Qiagen, Hilden,
Germany) was added for 5–10 min at room temperature. The
supernatant was collected by centrifugation at 12,000 rpm for 5 min
at 4°C followed by the addition of chloroform. Miscible liquids
were agitated and placed for 5–10 min at room temperature. The
miscible liquids were collected by centrifugation at 12,000 rpm for
15 min at 4°C. The supernatant was absorbed into the new centrifuge
tube. The isopropanol, which was the same volume as that of the
supernatant, was added to the centrifuge tube, and placed for 10
min at room temperature. The liquid was centrifuged at 12,000 rpm
for 10 min to obtain a sediment. One millimeter 75% ethyl alcohol
was added to the centrifuge tube. The liquid was centrifuged at
12,000 rpm for 5 min to obtain sediment again. The sediment
obtained was dried at room temperature, and then treated in 20–100
μl DEPC-H2O. Total RNA was maintained at
−80°C.
cDNA synthesis
Total RNA (1 μl), 1 μl 0.5
μg/μl oligo(dT)18 and 10 μl
DEPC-H2O was mixed, subjected to PCR at 70°C for 5 min
and cooled rapidly on ice. Four microliters 5X reaction buffer, 1
μl 20 U/μl RNA enzyme inhibitors and 2 μl 10
mmol/l dNTP were dissolved in miscible liquids and subjected to PCR
at 37°C for 5 min. One microliter 200 U/μl reverse
transcriptase was added and the mixture was subjected to PCR.
Cycling conditions used were: 42°C for 60 min and 70°C for 10 min.
cDNA synthesis was maintained at −80°C.
DNA extraction
The primers were designed according to gene coding
sequence (CDS) of OATP1B1 in GenBank. KpnI and NotI
restriction sites were subsequently inserted. The primers used
were: forward: 5′-TTAGCGGCCGCATGGACCAAAATCAACATT-3′ and reverse,
5′-GCCGGTACCTTAACAATGTGTTTCACTATCTG-3′.
cDNA (2–4 μl), 2 μl sense primer (10
pM), 2 μl reverse primer (10 pM), 4 μl dNTP (2 mM), 5
μl 10X PCR buffer, 1 μl Taq DNA Polymerase
(Takara, Otsu, Japan) and moderate DEPC-H2O were mixed
in the centrifuge tube, and the total volume was 50 μl.
Cycling conditions used were: 94°C for 10 min followed by 40 cycles
at 95°C for 30 sec, 58°C for 90 sec and 72°C for 30 sec, and then
72°C for 7 min. DNA was kept at −80°C. DNA OATP1B1 was analyzed by
electrophoresis of 0.5 or 1% agarose gel followed by gene
sequencing.
Construction of plasmids
KpnI and NotI, DNA OATP1B1 and
pcDNA3.1(-) plasmid were mixed at 37°C for 1–4 h and at 70°C for 10
min according to the manufacturer’s instructions (Takara). T4 DNA
ligase (Takara) was then added to the mixture at 16°C for 1–4 h and
at 70°C for 10 min. pcDNA3.1(-)-OATP1B1 plasmid was identified by
restriction enzyme digestion (KpnI and NotI), and
analyzed by electrophoresis of 0.5% agarose gel and gene
sequencing.
Site-directed mutagenesis of OATP1B1
388GG and 521CC
The site-directed mutagenesis primers of OATP1B1
388GG and OATP1B1 521CC were designed according to OATP1B1 gene CDS
in GenBank (Table I). The plasmid
of 1 μl pcDNA3.1 (-)-OATP1B1, 5 μl 10X Pfu polymerase
buffer (Mg2+), 2 μl sense primer (10 pM), 2
μl reverse primer (10 pM), 1 μl Pfu DNA polymerase (5
U) (Promega, Shanghai, China), and 39 μl DEPC-H2O
were mixed and subjected to PCR. Cycling conditions were as
follows: 95°C for 4 min followed by 25 cycles at 95°C for 60 sec,
62°C for 45 sec and 72°C for 30 sec, and then 72°C for 7 min. PCR
amplification products were digested with the restriction
endonuclease ClaI (Promega) at 37°C for 1–3 h. The plasmids
were maintained at −80°C. The plasmids were separated using 0.5 or
1% agarose gel followed by gene sequencing.
 | Table IThe site-directed mutagenesis primers
of OATP1B1 388GG and 521CC. |
Table I
The site-directed mutagenesis primers
of OATP1B1 388GG and 521CC.
| Name of primer | 5′→3′ |
|---|
| 388GG |
GAAAGAAACTAATATCGATTCATCAGAAAATTC |
| 388GG |
GATTATAGGTAAGTAGTCTTTTAAGTTGTAGC |
| 521CC |
ACATGTGGATATATGCGTTCATGGGTAATAT |
| 521CC |
TATTACCCATGAACCCATATATCCACATG |
Plasmid transfection
The plasmids were constructed and transfected into
MCF-7 cells using Lipofectamine 2000 (Invitrogen) according to the
manufacturer’s instructions in a humidified atmosphere at 37°C with
5% CO2. After 6 h, the transfection medium was replaced
with Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal
bovine serum (FBS) without antibiotic in a humidified atmosphere at
37°C with 5% CO2. The OATP1B1 gene polymorphisms were
examined in MCF-7 cell lines by RT-PCR and western blot assay.
Cell culture
MCF-7 human breast cancer cells were obtained from
the Shanghai Institute of Cell Biology in the Chinese Academy of
Sciences for the present study. MCF-7 cells were maintained in DMEM
containing 10% FBS supplemented with 100 U penicillin/streptomycin
in a humidified atmosphere at 37°C with 5% CO2.
Experiments
The experiment was divided into six groups: A group
was MCF-7; B group was MCF-7 with TAM (10 μM); C group was
MCF-7 transfected with pcDNA3.1(-) plasmid and provided with TAM
(10 μM); D group was MCF-7 transfected with
pcDNA3.1(-)-OATP1B1 plasmid and provided with TAM (10 μM); E
group was MCF-7 transfected with pcDNA3.1(-)-OATP1B1 388GG plasmids
and provided with TAM (10 μM); and F group was MCF-7
transfected with pcDNA3.1(-)-OATP1B1 521CC plasmids and provided
with TAM (10 μM).
MCF-7 cell proliferation assay
MCF-7 cell proliferation was examined using the
colorimetric assay using
3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assays. Cells were plated at 2.5−5×105 cells/well in a
96-well plate and incubated for 24 h. The plasmids were transfected
into MCF-7 cells. The cells were treated with TAM solutions at
concentrations of 10 μM for 24 and 48 h. Phosphate-buffered
saline (PBS; pH 7.4) alone was used as the vehicle-group. TAM was
dissolved in PBS and was used as treat-groups. For the MTT assay,
the vehicle- and treat-groups were incubated with 150 μl MTT
for 4 h at 37°C. The medium was extracted from the plates. The
plates were incubated with 100 μl dimethyl sulf-oxide for 10
min at room temperature and agitated. The optic density was
determined using an ELISA reader at 540 nm.
Quantification of apoptotic cells by flow
cytometry
Apoptotic cells were measured by flow cytometry (BD
Biosciences, San Diego, CA, USA). The cells were stained using an
Annexin V-FITC Apoptosis Detection kit according to the
manufacturer’s instructions. Briefly, MCF-7 cells
(5.0×105 cells/well) were plated in 6-well plates and
incubated for 24 h. The plasmids were subsequently transfected into
MCF-7 cells. The cells were treated with TAM solutions at
concentrations of 10 μM for 24 and 48 h. PBS (pH 7.4) alone
was used as the vehicle-group. TAM was dissolved in PBS and was
used as treat-groups. For the apoptotic cell analysis, the cells of
the vehicle- and treat-groups were trypsinized without
ethylenediaminetetraacetic acid (EDTA) and centrifuged at 2,000 × g
for 5 min. The cells were collected, washed in PBS (pH 7.4), and
centrifuged at 2,000 ×x g for 5 min. The cell samples were
suspended with 500 μl combined buffer solution. Then, 5
μl fluorescein isothiocyanate (FITC)-Annexin V and 10
μl propidium iodide (PI) were added into the cell samples
and incubated for 5–15 min at room temperature in the dark.
Apoptotic cells were analyzed by flow cytometry (BD
Biosciences).
Cell cycle analysis by flow
cytometry
Cell cycle analysis of MCF-7 cells was examined
using the standard PI method by flow cytometry (BD Biosciences).
Briefly, MCF-7 cells (5.0×105 cells/well) were plated in
6-well plates and incubated for 24 h. The plasmids were
subsequently transfected into MCF-7 cells. The cells were treated
with TAM solutions at concentrations of 10 μM for 24 and 48
h. PBS (pH 7.4) alone was used as the vehicle-group. TAM was
dissolved in PBS and was used as treat-groups. For the cell cycle
analysis, the cells of the vehicle- and treat-groups were
trypsinized without EDTA and centrifuged at 2,000 × g for 5 min.
The cells were harvested, washed in PBS (pH 7.4) and fixed in 70%
with ice-cold ethanol at 0–4°C overnight. The cells were collected
and centrifuged at 2,000 × g for 5 min and the supernatant was
removed. The cell pellet was resuspended and washed in PBS (pH
7.4). The cell solution was transferred to a flow tube, and
incubated with 10 ml of RNase A (10 mg/ml) for 1 h at 37°C. The
cell samples were incubated with 50 ml of PI (250 mg/ml) for 30 min
in the dark at 0–4°C. The cell cycle proliferation index was
subsequently calculated using the formula: PI = (G2/M +
S)/ (G0/G1 + S + G2/M) ×100%. The
cell cycle distribution was analyzed by flow cytometry (BD
Biosciences).
Statistical analysis
The experiments were repeated at least three times.
Data are presented as the mean ± SEM. The differences between means
were analyzed using the two-tailed Student’s t-test. Statistical
analyses were performed using SPSS 17.0 software. Differences with
P<0.05 were considered to indicate a statistically significant
result.
Results
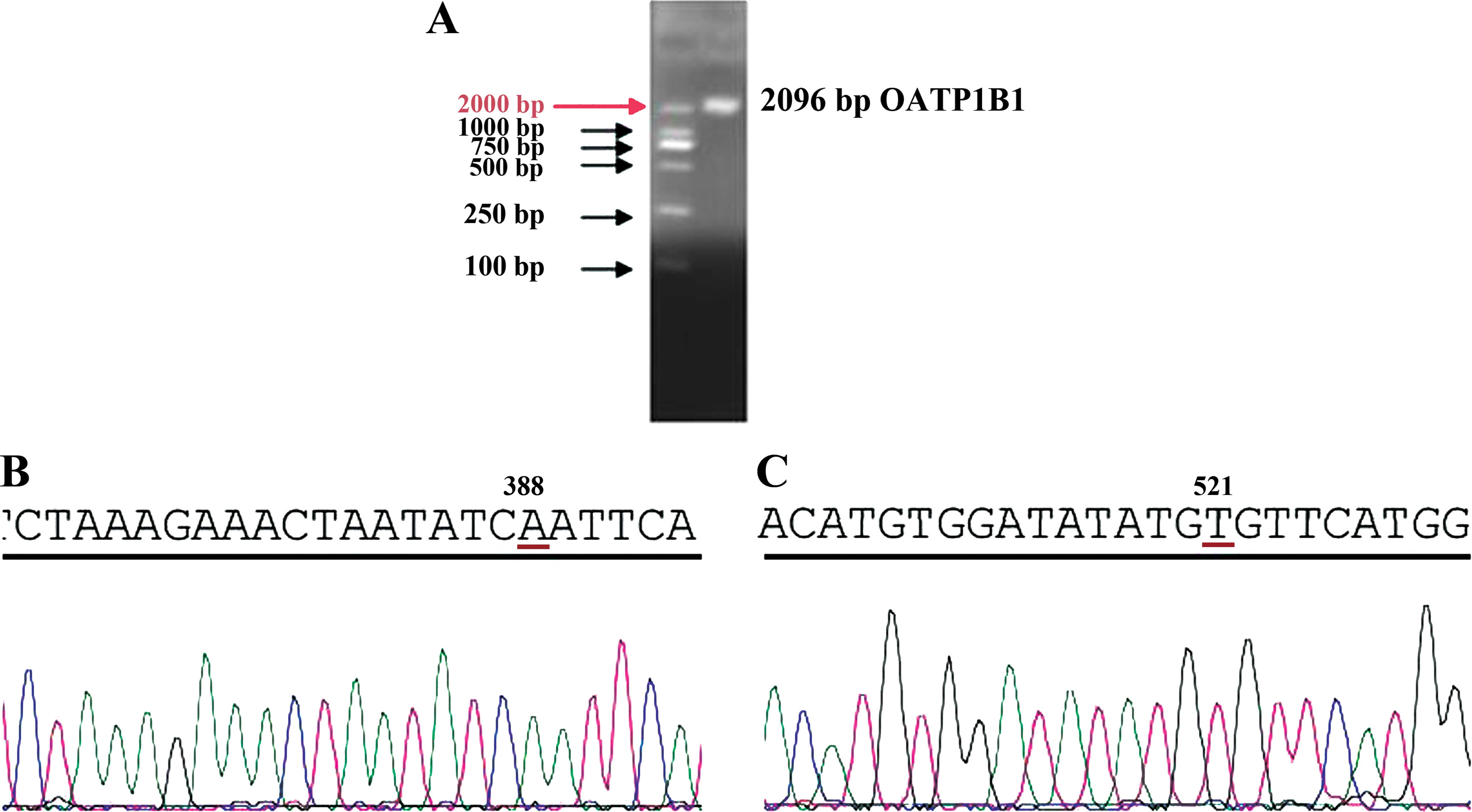
DNA fragmentation of OATP1B1
DNA fragmentation of OATP1B1 was analyzed by
electrophoresis of 1% agarose gel, which showed that the amplified
gene fragment was 2,096 bp (Fig.
1A). The DNA fragmentation of OATP1B1 was analyzed by gene
sequencing, which showed that the amplified gene fragment
identified the 388 site as A, and the 521 site as T (Fig. 1B and C).
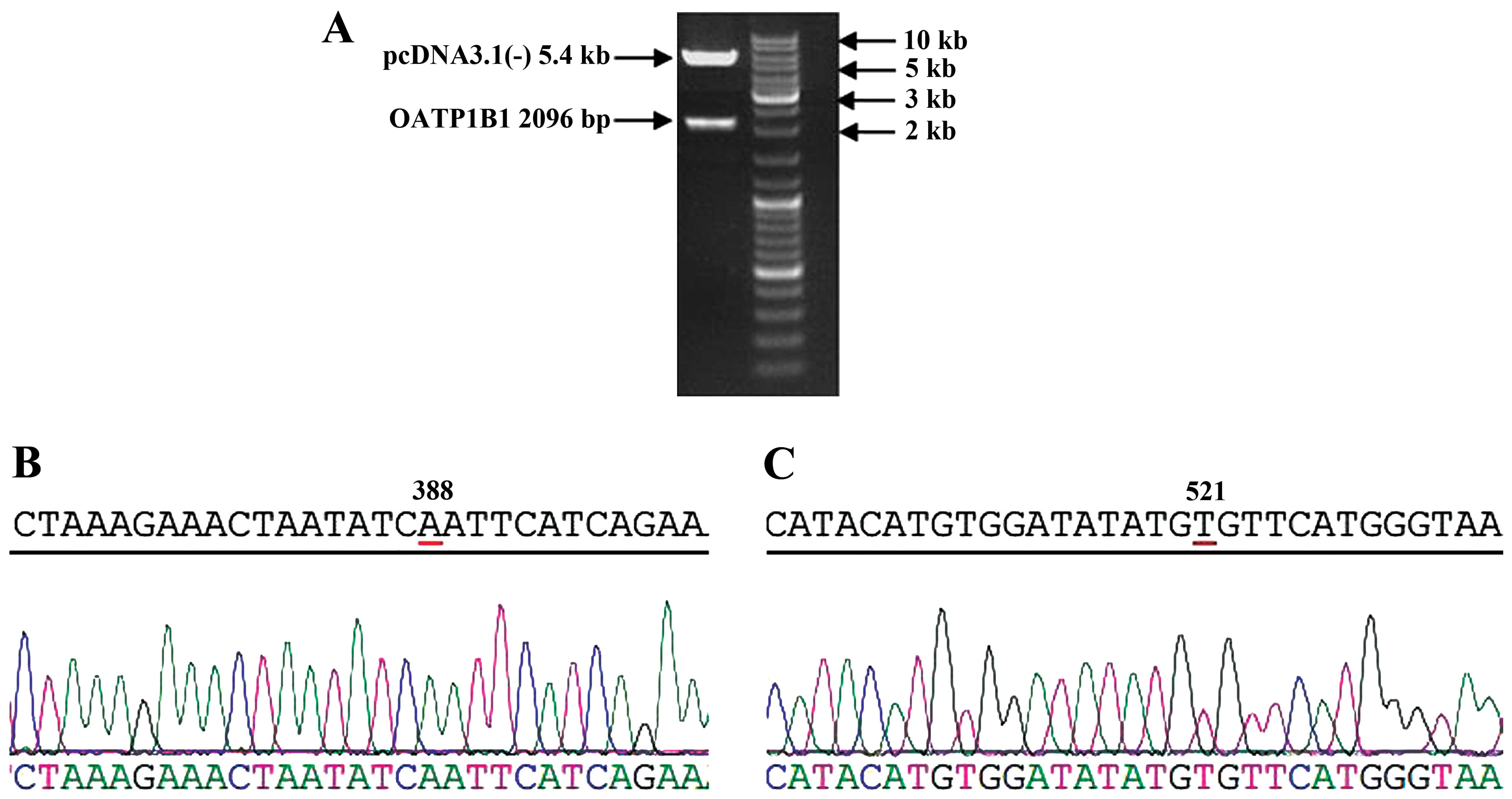
Restriction enzyme and gene sequence of
pcDNA3.1(-)-OATP1B1
The established plasmids of pcDNA3.1(-)-OATP1B1 were
digested by restriction enzymes (KpnI and NotI). The
products were analyzed by electrophoresis of 0.5% agarose gel,
which showed that pcDNA3.1(-) was 5.4 kb, while the OATP1B1
fragment was 2,096 bp (Fig. 2A).
Gel extraction of the OATP1B1 fragment was performed and the
fragment was analyzed by gene sequencing, which showed that the
amplified gene fragment identified the 388 site as A, and the 521
site as T (Fig. 2B and C).
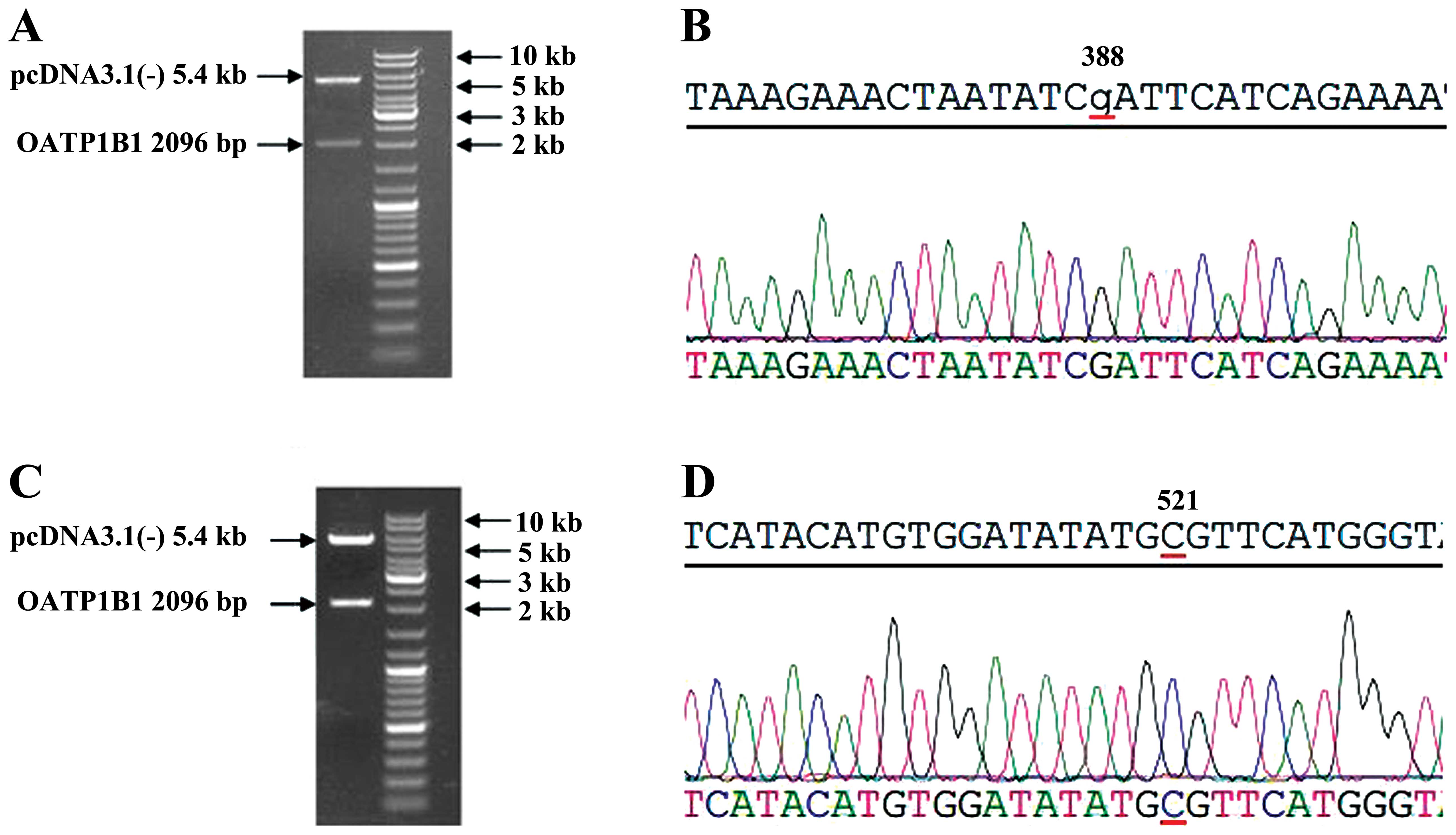
Restriction enzymes and gene sequencing
of pcDNA3.1(-)-OATP1B1 388GG and 521 CC
The established plasmids of pcDNA3.1(-)-OATP1B1
388GG were digested by restriction enzymes, KpnI and
NotI. These products were analyzed by electrophoresis of
0.5% agarose gel, which showed that pcDNA3.1(-) was 5.4 kb, and the
OATP1B1 fragment was 2,096 bp (Fig.
3A). The OATP1B1 fragment was extracted using gel extraction
and analyzed by gene sequencing. The amplified gene fragment
identified at the 388 site was G (Fig.
3B).
The established plasmids of pcDNA3.1(-)-OATP1B1
521CC were digested by restriction enzymes, KpnI and
NotI. The products were analyzed by electrophoresis of 0.5%
agarose gel, which showed that pcDNA3.1(-) was 5.4 kb, and the
OATP1B1 fragment was 2,096 bp (Fig.
3C). The OATP1B1 fragment was extracted using gel extraction
and was analyzed by gene sequencing, which showed that the
amplified gene fragment identified at the 521 site was C (Fig. 3D).
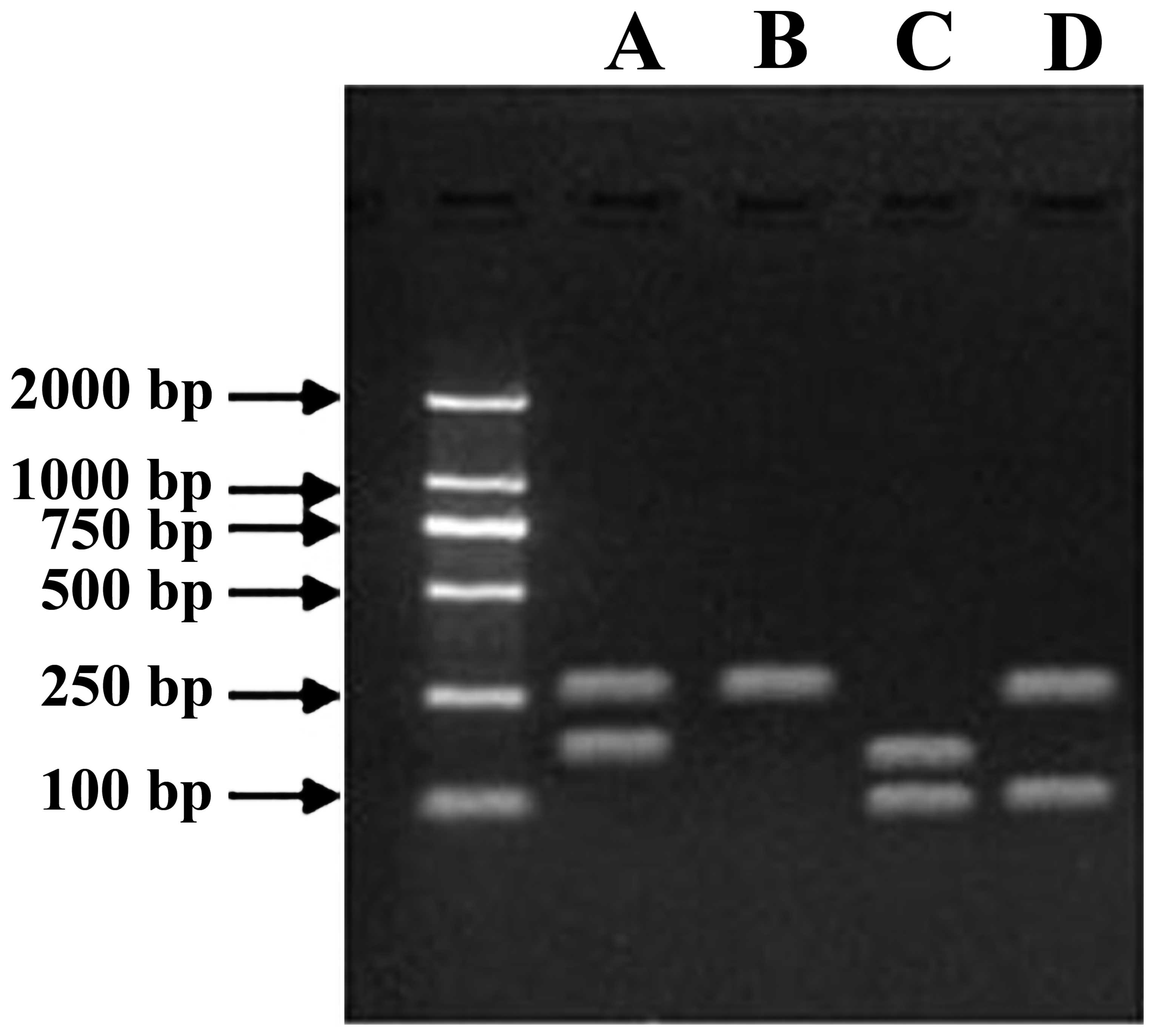
Cataphoresis of plasmid transfection
RNA was extracted from the transfection of MCF-7
cells of each group, and PCR was used to amplify double-stranded
DNA. The electrophoretic analysis was performed on the product
using 1% agarose gel, which showed that the plasmid was
successfully expressed (Fig.
4).
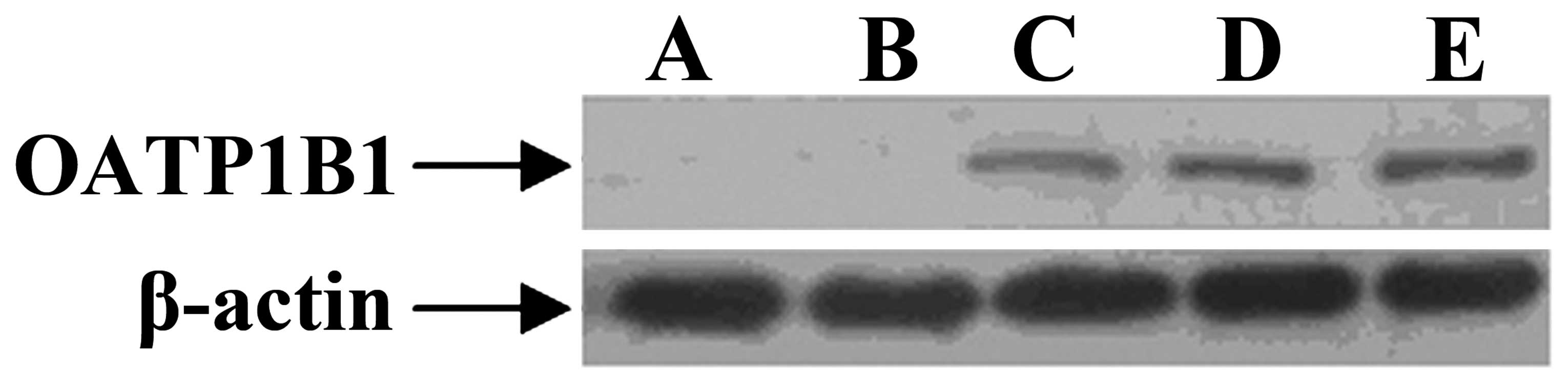
Western blotting of plasmid
transfection
The OATP1B1 gene polymorphisms in MCF-7 cell lines
were examined by western blot assay. A was MCF-7 cell, B was
pcDNA3.1(-) was transfected into MCF-7 cell, C was
pcDNA3.1(-)-OATP1B1 was transfected into MCF-7 cell, D was
pcDNA3.1(-)-OATP1B1 388GG was transfected into MCF-7 cell, E was
pcDNA3.1(-)-OATP1B1 521CC was transfected into MCF-7 cell. A and B
did not express OATP1B1 protein, whereas C-E expressed OATP1B1
protein (Fig. 5).
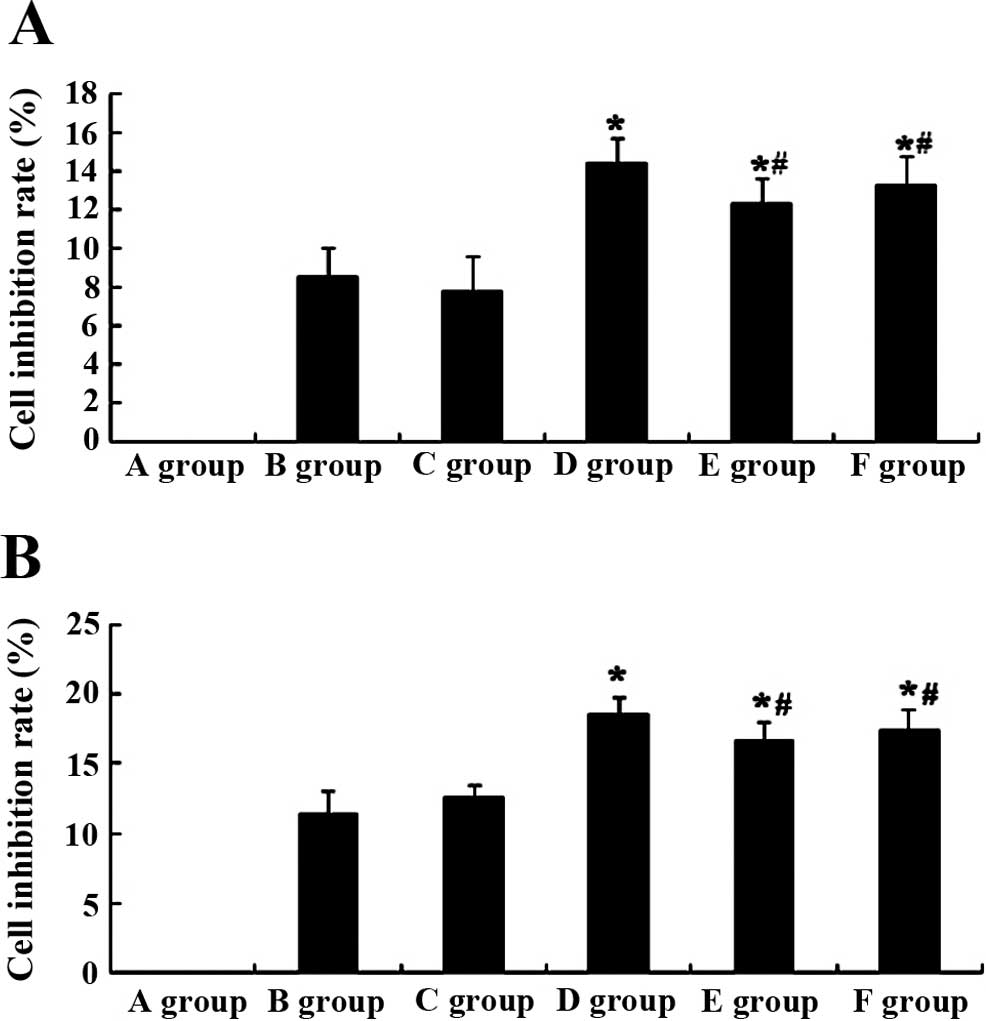
TAM effect on the cell inhibition rate of
MCF-7 by plasmid transfection
We established expression platforms of OATP1B1
genetic polymorphisms to assess the role of OATP1B1 genetic
polymorphisms of the effect of TAM (10 μM) on the inhibition
rate of MCF-7 cells. The results showed that at 24 h, the
inhibition rate of the D group was markedly increased (P<0.05,
n=3), compared to the B group (Fig.
6A). However, the inhibition rate of MCF-7 cells of the E and F
groups was slightly decreased (P>0.05, n=3) compared to the D
group, although the difference was not statistically significant
(Fig. 6A).
After the effect of TAM (10 μM) on the
proliferation of MCF-7 cells at 48 h, the inhibition rate of D
group was markedly increased (P<0.05, n=3), compared to the B
group (Fig. 6B). However, the
inhibition rate of MCF-7 cells of the E and F groups were slightly
decreased (P>0.05, n=3) compared to the D group, although the
difference was not statistically significant (Fig. 6B).
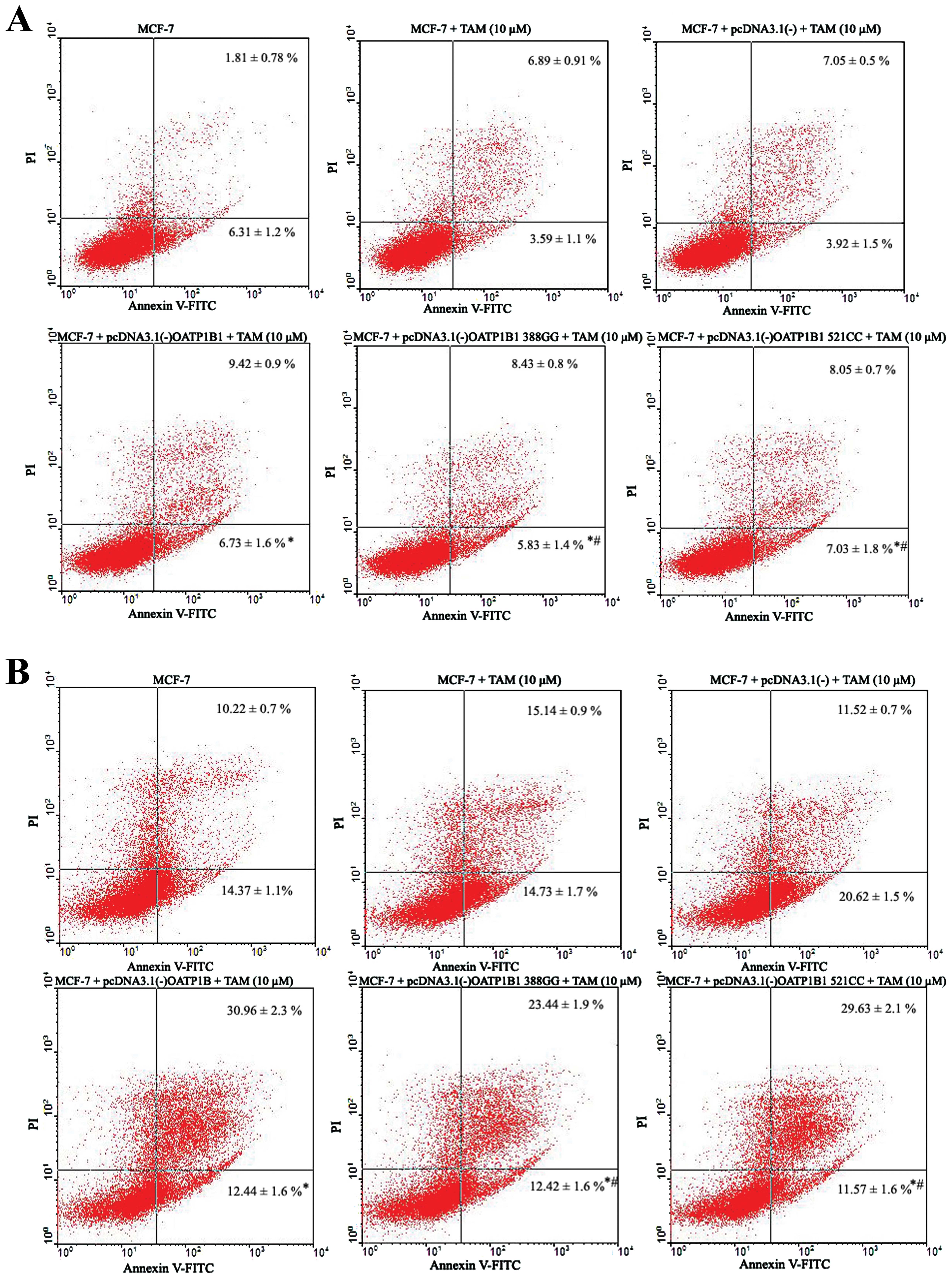
TAM effect on the cell apoptotic rate of
MCF-7 by plasmid transfection
We established expression platforms of OATP1B1
genetic polymorphisms to assess the role of OATP1B1 genetic
polymorphisms on TAM (10 μM) with regard to the apoptosis of
MCF-7 cells. The results showed that at 24 h, apoptosis in the D
group was markedly increased (P<0.05, n=3), compared to the B
group (Fig. 7A and B). However,
apoptosis of MCF-7 cells of the E and F groups was slightly
decreased (P>0.05, n=3) compared to the D group, although the
difference was not statistically significant (Fig. 7A and B).
After the effect of TAM (10 μM) on the
apoptosis of MCF-7 cells at 48 h, apoptosis of the D group was
markedly increased (P<0.05, n=3), compared to the B group
(Fig. 7C and D). However, the
apoptosis of MCF-7 cells of the E and F groups was slightly
decreased (P>0.05, n=3) compared to the D group, although the
differences were not statistically significant (Fig. 7C and D).
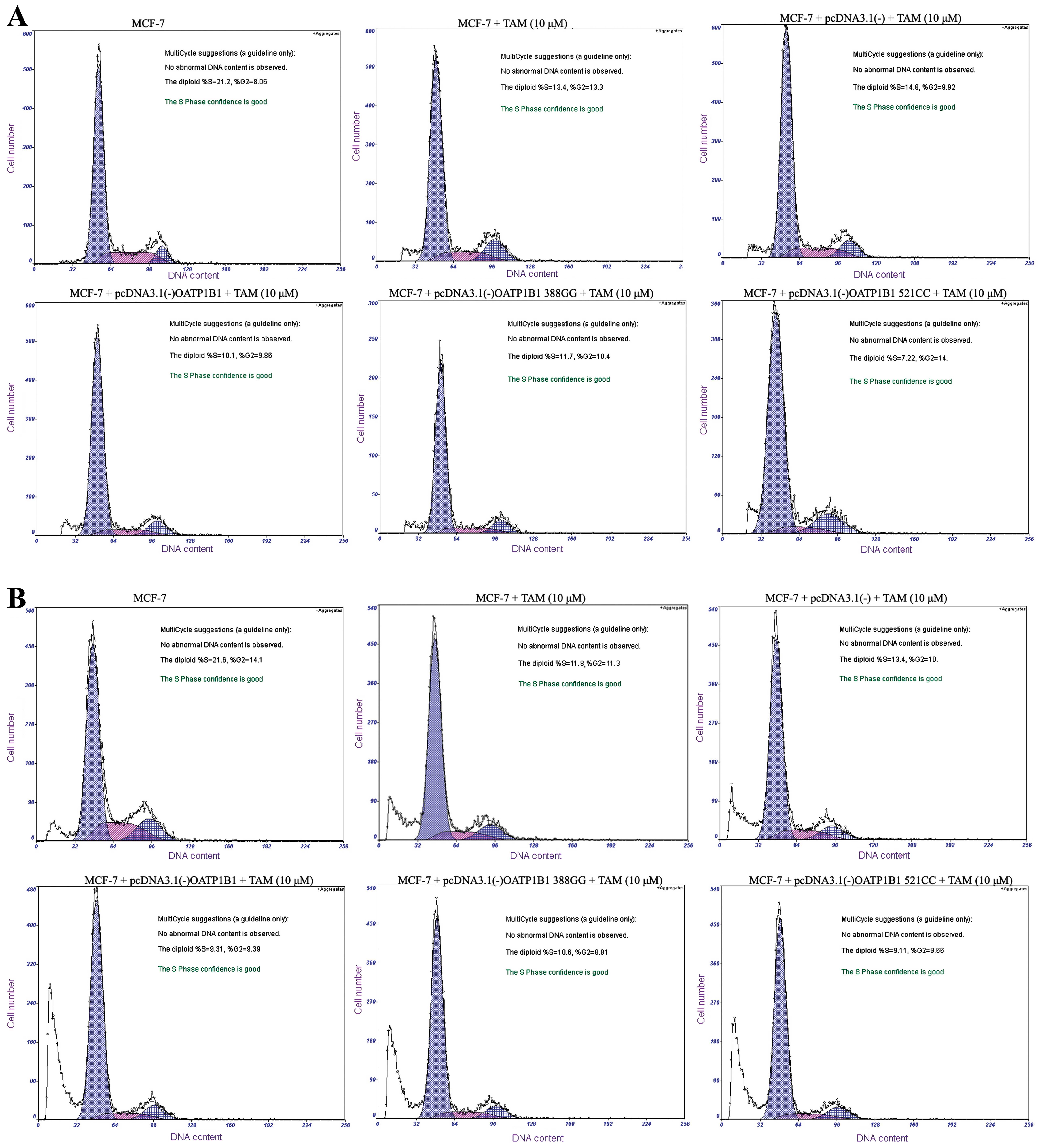
TAM effect on the cell cycle of MCF-7 by
plasmid transfection
We established expression platforms of OATP1B1
genetic polymorphisms to assess the role of OATP1B1 genetic
polymorphisms concerning the effect of TAM (10 μM) on the
cell cycle of MCF-7 cells. After the effect of TAM (10 μM)
on the cell cycle of MCF-7 cells at 24 h, the
G0/G1 phase in D group was markedly increased
(P<0.05, n=3), the S and G2M phases were markedly
decreased (P<0.05, n=3), compared to the B group (Table II, Fig.
8A). In E and F groups, the G0/G1 phase
was slightly decreased (P>0.05, n=3), while the S and
G2M phases were slightly increased (P>0.05, n=3),
respectively, compared to the D group, although the differences
were not statistically significant (Table II, Fig.
8A).
 | Table IITAM effect on the cell cycle of MCF-7
by plasmid transfection. |
Table II
TAM effect on the cell cycle of MCF-7
by plasmid transfection.
| Time | Group |
G0/G1 (%) | S (%) | G2M
(%) | PI-value |
|---|
| 24 h | A | 68.71±3.12 | 22.24±1.21 | 5.78±1.02 | 28.97 |
| B | 74.33±2.24 | 12.71±1.06 | 12.84±1.01 | 25.58 |
| C | 74.79±2.31 | 14.81±1.11 | 9.92±1.03 | 24.85 |
| D | 79.57±1.69 | 10.13±0.77 | 9.86±0.59 | 20.08a |
| E | 77.76±1.78 | 11.74±1.03 | 10.4±1.00 | 22.16a,b |
| F | 79.02±1.89 | 6.88±0.98 | 14.00±0.76 | 20.90a,b |
| 48 h | A | 64.32±3.53 | 21.45±2.31 | 13.93±2.13 | 35.49 |
| B | 75.90±2.72 | 12.3±1.58 | 11.06±1.35 | 23.53 |
| C | 76.63±2.69 | 13.00±1.62 | 10.33±1.40 | 23.34 |
| D | 81.21±2.09 | 9.11±1.18 | 9.66±1.21 | 18.78a |
| E | 80.15±2.11 | 10.66±1.35 | 8.81±1.25 | 19.54a,b |
| F | 81.00±2.10 | 9.31±1.20 | 9.39±1.17 | 18.76a,b |
After the effect of TAM (10 μM) on the cell
cycle of MCF-7 cells at 48 h, the G0/G phase in the D
group was markedly increased (P<0.05, n=3), while the S and
G2M phases were markedly decreased (P<0.05, n=3),
compared to the B group (Table II,
Fig. 8B). In the E and F groups,
the G0/G1 phase was slightly decreased
(P>0.05, n=3), while the S and G2M phases were
slightly increased (P>0.05, n=3), respectively, compared to D
group, although the differences were not statistically significant
(Table II, Fig. 8B).
Discussion
The functions of OATP1B1 are numerous, of which the
specificity is distributed in the basolateral membrane of liver
cells of the human. A variety of endogenous substances and drugs
enter the liver cells via the portal system, including thyroid
hormones, prostaglandin, bile acid, and statins. The OATP1B1
gene is located on chromosome 12, with the full-length 10.86 kb
(7). The encoding gene of SLCO1B1
includes 2,076 bases, encoding 691 amino acids, including 15 exons
and 14 introns, as well as 20 function-genetic polymorphisms. The
OATP1B1 gene has multiple single-nucleotide polymorphisms
that are associated with abnormal uptake in in vitro or
in vivo experiments, of which the OATP1B1 gene
388A>G and 521T>C are the most common mutations (19). In a previous study, it was found
that the SLCO1B1 gene 388A>G polymorphism was not
statistically significantly associated with gallstone formation in
a north Indian population (20).
SLCO1B1 521 T>C plays a significant role in susceptibility to
colorectal cancer risk in the Turkish population (14). SLCO1B1 388A>G and 521T>C
polymorphisms were also not associated with response to
atorvastatin or simvastatin (21).
In the present study, a molecular biology technique
was applied to construct the pcDNA3.1(-)-OATP1B1 plasmid. The
site-directed mutagenesis method was then used to establish
pcDNA3.1(-)-OATP1B1 388GG and 521CC plasmids successfully. The
pcDNA3.1(-)-OATP1B1-MCF-7 gene polypeptide expression platform
demonstrated that OATP1B1 388GG and 521CC mutations lead to a
decrease of the inhibition and apoptotic rate of breast cancer
cells, compared to the OATP1B1 group, although the differences were
not statistically significant.
Breast cancer cells in the
G0/G1 phase were reduced in the OATP1B1 388GG
and 521CC groups, compared to the OATP1B1 group. Breast cancer
cells in the S and G2M phases were increased in the
OATP1B1 388GG and 521CC groups, compared to the OATP1B1 group,
although the differences were not statistically significant.
Therefore, pre-synthesis DNA of breast cancer cells in the OATP1B1
388GG and 521CC group were reduced, while DNA increased in the
synthesis and post-synthesis phase, causing the decreased
inhibition rate of MCF-7 cells, thereby reducing the effect of TAM
on MCF-7 cells. This conclusion is consistent with the inhibition
rate and apoptosis results.
In summary, our present investigation has shown that
OATP1B1 388GG and 521CC mutations may inhibit the activity of the
OATP1B1 protein. OATP1B1 388GG and 521CC mutations may lead to
restraining of the turn-over capacity of OATP1B1 and a reduction of
TAM that is internalized into MCF-7 cells, resulting in weakened
TAM treatment for breast cancer.
Nozawa et al (22) and Iwai et al (23) employed HEK293 cells and used estrone
sulfate and estradiol-17β-glucuronide as a substrate, finding
OATP1B1 388GG and 521CC had no significant effect on OATP1B1
absorbing ability, consistent with the results of the present
study. OATP1B1 388GG and 521CC mutations cause decreased capacity
of OATP1B1 in absorbing TAM, without statistical significance
compared with the gene groups that have not mutated. This occurs
due to OATP1B1 388GG and 521CC mutations being single mutation
sites, and these changes of the DNA sequence result in such limited
change of the protein structure and composition of OATP1B1 protein
that effective change for the turn-over capacity cannot be markedly
inhibited. This phenomenon is extremely common in gene mutations.
The results of that study found that CYP2D6*4 or *10 mutations have
a limited impact on the efficacy of TAM, and the difference was not
statistically significant (24).
Previous findings have shown that CYP2C19*2 and *3 mutations have
limited impact on the efficacy of TAM, and the difference was not
statistically significant (25).
The ability of SLCO1B1*15 (388G/521C) in absorbing
estrone sulfate and estradiol-17β-glucuronide was found to be
significantly reduced (26),
indicating that the double mutation (SLCO1B1*15) may have greater
significance on the transport of TAM. In summary, 388GG and 521CC
of OATP1B1 gene polymorphism expression platforms were established
in this experiment. The results show that OATP1B1 388GG and 521CC
mutations lead to the decreased turn-over capacity of OATP1B1 and a
reduction of TAM in the treatment of breast cancer, although the
difference was not statistically significant. Thus construction of
a OATP1B1*15 double mutation plasmid to study the changes in drug
absorption should be carried out in future investigations.
Acknowledgments
This study was supported by the Key Clinical
Medicine Application Technology Item of Anhui Provincial Health
Department (no. 2010A013), and the Natural Science Foundation of
China (grant no. 81173134).
References
|
1
|
Knaul FM, Bhadelia A, Gralow J, et al:
Meeting the emerging challenge of breast and cervical cancer in
low- and middle-income countries. Int J Gynaecol Obstet. 119(Suppl
1): S85–S88. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Harbeck N: American Society of Clinical
Oncology highlights 2013: breast cancer and gynecological
malignancies. Future Oncol. 9:1433–1436. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Karaoz B, Aksu H and Küçük M: A
qualitative study of the information needs of premenopausal women
with breast cancer in terms of contraception, sexuality, early
menopause, and fertility. Int J Gynaecol Obstet. 109:118–120. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kattan J and Kourie HR: The use of
everolimus to reverse tamoxifen resistance in men with metastatic
breast cancer: a case report. Invest New Drugs. 32:1046–1047. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yaacob NS, Kamal NN and Norazmi MN:
Synergistic anticancer effects of a bioactive subfraction of
Strobilanthes crispus and tamoxifen on MCF-7 and MDA-MB-231 human
breast cancer cell lines. BMC Complement Altern Med. 14:2522014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alcazar-González GA, Calderón-Garcidueñas
AL, Garza-Rodríguez ML, et al: Comparative study of polymorphism
frequencies of the CYP2D6, CYP3A5, CYP2C8 and IL-10 genes in
Mexican and Spanish women with breast cancer. Pharmacogenomics.
14:1583–1592. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hsiang B, Zhu Y, Wang Z, et al: A novel
human hepatic organic anion transporting polypeptide (OATP2).
Identification of a liver-specific human organic anion transporting
polypeptide and identification of rat and human
hydroxymethylglutaryl-CoA reductase inhibitor transporters. J Biol
Chem. 274:37161–37168. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Niemi M, Schaeffeler E, Lang T, et al:
High plasma pravastatin concentrations are associated with single
nucleotide polymorphisms and haplotypes of organic anion
transporting polypeptide-C (OATP-C, SLCO1B1). Pharmacogenetics.
14:429–440. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mwinyi J, Johne A, Bauer S, et al:
Evidence for inverse effects of OATP-C (SLC21A6) *5 and *1b
haplotypes on pravastatin kinetics. Clin Pharmacol Ther.
75:415–421. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Niemi M, Neuvonen PJ, Hofmann U, et al:
Acute effects of pravastatin on cholesterol synthesis are
associated with SLCO1B1 (encoding OATP1B1) haplotype *17.
Pharmacogenet Genomics. 15:303–309. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Igel M, Arnold KA, Niemi M, et al: Impact
of the SLCO1B1 polymorphism on the pharmacokinetics and
lipid-lowering efficacy of multiple-dose pravastatin. Clin
Pharmacol Ther. 79:419–426. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chung JY, Cho JY, Yu KS, et al: Effect of
OATP1B1 (SLCO1B1) variant alleles on the pharmacokinetics of
pitavastatin in healthy volunteers. Clin Pharmacol Ther.
78:342–350. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ieiri I, Suwannakul S, Maeda K, et al:
SLCO1B1 (OATP1B1, an uptake transporter) and ABCG2 (BCRP, an efflux
transporter) variant alleles and pharmacokinetics of pitavastatin
in healthy volunteers. Clin Pharmacol Ther. 82:541–547. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ozhan G, Kara M, Sari FM, et al: Influence
of the functional polymorphisms in the organic anion transporting
polypeptide 1B1 in the susceptibility to colorectal cancer. Genet
Test Mol Biomarkers. 17:214–218. 2013. View Article : Google Scholar
|
|
15
|
D’Avolio A, Carcieri C, Cusato J, et al:
Intracellular accumulation of atazanavir/ritonavir according to
plasma concentrations and OATP1B1, ABCB1 and PXR genetic
polymorphisms. J Antimicrob Chemother. 69:3061–3066. 2014.
View Article : Google Scholar
|
|
16
|
Hua WJ, Hua WX, Nan FY, et al: The
influence of herbal medicine ursolic acid on the uptake of
rosuvastatin mediated by OATP1B1*1a and *5. Eur J Drug Metab
Pharmacokinet. 39:221–230. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rose RH, Neuhoff S, Abduljalil K, et al:
Application of a physiologically based pharmacokinetic model to
predict OATP1B1-related variability in pharmacodynamics of
rosuvastatin. CPT Pharmacometrics Syst Pharmacol. 3:e1242014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rohrbacher M, Kirchhof A, Skarke C, et al:
Rapid identification of three functionally relevant polymorphisms
in the OATP1B1 transporter gene using Pyrosequencing™.
Pharmacogenomics. 7:167–176. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nishizato Y, Ieiri I, Suzuki H, et al:
Polymorphisms of OATP-C (SLC21A6) and OAT3 (SLC22A8) genes:
consequences for pravastatin pharmacokinetics. Clin Pharmacol Ther.
73:554–565. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jindal C, Kumar S, Choudhari G, et al:
Organic anion transporter protein (OATP1B1) encoded by SLCO1B1 gene
polymorphism (388A>G) & susceptibility in gallstone disease.
Indian J Med Res. 129:170–175. 2009.PubMed/NCBI
|
|
21
|
Giannakopoulou E, Ragia G, Kolovou V, et
al: No impact of SLCO1B1 521T>C, 388A>G and 411G>A
polymorphisms on response to statin therapy in the Greek
population. Mol Biol Rep. 41:4631–4638. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nozawa T, Nakajima M, Tamai I, et al:
Genetic polymorphisms of human organic anion transporters OATP-C
(SLC21A6) and OATP-B (SLC21A9): allele frequencies in the Japanese
population and functional analysis. J Pharmacol Exp Ther.
302:804–813. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iwai M, Suzuki H, Ieiri I, et al:
Functional analysis of single nucleotide polymorphisms of hepatic
organic anion transporter OATP1B1 (OATP-C). Pharmacogenetics.
14:749–757. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sirachainan E, Jaruhathai S, Trachu N, et
al: CYP2D6 polymorphisms influence the efficacy of adjuvant
tamoxifen in Thai breast cancer patients. Pharmgenomics Pers Med.
5:149–153. 2012.PubMed/NCBI
|
|
25
|
Okishiro M, Taguchi T, Jin Kim S, et al:
Genetic polymorphisms of CYP2D6*10 and CYP2C19*2, *3 are not
associated with prognosis, endometrial thickness, or bone mineral
density in Japanese breast cancer patients treated with adjuvant
tamoxifen. Cancer. 115:952–961. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tirona RG, Leake BF, Merino G and Kim RB:
Polymorphisms in OATP-C: identification of multiple allelic
variants associated with altered transport activity among European-
and African-Americans. J Biol Chem. 276:35669–35675. 2001.
View Article : Google Scholar : PubMed/NCBI
|